Abstract
Nuclear reprogramming requires the removal of epigenetic modifications imposed on the chromatin during cellular differentiation and division. The mammalian oocyte can reverse these alterations to a state of totipotency, allowing the production of viable cloned offspring from somatic cell nuclei. To determine whether nuclear reprogramming is complete in cloned animals, we assessed the telomerase activity and telomere length status in cloned embryos, fetuses, and newborn offspring derived from somatic cell nuclear transfer. In this report, we show that telomerase activity was significantly (P < 0.05) diminished in bovine fibroblast donor cells compared with embryonic stem-like cells, and surprisingly was 16-fold higher in fetal fibroblasts compared with adult fibroblasts (P < 0.05). Cell passaging and culture periods under serum starvation conditions significantly decreased telomerase activity by approximately 30–50% compared with nontreated early passage cells (P < 0.05). Telomere shortening was observed during in vitro culture of bovine fetal fibroblasts and in very late passages of embryonic stem-like cells. Reprogramming of telomerase activity was apparent by the blastocyst stage of postcloning embryonic development, and telomere lengths were longer (15–23 kb) in cloned fetuses and offspring than the relatively short mean terminal restriction fragment lengths (14–18 kb) observed in adult donor cells. Overall, telomere lengths of cloned fetuses and newborn calves (≈20 kb) were not significantly different from those of age-matched control animals (P > 0.05). These results demonstrate that cloned embryos inherit genomic modifications acquired during the donor nuclei's in vivo and in vitro period but are subsequently reversed during development of the cloned animal.
The production of viable, fertile cloned animals by nuclear transplantation of somatic cells from cultured cell lines and adult tissues (1, 2) has challenged our understanding of terminal cell differentiation, cellular aging, and the proliferative capacity of cells. Successful cloning requires the reprogramming of the donor nuclei from pluripotent or differentiated cells to an undifferentiated state to permit the temporal and spatial reexpression of genes involved in embryo and fetal development. Somatic nuclei progressively acquire differentiated functions through the gradual implementation of epigenetic chromatin modifications during embryogenesis and postembryonic development (3). Amazingly, the cytoplasm of the mammalian oocyte can, under optimal conditions, reverse the changes to the chromatin structure and function of the differentiated transplanted nucleus to a state of totipotency. The inefficiency in the nuclear reprogramming has been suggested as one of the major causes of the high rate of embryonic, fetal, and neonatal failures observed after nuclear transplantation (4).
One characteristic structural DNA change of most dividing in vivo and in vitro somatic cells is the shortening of the telomeres, the long tandem arrays of hexameric DNA sequences (TTAGGG)n at the ends of mammalian chromosomes, during DNA replication (5–7). Telomeres are critical structures that function in the stability, replication, and segregation of the chromosome during mitosis (8), and the gradual loss of telomeric sequence has been proposed as a “mitotic clock” leading to cell cycle arrest or cellular senescence (9). Conventional DNA polymerases cannot replicate the extreme 5′ ends of chromosomes because removal of the most terminal RNA primer in the lagging strand leaves a small region of uncopied DNA (10). This telomeric DNA loss has been calculated to be between 50 and 200 bp per cell division (11, 12). Once telomeres shorten below a critical length, they lose the capacity to cap chromosomes effectively and are thought to activate a DNA damage response pathway that causes cell cycle arrest (13).
Cells can overcome this “end-replication problem” by the activation/expression of the ribonucleoprotein telomerase, which synthesizes TTAGGG repeats de novo onto the ends of chromosomes (14). This multisubunit, reverse transcriptase, uses its RNA component as a template for the synthesis of telomeric DNA. Normal human somatic cells do not express telomerase activity and therefore have a limited replicative life span in vitro (15). Telomerase activity and maintenance of telomere length has been observed in the germ line and in most immortal cell lines and tumor samples analyzed thus far (16, 17). The introduction of the telomerase catalytic subunit (hTERT) into normal human diploid cells displayed normal growth controls and normal karyotypes and succeeded in extending the life span of the cells (18, 19). Late-generation mice lacking the telomerase RNA (mTR−/−) component displayed shortened telomeres and chromosome abnormalities and exhibited defective spermatogenesis, increased apoptosis, and decreased proliferation in the testis, bone marrow, and spleen (20). Thus, telomerase activity in the germ line is thought to prevent cumulative telomere shortening from generation to generation.
The telomere hypothesis of aging can be tested in vivo by the use of nuclear transfer technology because it allows the production of cloned animals from adult and cultured somatic cells, without the involvement of the germ line. Sheep cloned by nuclear transfer of cultured cells from embryonic or fetal tissue showed telomere shortening of approximately 10–15% compared with age-matched controls (21, 22). Furthermore, Dolly, cloned from a cultured mammary cell from a 6-year-old ewe, displayed telomere loss of approximately 20% (21, 22). Therefore, both the age of the donor nucleus and the proliferation in culture contributed to the telomeric loss observed in this small sample size of cloned sheep. In contrast, Lanza et al. (23) have demonstrated the reversal of cellular aging with the use of senescent donor somatic cells as the nuclear donor. Fibroblasts from bovine fetuses cloned from these cells displayed an extended replicative life span and rebuilding of telomere length compared with control fetal fibroblasts (FFs) and senescent fibroblasts, respectively (23). Moreover, nucleated blood cells from 5- to 10-month-old cloned cattle appeared to have longer telomere lengths compared with newborn and age-matched control animals (23).
