Abstract
Fertile and diploid nuclear transplants were successfully generated by using embryonic cells as donors in a small laboratory fish, medaka (Oryzias latipes). Embryonic cell nuclei from transgenic fish carrying the green fluorescent protein (GFP) gene were transplanted into unfertilized eggs enucleated by x-ray irradiation. In this study, 1 out of 588 eggs transplanted in the first experiment and 5 out of 298 eggs transplanted in the second experiment reached the adult stage. All of these nuclear transplants were fertile and diploid, and the natural and GFP markers of the donor nuclei were transmitted to the F1 and F2 offspring in a Mendelian fashion. This systematic study proves the feasibility of generating nuclear transplants by using embryonic cells from fish as donors, and it is supported by convincing evidence.
Fish are an important animal resource not only for biomedical research, but also for food production. However, the usefulness of fish as an animal resource is limited to some extent, because modern techniques of genetic modification and cloning, although established in mammalian species (1, 2), are not yet available in fish.
Nuclear transplantation is a key technique in genetic modification and cloning. In fish, nuclear transplantation was first reported in the 1960s by Chinese researchers, although they obtained no conclusive results (reviewed in ref. 3). Gasaryan et al. (4) transplanted embryonic nuclei into nonenucleated and enucleated eggs of the loach and obtained nuclear transplants that developed into feeding larvae. Several groups (reviewed in refs. 5 and 6) generated nucleo-cytoplasmic hybrids by using mainly cyprinid fish and transplanting the embryonic nuclei of one species into the enucleated eggs of another. However, these achievements of nuclear transplantation in fish have not been widely adopted by other groups and have not yet been put to any practical use.
To develop basic techniques of nuclear transplantation in fish, we transplanted nuclei from embryonic cells into nonenucleated eggs of a small laboratory fish, medaka (Oryzias latipes), and obtained nuclear transplants that developed into adult fish but were triploid and infertile (7). In the triploid nuclear transplants, not only natural marker genes but also an introduced gene from the donor nuclei were seen to be expressed in accord with the characteristics of the promoter (8). The generation of diploid and fertile nuclear transplants with embryonic cells as donors is considered to be the next step toward the establishment of a nuclear transplantation technique using somatic or cultured cells. Here, we report the successful development of this step.
Materials and Methods
Fish.
Breeding conditions of the fish have been described (7). Developmental stages of the medaka embryos were determined according to Iwamatsu's description (9).
Donor transgenic fish.
Two series of experiments were performed. In the first experiment, a transgenic fish carrying the green fluorescent protein gene (GFP) driven by the medaka elongation factor 1a-A gene promoter (EF-1a-A/GFP) was used as the donor (Fig. 1 A and B). The plasmid pEF-1a-A/GFP was prepared by replacing the promoter region of cytomegalovirus (CMV) in pCMX/GFP-1 (10) with that of the medaka EF-1a-A gene (11). This DNA was microinjected into one-cell-stage embryos of the host strain, HNI-I, which is an inbred strain with a wild-type body color (12). Homozygotes for EF-1a-A/GFP were generated by crossing transgene carriers.
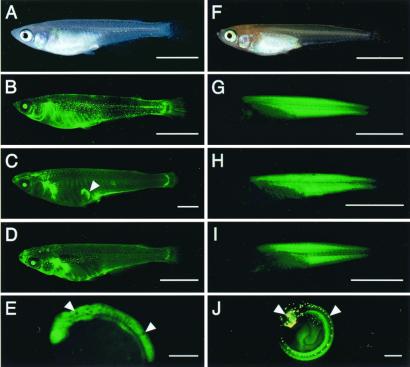
Figure 1.
Expression of the GFP in nuclear transplants and their offspring. (A) A 1.5-month-old donor transgenic fish, carrying EF-1a-A/GFP, under visible light. The body color is that of the wild type. (B) A fluorescent image of A. An intense fluorescence in the eyes and a weak one throughout the skin is observed. Fluorescence in the belly and a dot-like one in the trunk are autofluorescence of pigment cells. (C) A 7-month-old nuclear transplant (1NT1). The arrowhead indicates an artifact. (D) A 1-month-old F1 offspring derived from 1NT1. (E) A 1.5-day-old embryo from an F2 offspring derived from 1NT1, with the wild-type body color showing densely pigmented melanophores in the embryonic body (arrowheads). (F) A 3-week-old donor transgenic fish, carrying β-Act/GFP-N, under visible light. The body color is orange-red. (G) A fluorescent image of F. An intense fluorescence is observed in the muscle tissue. (H) A 3-week-old nuclear transplant produced in a separate experiment. In the second experiment, no photograph of the nuclear transplants was taken to avoid accidental killing of the fish. (I) A 1-month-old F1 offspring derived from 2NT1. (J) A 5-day-old embryo from an F2 offspring derived from 2NT4. Yellow spots in the head and on the dorsal midline are autofluorescence of leucophores (arrowheads). [Bars = 5 mm (A–D and F–I) and 0.3 mm (E and J).]
In this transgenic fish, GFP fluorescence, as expression of the transgene, was observed in the entire body from the early gastrula stage to the 5-day-old embryo stage, and in various tissues except for the muscle throughout the subsequent developmental stages to the adult stage (Fig. 1B) (13).
In the second experiment, a different transgenic stock of fish was used as the donor (Fig. 1 F and G). The fish carried the GFP gene with the nuclear localization signal, driven by the medaka β-actin gene promoter (β-Act/GFP-N). The plasmid pβ-Act/GFP-N was prepared by replacing the promoter region of CMV in pCMX/GFP-1 with that of the medaka β-actin gene (14) and ligating the nuclear localization signal derived from simian virus 40 (SV40) to the 3′ end of the GFP gene (15). Introduction of the plasmid to the host orange-red fish (OR), which is a commercially available variety and lacks melanized melanophores in the skin, and generation of homozygotes for β-Act/GFP-N were performed by the methods described above.
In the donor transgenic fish carrying β-Act/GFP-N, the GFP expression was first observed weakly in the somites at the 2-day-old embryo stage. An intense fluorescence was restricted to the skeletal muscle throughout the subsequent developmental stages to the adult stage (Fig. 1G) (15).
Fish used as recipients and parents for crossing.
Nontransgenic OR was used as the source of recipient eggs in both the first and second experiments. The same variety and an inbred strain originated from the variety, HO5 (16), were used as parents that were crossed with nuclear transplants and their progeny.
Enucleation of Recipient Eggs.
Recipient eggs were enucleated by x-ray irradiation. Unfertilized eggs collected from OR females were placed in a balanced salt solution (BSS) for medaka (17) supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin [(BSS plus penicillin and streptomycin (PS)] in a 6-cm plastic dish coated with 0.1% BSA solution. The dish was filled with BSS plus PS to 0.5 mm higher than the upper border of the eggs. X-ray irradiation was performed by an 80-kV and 8-mA x-ray facility (OM-100RE; OHMIC, Tokyo). Eggs were exposed to a total x-ray dose of 100 or 200 Gy at an exposure rate of 8.51 Gy/min through a 0.2-mm-thick aluminum filter. After x-ray irradiation, the eggs were stored in BSS plus PS containing 0.5% BSA (BSS plus PS and BSA) at 18°C for up to 7 h until use.
Nuclear Transplantation.
Nuclear transplantation was basically conducted by the methods described previously (7). Briefly, blastoderms were collected from about 20 donor embryos in the mid-blastula stage and dissociated into single cells by being pipetted in Ca2+- and Mg2+-free PBS. The cells were collected by centrifugation and stored until use for up to 6 h at 4°C in a buffer solution, which is a modification of the transplantation buffer for Xenopus (18). It consisted of 0.25 M sucrose, 120 mM NaCl, 0.5 mM spermidine trihydrochloride (Sigma), 0.15 mM spermine tetrahydrochloride (Sigma), and 15 mM Hepes (pH 7.3).
Single donor cells were slightly ruptured by being sucked into glass microcapillaries, and they were transplanted into the cytoplasm of the recipient eggs at the animal pole through the micropyle. Eggs were held in a V-shaped groove on a 2% agar plate in a 6-cm plastic dish filled with BSS plus PS and BSA. The eggs were automatically activated when they were pricked with microcapillaries for nuclear transplantation (19). Nuclear transplantation was carried out by using a hydraulic injector (CellTram Oil; Eppendorf, Hamburg) connected to a micromanipulator (MO-202; Narishige, Tokyo) under a stereoscopic microscope (MZAPO; Leica, Heerbrugg, Switzerland) at approximately 10°C by placing the dish on a cooling plate (Thermo Plate; Tokai Hit, Shizuoka, Japan).
Each of the operated eggs was transferred into a well of 24-well-plastic plates filled with BSS containing 2 ppm methylene blue and cultured at 18°C for the first 24 h and then at 26°C until hatching. Hatched larvae were reared to the adult stage.
Observation and Imaging of GFP Fluorescence.
GFP fluorescence was observed by using a fluorescence stereoscopic microscope (MZFLII; Leica). Fluorescent images were obtained by using a microscopy camera (MPS60, Leica) mounted on the stereoscopic microscope.
Chromosome Preparation.