The aims of this study were as follows: (i) to determine the effects of in vitro culture and serum deprivation on telomerase activity and telomere length in bovine cell types used for cloning and (ii) to determine whether telomerase activity is reprogrammed and telomere length is restored after nuclear transfer (cloning) of cultured somatic cells. To accomplish these goals we have compared telomerase activity and telomere lengths in early- and late-passage bovine fibroblasts and embryonic stem (ES)-like cells, cloned embryos reconstructed with the use of various donor cell types, cloned fetuses, and live-born offspring with their original donor cell cultures and age-matched controls. Here we report that although telomere length and telomerase activity decline in bovine cells during in vitro culture, telomerase activity is reprogrammed as early as the blastocyst stage, and the telomere length is rebuilt in cloned cattle reconstructed with the use of donor nuclei from cultured adult and fetal cells.
Materials and Methods
Cells and Primary Tissues.
Bovine fetal tissues and fibroblasts were prepared either from 40- to 50-day-old fetuses or from cloned fetuses surgically removed from the recipients' uteri. Tissues were minced, and either they were snap frozen (−80°C) for later analysis or the cells were isolated by multiple trypsin/EDTA treatments (0.25% trypsin/0.02% EDTA; GIBCO/BRL) for 10 min at 37°C. Cells were washed and plated in tissue culture flasks with DMEM (GIBCO/BRL) + 10% FCS and 0.5% antibiotics (10,000 units of penicillin per ml and 10,000 μg of streptomycin per ml; GIBCO/BRL). The calf and adult bovine fibroblast cell lines were derived from surgical excisional biopsies. Thin (1–3 mm) pieces of the s.c. tissue were transferred into 25 mm2 flasks containing DMEM + 10% FBS + 1% (vol/vol) penicillin/streptomycin and cultured at 37°C in air containing 5% CO2.
To establish ES-like cell lines, the inner cell mass of bovine blastocysts was mechanically isolated and placed onto mitomycin C (Sigma; 10 μg/ml, 3 h)-arrested mouse fibroblasts plated onto 0.1% gelatin (Sigma)-coated 60-mm Petri dishes (Nunc). The cells were cultured in DMEM supplemented with 20% charcoal-treated FCS, nucleosides (Sigma; 0.03 mM adenosine, 0.03 mM cytidine, 0.03 mM guanosine, 0.03 mM uridine, and 0.01 mM thymidine), nonessential amino acids (Sigma), 2 mM l-glutamine (Sigma), 0.1 mM 2-mercaptoethanol (Sigma), and antibiotics. Culture medium was replaced every 3 days, and growing cells were mechanically replated every 7–12 days. Cells were individualized by treatment with 0.3% protease E (Sigma) in PBS. Both ES-like cells and fibroblasts were either serum starved (0.5% FCS + DMEM, 1–10 days) or serum fed (10% FCS + DMEM) before nuclear transfer or analysis.
Granulosa cells were recovered by transvaginal follicular aspiration from a 5-year-old Holstein. Cells were separated by shaking in PBS, washed in DMEM with 10% FCS + antibiotics, and plated in tissue culture flasks with the same medium. Trypsin/EDTA treatment was used for cell passaging once every 7 days and to obtain donor cells for oocyte reconstruction.
In Vitro Bovine Oocyte and Embryo Culture.
Cumulus–oocyte complexes were aspirated from follicles 2–10 mm in diameter, and in vitro oocyte and embryo culture methods (24) were used for the production of in vitro fertilized and cloned bovine preattachment embryos. Embryos were cocultured in either TCM-199 (GIBCO/BRL) or Menezo's B2 medium (Pharmascience, Paris, France) supplemented with 10% FCS in the presence of bovine oviduct epithelial cells or BRL cells in a humidified atmosphere of 5% CO2 in air at 39°C.
Oocyte Reconstruction by Nuclear Transfer.
The methods for oocyte enucleation, somatic cell–oocyte fusion, embryo activation, cloned embryo culture, and embryo transfer into synchronized recipients were carried out as described (25, 26). Bovine oocytes were reconstructed by nuclear transfer with the use of bovine adult fibroblasts, FFs, or granulosa cells as the nuclear donor.
Measurement of Telomerase Activity and Telomere Length.