The chromosome number was determined in the nuclear transplants that reached the adult stage and in their F1 progeny, both male and female, in both experiments. In one nuclear transplant obtained in the first experiment, finely minced tissue fragments of the tail fin were cultured in a gelatin-coated well of a four-well plastic plate containing Leiboviz's L-15 medium supplemented with FCS and antibiotics. Fibroblastic cells, arising from the fragments, were passaged up to four times and treated with colcemid dissolved in the culture medium to a final concentration of 1 μg/ml for 4 h before harvest. The remaining nuclear transplants and F1 offspring were placed in 0.01% colchicine solution for 15 h. The spleen and gill were removed and finely minced together. Chromosomal preparations were made according to standard techniques (20). At least ten cells at metaphase were examined for each fish.
Allozyme Analyses.
Phosphoglucomutase (PGM) allozyme analyses were carried out in nuclear transplants and F1 offspring in the first experiment, as previously described (7). In brief, a liver tissue sample was homogenized with an equal volume of distilled water containing a mixture of protease inhibitors, including 30 μM (p-amidinophenyl) methanesulfonyl fluoride (Wako Pure Chemical, Osaka), 100 μg/ml trans-epoxysuccinyl-l-leucylamido (4-guanidino) butane (Sigma) and 1 mM EDTA. The homogenized tissue was centrifuged and electrophoresed on 10% polyacrylamide gel. The gels were stained for PGM for 5 min with 0.1 M Tris⋅HCl solution (pH 7.1) containing 6 mg/ml sodium glucose monophosphate, 2 mg/ml MgCl2, 0.1 mg/ml NADP, 0.1 mg/ml 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-tetrazolium bromide, 0.025 mg/ml phenazine methosulfate, and 1 unit/ml glucose-6-phosphate dehydrogenase at 37°C.
PCR Analyses.
Genomic DNA was extracted from the fins of adult fish or from hatched fry by a modified method of Blin and Stafford (21). Samples were digested with 100 μl of TNES solution (10 mM Tris⋅HCl, pH 8.0/150 mM NaCl/25 mM EDTA/0.5% SDS) containing 10 μg each of proteinase K and RNase A. The DNA was recovered by serial extractions with phenol, chloroform and isoamylalcohol, and ethanol precipitation. PCR was performed using 0.5 unit of ExTaq DNA polymerase (Takara, Osaka), 100 nM of each primer, and 30–40 ng of DNA as a template in a volume of 20 μl. Primers specific for the GFP gene (forward: 5′-TGCCACCTACGGCAAGCTGA- 3′ and reverse: 5′-TGTTGCCGTCCTCCTTGAAG-3′) were used for the detection of the EF-1a-A/GFP transgene (expected size: 299 bp). A forward primer 5′-ACAAGAGAATGCAGCCCA-3′ (specific for the promoter region of the β-actin gene) and a reverse primer 5′-TGAAGTTGTACTCCAGCTTG-3′ (specific for the GFP gene) were used for the detection of the β-Act/GFP-N transgene (expected size: 652 bp). To confirm the success of DNA extraction and the following PCR, the endogenous EF-1a-A gene was detected by using two primers specific for the medaka EF-1a-A gene (forward: 5′-CAGGACGTCTACAAAATCGG-3′ and reverse: 5′-AGCTCGTTGAACTTGCAGGCG-3′). The expected size of the PCR product was 519 bp. The PCR parameters were as follows: 94°C for 3 min, followed by 32 (for the EF-1a-A/GFP) or 35 (for the β-Act/GFP-N) cycles of 94°C, 30 s; 55°C, 45 s; and 72°C, 1 min. All PCR products were fractionated on 1.5% agarose gels.
Results
Nuclear Transplantation.
In the first experiment, 588 eggs were operated upon. Of these, 573 (97.4%) were activated, as shown by breakdown of the cortical alveoli and formation of a wide perivitelline space. Three hundred eighty-six (65.6%) reached the blastula stage, 29 (4.9%) reached the stage of embryonic body formation (1-day-old embryo stage), and 2 (1NT1 and 1NT2) (0.3%) hatched (Table 1). One (1NT2) of the hatched fry, however, exhibited an abnormal swelling of the intestine caused by indigestion 50 days after hatching, i.e., before sexual maturation. This abnormality in 1NT2 does not appear to be a result of the transplantation, because a similar phenomenon also occurred at the same time among the control fish. 1NT2 was confirmed to be female by the presence of an immature ovary found by dissection immediately after its death. The entire body was stored at −80°C for further studies. 1NT1 matured sexually within 2.5 months of hatching, as in the case of the donor transgenic fish. It was female and produced offspring daily with a normal brood size of about 30 eggs per spawning when it was crossed with a male HO5.
Table 1.
Development of nuclear transplants generated by transplantation of blastula nuclei into enucleated eggs*
| Experiment | No. of
individuals
|
||||||
|---|---|---|---|---|---|---|---|
| Eggs operated | Activated | Cleaved | Blastula | Embryonic body formation | Hatched | Adult | |
| 1 | 588 | 573 (97.4)† | 409 (69.6) | 386 (65.6) | 29 (4.9) | 2 (0.3) | 1 (0.2) |
| 2 | 298 | 291 (97.7) | 209 (70.1) | 203 (68.1) | 48 (16.1) | 7 (2.3) | 5 (1.7) |
Total numbers in 17 and 10 experiments in the first and second experiments, respectively, are shown.
Numbers in parentheses represent the percentage of the total number of transplants.
In the second experiment, 298 eggs were operated. The developmental process from activation to the blastula stage of the operated eggs occurred at levels of success similar to those in the first experiment. More transplants, that is, 48 (16.1%) and 7 (2.3%), reached the stages of embryonic body formation and hatching, respectively, in this experiment than in the first experiment. Two of the hatched larvae died immediately. The remaining five (1.7%), namely 2NT1, 2NT2, 2NT3, 2NT4, and 2NT5 (2NT1 through 2NT5), matured sexually within 1.5 months, as in the case of the donor transgenic fish. They were all female and produced offspring daily with a normal brood size when they were crossed with a male OR.
Characterization of Nuclear Transplants.
In the first experiment, 1NT1 and 1NT2 exhibited the wild-type body color that is a natural genetic marker of the donor fish and genetically dominant to the orange-red body color. Melanophores appeared on the yolk sphere at the anticipated developmental stage, the 9-somite stage, in the nuclear transplants.
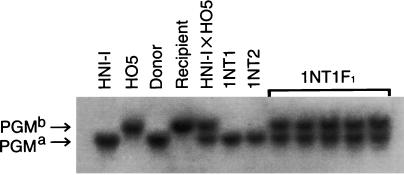
Allozymic polymorphism is known in various enzymes among wild populations (22) and laboratory strains (12) of medaka. HNI-I and HO5 are known to carry different allozyme markers of PGM, namely, PGMa and PGMb, respectively (12). OR has been previously shown to carry PGMb (7). PGMa, but not PGMb, was detected in both 1NT1 and 1NT2 by allozyme analyses (Fig. 2).
Figure 2.
PGM allozymes in nuclear transplants and F1 offspring in the first experiment. HNI-I, the host strain of the donor transgenic fish; HO5, an inbred strain of OR; donor, donor transgenic fish carrying pEF-1a-A/GFP; recipient, OR; HNI-I x HO5, an F1 hybrid produced by crossing HNI-I and HO5; 1NT1 and 1NT2, nuclear transplants; 1NT1F1, five F1 offspring obtained by crossing 1NT1 and HO5.
Both of the transplants expressed GFP. The expression pattern of GFP in them was the same as that in the donor fish throughout the course of the embryonic development and growth to the adult stage (Fig. 1C). The fluorescence in the internal organs of 1NT1 at the adult stage was observed in the frozen specimens that were kept at −80°C after the removal of tissues of the liver and the fins for biochemical and chromosomal analyses; it was also confirmed that the expression pattern of GFP in the internal organs of 1NT1 was the same as that of the donor transgenic fish (data not shown). A similar expression pattern of GFP was observed in the frozen specimen of 1NT2 (data not shown).
In the second experiment, the five nuclear transplants reached the adult stage (2NT1 through 2NT5) exhibited the orange-red body color and expressed GFP. Their GFP expression patterns were the same as those of the donor fish throughout the embryonic and subsequent developmental stages to the adult stage (Fig. 1H).
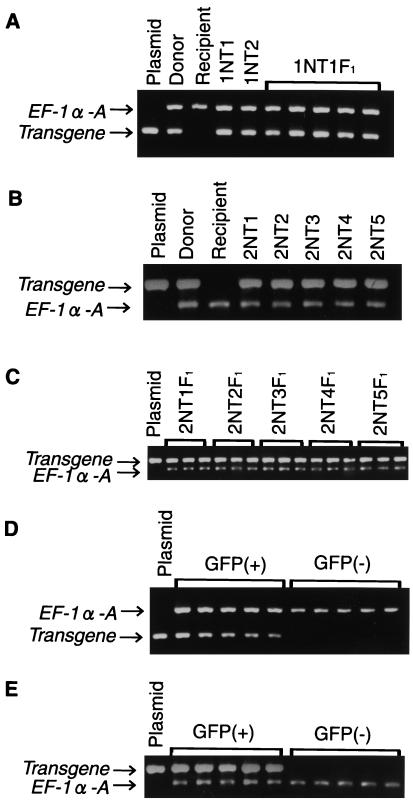
The transgenes were detected in all of the seven nuclear transplants (1NT1 and 1NT2, and 2NT1 through 2NT5) in both experiments (Fig. 3 A and B). The diploid chromosome number, 48, was detected in all of the six surviving nuclear transplants (1NT1 and 2NT1 through 2NT5). These results are summarized in Table 2.
Figure 3.
Detection of the transgene in nuclear transplants and their offspring. (A) EF-1a-A/GFP in nuclear transplants and F1 offspring. Plasmid, pEF-1a-A/GFP; donor, donor transgenic fish; recipient, OR; 1NT1 and 1NT2, nuclear transplants; 1NT1F1, five F1 offspring derived from 1NT1. (B) β-Act/GFP-N in nuclear transplants. Plasmid, pβ-Act/GFP-N; donor, donor transgenic fish; recipient, OR; 2NT1 through 2NT5, nuclear transplants. (C) β-Act/GFP-N in F1 offspring. Plasmid, pβ-Act/GFP-N; 2NT1F1 through 2NT5F1, three each of F1 offspring obtained by crossing 2NT1, 2NT2, 2NT3, 2NT4, and 2NT5 with OR. (D) EF-1a-A/GFP in F2 offspring derived from 1NT1. Plasmid, pEF-1a-A/GFP; GFP (+) and GFP (−), five each F2 offspring exhibiting fluorescence and no fluorescence, respectively. (E) β-Act/GFP-N in F2 offspring derived from 2NT1; plasmid, pβ-Act/GFP-N; GFP (+) and GFP (−), five each F2 offspring exhibiting fluorescence and no fluorescence, respectively. The endogenous EF-1a-A gene is amplified and used as a control in all PCR analyses.
Table 2.
Characterization of nuclear transplants
| Experiment | Nuclear transplants | Body color | PGM allozyme | GFP fluorescence | GFP gene | Fertility | Sex | No. of chromosomes | Ploidy |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1NT1 | Wild type | PGMa | + | + | + | F | 48 | Diploid |
| 1NT2 | Wild type | PGMa | + | + | ND | F | ND | ND | |
| 2 | 2NT1 | Orange-red | ND | + | + | + | F | 48 | Diploid |
| 2NT2 | Orange-red | ND | + | + | + | F | 48 | Diploid | |
| 2NT3 | Orange-red | ND | + | + | + | F | 48 | Diploid | |
| 2NT4 | Orange-red | ND | + | + | + | F | 48 | Diploid | |
| 2NT5 | Orange-red | ND | + | + | + | F | 48 | Diploid |
F, female; ND, not determined.
Inheritance of Genetic Markers.
The inheritance patterns of the genetic markers in the F1 and F2 offspring derived from the nuclear transplants were analyzed by crossing experiments.
F1 generation.
In the first experiment, all of the 107 F1 offspring obtained by the crossing of 1NT1 and HO5 exhibited the wild-type body color and expressed GFP. The expression pattern of GFP was the same as that of the donor fish throughout the course of the embryonic development and growth to the adult stage (Fig. 1D). All of the five individuals examined exhibited both PGMa and PGMb (Fig. 2).
In the second experiment, 2NT1 through 2NT5 were each crossed with OR by pair mating, and 50–115 F1 individuals were obtained from each crossing. All of the F1 progeny exhibited the orange-red body color and expressed GFP. The GFP expression pattern of the F1 offspring was the same as that of the donor transgenic fish from the embryonic through the postembryonic development to the adult stage (Fig. 1I).
The presence of the transgene was examined in 15 F1 individuals derived from 1NT1 and 13 F1 individuals derived from each of 2NT1 through 2NT5 by PCR analyses. The transgene specific to the corresponding donor fish was detected in all of them. Part of the results are shown in Fig. 3 A and C.
The chromosome number was examined in five F1 individuals derived from each of the six surviving nuclear transplants in both experiments. All of them had the diploid number of chromosomes (data not shown). These results obtained by genetic analyses of the F1 generation revealed that all of the surviving nuclear transplants are homozygous in terms of the natural and introduced marker genes.
F2 generation.
For further analyses of the F2 generation, five F1 individuals, both male and female, derived from 1NT1 were crossed with HO5 or OR by pair mating. One hundred five to 137 F2 offspring obtained in each crossing were examined with respect to their body color and GFP fluorescence throughout the embryonic development up to the hatching stage. They were segregated into individuals with melanophores (wild type) and those without melanophores (orange-red), in terms of body color, at a 1:1 ratio. Similarly, they showed segregation in terms of GFP expression into positive and negative at a 1:1 ratio (Table 3). The body color and GFP expression were inherited independently (data not shown). The expression pattern of GFP of the offspring was the same as that of the donor fish (Fig. 1E). The presence of the transgene EF-1a-A/GFP was examined in five each of the GFP-expressing and -nonexpressing hatched fry by PCR analyses. The transgene was specifically detected in individuals expressing GFP (Fig. 3D).
Table 3.
Segregation of the body color and GFP fluorescence expression in the F2 generation
| Experiment | Nuclear transplants | No. of F1 offspring used for crossing | Phenotypes*
|
|||
|---|---|---|---|---|---|---|
| Body color
|
GFP fluorescence
|
|||||
| Wild type | Orange-red | Present | Absent | |||
| 1 | 1NT1 | 5 | 51.5 ± 3.4† | 48.5 ± 3.4 | 52.8 ± 3.2 | 47.3 ± 3.2 |
| 2 | 2NT1 | 6 | 0 | 100 | 49.7 ± 4.9 | 50.3 ± 4.9 |
| 2NT2 | 5 | 0 | 100 | 48.7 ± 3.6 | 51.3 ± 3.6 | |
| 2NT3 | 5 | 0 | 100 | 48.7 ± 6.6 | 51.3 ± 6.6 | |
| 2NT4 | 6 | 0 | 100 | 46.8 ± 4.7 | 53.3 ± 4.7 | |
| 2NT5 | 5 | 0 | 100 | 51.7 ± 3.1 | 48.3 ± 3.1 | |
The number expresses percentages of fish that exhibit each phenotype.
Mean ± SD.
In the second experiment, five or six F1 individuals, both male and female, derived from each five nuclear transplants (2NT1 through 2NT5) were crossed with OR by pair mating, yielding 103 to 163 F2 offspring from each crossing. The body color and GFP expression of the F2 individuals were observed throughout the course of the embryonic development up to the hatching stage. All of the F2 offspring showed a lack of densely pigmented melanophores in their skin, as expected of embryos with the orange-red body color. Half of them were positive while the other half were negative for the GFP expression (Table 3). The GFP expression pattern during the course of embryonic development was the same as that of the donor transgenic fish (Fig. 1J). The presence of the transgene β-Act/GFP-N was examined in five each of the GFP-expressing and -nonexpressing hatched fry by PCR analyses. The transgene was detected in all of the GFP-expressing individuals but not in all of the GFP-nonexpressing ones (Fig. 3E).
Discussion
Establishment of the Nuclear Transplantation Technique in Fish.
In the present study, embryonic cell nuclei were transplanted into unfertilized eggs of the medaka enucleated with x-ray irradiation. A total of six nuclear transplants that grew to the adult stage were obtained from the two experiments. They were diploid and fertile, and homozygous for the natural and introduced marker genes from the donor nuclei. These genetic markers were transmitted to the subsequent generations in a Mendelian fashion. Thus far, there have been no reliable data for the production of adult fertile nuclear transplants transmitting genetic markers to their progeny in fish (reviewed in ref. 3). The present study demonstrates successful nuclear transplantation in fish using embryonic cells. This is an important step toward the generation of cloned fish from somatic or cultured cells.
Same Expression Patterns of the GFP Transgenes in Nuclear Transplants and in the Donor Transgenic Fish.
In the first experiment, the GFP gene was driven by the promoter of the medaka EF-1a-A gene. The pattern of GFP expression in the nuclear transplants and their offspring was the same as that of the donor fish throughout the course of the embryonic development and subsequent growth to the adult stage. This finding indicates that the transgene derived from the donor nuclei was expressed faithfully in accord with the characteristics of the promoter in the nuclear transplants and their offspring. A similar expression pattern of the same transgene was shown in the triploid nuclear transplants in a previous study (8). In the second experiment, the GFP transgene was also expressed in the same pattern as that of the donor transgenic fish.
The successful expression of introduced genes in the nuclear transplants is an important step toward the development of nuclear transplantation techniques for the genetic modification of fish. However, the introduced gene used in the present study originated from the transgenic fish from which the donor nuclei were obtained, and not directly introduced into the donor cells. In mammalian species, transgenic animals are cloned by nuclear transplantation by using cultured cells carrying transgenes that are directly introduced (23, 24) or targeted (25, 26). In Xenopus, nuclei from cell lines carrying reporter genes were transplanted into unfertilized eggs. The nuclear transplants reached the respective embryonic stages and expressed the reporter genes (27, 28). These results in mammals and Xenopus may be helpful in outlining the next step of nuclear transplantation in the medaka using cultured cells.
Totipotency of Nuclei in Fish Embryonic Cells.
The first nuclear transplantation was achieved by Briggs and King (29) using a frog, Rana pipiens; they showed that nuclei from blastula embryos are totipotent. Their work led many researchers to investigate whether nuclei of differentiated cells are also totipotent in amphibians (reviewed in refs. 3 and 30). Recent studies of nuclear transplantation in mammals (reviewed in ref. 31) clearly showed that nuclei from adult differentiated cells are totipotent. However, there have been few studies of the genetic potential of fish nuclei. In the present study, it was demonstrated that nuclei of medaka blastula cells are totipotent, as suggested by the production of germline chimeras following transplantation of blastula cells into embryos (32). The genetic potential of fish differentiated cells remains to be determined.
In fish, cryopreservation of fertilized eggs is difficult because of the presence of a large yolk mass, although sperm cryopreservation is possible. The nuclear transplantation technique established here would be a useful tool for the preservation of diploid fish, which could be reproduced by nuclear transplantation by using cryopreserved embryonic cells.
Contribution of Genetic Background of Donor and Recipient Fish to the Efficiency of Nuclear Transplantation.
It was noted that the survival rate of nuclear transplants after the blastula stage was considerably lower in the first experiment than in the second one. The donor transgenic fish strain (HNI-I) used in the first experiment and the recipient OR stock are known to belong to different genetic populations, from the northern and southern regions of Japan, respectively (33). The contribution of genetic background to the efficiency of nuclear transplantation was recently demonstrated in mice (25). Some genetic incompatibility between the nuclei and eggs might be more marked in the first experiment, where two different genetic populations were used as the donor and recipient fish, than in the second experiment where the donor and recipient fish were the same, OR.
Effects of Female Pronuclei in the Eggs on Nuclear Transplantation.
The survival rate of the nuclear transplants after the blastula stage in the first experiment was also lower than that in a previous experiment using nonenucleated eggs as recipients, although the same combination of donor and recipient fish was used (8). Similar results were reported in nuclear transplants of Xenopus (27). Female pronuclei in recipient eggs may have some ability to promote the embryonic development of nuclear transplants; in addition, x-ray irradiation used for enucleation may result in some harmful effects on maternal factors of the recipient eggs.
Imbalance of Sex Ratio in Nuclear Transplants.
All of the seven nuclear transplants (1NT1, 1NT2, and 2NT1 through 2NT5) obtained were female. In our previous studies, the sex ratio showed an extreme imbalance in the nuclear transplants in one experiment (8) but was balanced in the other one (7). It is unlikely that the imbalance in the sex ratio was attributable to the sex ratio of the donor embryos, because no special selection was carried out in the collection of donor embryos in any of the previous and present studies. In medaka, genetic sex is determined by the system of X and Y chromosomes (34). However, sex reversal in medaka following by hormonal treatment is known to occur during the process of sex differentiation (35). Some unknown mechanisms that promote female differentiation may be involved in the unbalanced sex ratio of the nuclear transplants.
Successful Inactivation of Egg Nuclei by X-Ray Irradiation.
The PGM allozyme marker of the recipient eggs was not detected in the nuclear transplants in the first experiment. Furthermore, triploid individuals and individuals with genetic markers of the recipient fish were not obtained in the first or second experiment, whereas such individuals were obtained by transplantation of embryonic cell nuclei into nonenucleated eggs (7, 8). These findings demonstrate that x-ray irradiation is useful for the inactivation of the recipient egg nuclei in medaka as in loach (4), although enucleation in mammals (1), Rana pipiens (36), and many fish (6) has been done by micropipettes, and in Xenopus (37) by UV irradiation.
Acknowledgments
We thank C. Inoue, H. Torihara, Y. Ishiguro, and S. Pristyaznhyuk for their technical assistance, and Y. Nishida for allowing us to use the x-ray facility. This study was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan, the Fishery Agency of Japan, and the Research for the Future Project of the Japan Society for the Promotion of Science.
Abbreviations
- β-Act, β-actin gene promoter
BSS, balanced salt solution
- EF1a-A
elongation factor 1a-A
- GFP
green fluorescent protein
- OR
orange-red fish
- PGM
phosphoglucomutase
- PS
penicillin and streptomycin
References
- 1.Campbell K H S, McWhir J, Ritchie W A, Wilmut I. Nature (London) 1996;380:64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 2.Wilmut I, Schnieke A E, McWhir J, Kind A J, Campbell K H S. Nature (London) 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 3.Di Berardino M A. Genomic Potential of Differentiated Cells. New York: Columbia Univ. Press; 1997. [Google Scholar]
- 4.Gasaryan K G, Hung N M, Neyfakh A A, Ivanenkov V V. Nature (London) 1979;280:585–587. doi: 10.1038/280585a0. [DOI] [PubMed] [Google Scholar]
- 5.Yan S Y. In: Cytoplasmic Organization Systems: A Primer in Developmental Biology. Malacinski G M, editor. New York: McGraw-Hill; 1989. pp. 61–81. [Google Scholar]
- 6.Yan S Y. Cloning in Fish: Nucleocytoplasmic Hybrids. Hong Kong: Educational and Cultural Press; 1998. [Google Scholar]
- 7.Niwa K, Ladygina T, Kinoshita M, Ozato K, Wakamatsu Y. Dev Growth Differ. 1999;41:163–172. doi: 10.1046/j.1440-169x.1999.00423.x. [DOI] [PubMed] [Google Scholar]
- 8.Niwa K, Kani S, Kinoshita M, Ozato K, Wakamatsu Y. Cloning. 2000;2:23–34. doi: 10.1089/15204550050145102. [DOI] [PubMed] [Google Scholar]
- 9.Iwamatsu T. Zool Sci. 1994;11:825–839. [Google Scholar]
- 10.Ogawa H, Umezono K. Acta Histochem Cytochem. 1998;31:303–308. [Google Scholar]
- 11.Kinoshita M, Nakata T, Yabe T, Adachi K, Yokoyama Y, Hirata T, Takayama E, Mikawa S, Kioka N, Takahashi M, Toyohara H, Sakaguchi M. Fish Sci. 1999;65:765–771. [Google Scholar]
- 12.Hyodo-Taguchi Y. Fish Biol J Medaka. 1996;8:11–14. [Google Scholar]
- 13.Kinoshita M, Kani S, Ozato K, Wakamatsu Y. Dev Growth Differ. 2000;42:469–478. doi: 10.1046/j.1440-169x.2000.00530.x. [DOI] [PubMed] [Google Scholar]
- 14.Takagi S, Sasado T, Tamiya G, Ozato K, Wakamatsu Y, Takeshita A, Kimura M. Mol Mar Biol Biotech. 1994;3:192–199. [PubMed] [Google Scholar]
- 15.Yamauchi M, Kinoshita M, Sasanuma M, Tsuji S, Terada M, Morimyo M, Ishikawa Y. J Exp Zool. 2000;287:285–293. [PubMed] [Google Scholar]
- 16.Hyodo-Taguchi Y, Egami N. Zool Sci. 1985;2:305–316. [Google Scholar]
- 17.Iwamatsu T. J Exp Zool. 1983;228:83–89. doi: 10.1002/jez.1402270115. [DOI] [PubMed] [Google Scholar]
- 18.Gurdon J B. J Embryol Exp Morphol. 1976;36:523–540. [PubMed] [Google Scholar]
- 19.Yamamoto T. Annot Zool Jpn. 1944;22:109–125. [Google Scholar]
- 20.Ladygina T, Wakamatsu Y. Fish Biol J Medaka. 1999;10:33–34. [Google Scholar]
- 21.Blin N, Stafford D W. Nucleic Acids Res. 1976;3:2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakaizumi M, Moriwaki K, Egami N. Copeia. 1983;1983:311–318. [Google Scholar]
- 23.Schnieke A E, Kind A J, Ritchie W A, Mycock K, Scott A R, Ritchie M, Wilmut I, Colman A, Campbell K H S. Science. 1997;278:2130–2133. doi: 10.1126/science.278.5346.2130. [DOI] [PubMed] [Google Scholar]
- 24.Cibelli J B, Stice S L, Golueke P J, Kane J J, Jerry J, Blackwell C, Ponce de Leon F A, Robl J M. Science. 1998;280:1256–1258. doi: 10.1126/science.280.5367.1256. [DOI] [PubMed] [Google Scholar]
- 25.Reideout W M, III, Wakayama T, Wutz A, Eggan K, Jackson-Grusby L, Dausman J, Yanagimachi R, Jaenisch R. Nat Genet. 2000;24:109–110. doi: 10.1038/72753. [DOI] [PubMed] [Google Scholar]
- 26.McCreath K J, Howcroft J, Campbell K H S, Colman A, Schnieke A E, Kind A J. Nature (London) 2000;405:1066–1069. doi: 10.1038/35016604. [DOI] [PubMed] [Google Scholar]
- 27.Kroll K L, Gerhart J C. Science. 1994;266:650–653. doi: 10.1126/science.7939720. [DOI] [PubMed] [Google Scholar]
- 28.Chan A P, Gurdon J B. Int J Dev Biol. 1996;40:441–451. [PubMed] [Google Scholar]
- 29.Briggs R, King T J. Proc Natl Acad Sci USA. 1952;38:455–463. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gurdon J B. J Cell Sci. 1986;4,(Suppl.):287–318. doi: 10.1242/jcs.1986.supplement_4.17. [DOI] [PubMed] [Google Scholar]
- 31.McLaren A. Science. 2000;288:1775–1780. doi: 10.1126/science.288.5472.1775. [DOI] [PubMed] [Google Scholar]
- 32.Wakamatsu Y, Hashimoto H, Kinoshita M, Sakaguchi M, Iwamatsu T, Hyodo-Taguchi Y, Tomita H, Ozato K. Mol Mar Biol Biotech. 1993;2:325–332. [Google Scholar]
- 33.Sakaizumi M. Genetica. 1986;69:119–125. [Google Scholar]
- 34.Aida T. Genetics. 1921;6:554–573. doi: 10.1093/genetics/6.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto T. J Exp Zool. 1953;123:571–594. [Google Scholar]
- 36.Di Berardino M A, Orr N H, McKinnell R G. Proc Natl Acad Sci USA. 1986;83:8231–8234. doi: 10.1073/pnas.83.21.8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurdon J B. Q J Microsc Sci. 1960;101:299–312. [Google Scholar]