Telomerase activity was measured by telomere repeat amplification protocol (TRAP) with the TRAPEZE telomerase detection kit (Intergen Company, Purchase, NY) as described (24). Cultured bovine cells, embryos, and tissues were extracted in Nonidet P-40 lysis buffer [10 mM Tris (pH 7.5)/1 mM MgCl2/1 mM EGTA/150 mM NaCl/10% glycerol/0.1 M PMSF/0.01% 2-mercaptoethanol/0.12 M sodium deoxycholate/1% Nonidet P-40] and 100 units/ml RNase inhibitor (Roche Molecular Biochemicals). To compare relative telomerase activities in the linear range the optimal embryo equivalents or protein content used for each TRAP assay was determined by serial dilutions of each cell extract. The extended TRAP products were amplified by PCR, resolved on 12% polyacrylamide gels, and revealed by exposure to a PhosphorImager cassette (molecular analyst Software; Bio-Rad). The relative telomerase activity was determined for each sample by the densitometric analysis of TRAP reaction products in relation to its 36-bp internal standard and the TRAP signals for the positive cell extract (immortalized 293 cells) and its 36-bp internal standard.
Mean telomere length was determined by terminal restriction fragment (TRF) analysis with the TeloQuant Telomere Length Assay Kit (PharMingen). Isolated DNA (2.5–10 μg) was digested with a HinfI/RsaI enzyme mixture (4 units/μg of DNA; GIBCO/BRL and Life Technologies, Grand Island, NY) for 12–16 h at 37°C. Undigested and digested DNA samples were resolved by 0.6% agarose gel electrophoresis at 5 V/cm for 2–3 h to check the integrity and complete digestion of genomic DNA, respectively. Digested DNA samples were then resolved by 0.6% agarose gel electrophoresis at 1 V/cm overnight (24 h). Gels were denatured, neutralized, and Southern transferred to a positively charged nylon membrane (Amersham Hybond-N+), which was then hybridized with a biotinylated telomere probe (TeloQuant; PharMingen). Mean TRF lengths were analyzed from a densitometric scan of the autoradiogram and calculated as described (11). Experiments were repeated a minimum of three times on different cell, embryo, and tissue samples. Statistical analyses were performed with the use of a post hoc test (Tukey–Kramer) for multiple comparisons to determine the significance of the differences (P < 0.05) between mean relative telomerase activities and mean TRF lengths (27).
Results
Telomerase Activity in Bovine Cell Lines.
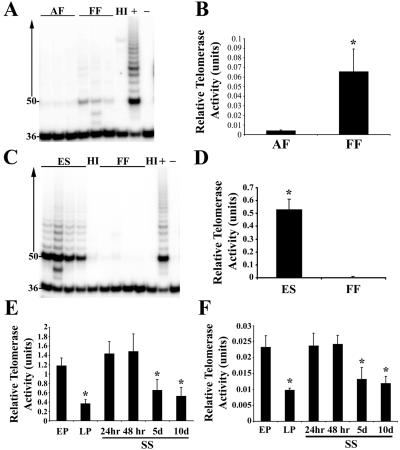
Telomerase activity in bovine cell lines displayed a profile similar to that of the same cell types of other species (7, 28, 29). Bovine ES-like cells exhibited a considerably higher level of telomerase activity in relation to fibroblast cells (P < 0.05), the levels of which were barely detectable (Fig. 1 C and D). Similarly, bovine FFs (first passage) displayed a significantly (P < 0.05) higher relative telomerase activity compared with adult fibroblasts (fourth passage). Although telomerase processivity was low, telomerase activity was almost 16 times greater in FFs compared with the adult cells (Fig. 1 A and B). The relatively low levels of telomerase activity observed in bovine fibroblasts are probably due to the lack of active telomerase enzyme rather than the presence of telomerase inhibitors because mixing experiments of ES extracts with fibroblast extracts did not significantly (P > 0.05) diminish TRAP products compared with controls (data not shown).
Figure 1.
The effect of cell passage number and serum starvation on telomerase activity. Telomerase activity was measured in protein extracts (0.5 μg) from adult fibroblast (AF) (passage 4) and FF (passage 1) cell lines (A) and in protein extracts (0.25 μg) from bovine ES-like cells (passage 8) and FF (passage 1) cell lines (C), with the use of the TRAP assay. Heat-inactivated (HI) control, positive 293 cell extract control (+), and negative lysis buffer control (−), are displayed. (B–F) Densitometric analyses of TRAP reaction products displaying the relative telomerase activities between bovine cell samples. Telomerase activity was measured in protein extracts (0.25 μg) from (E) bovine ES cell-like cells and (F) FFs at early passage (EP) (passages 8 and 1, respectively) and late passage (LP) (passages 34 and 24, respectively) and early-passage cells after increasing incubation times under serum starvation (SS) conditions (24 h, 48 h, 5 days, 10 days). Asterisks (*) denote significant differences (P < 0.05) between mean telomerase activities.
The Effect of Cell Passage and Serum Starvation on Telomerase Activity.
Serum starvation of cultured somatic cells before nuclear transfer is used to increase the percentage of cells in the G0/G1 cell cycle stage and is thought to improve the pregnancy rates of cloned embryos (30). To assess the impact of cell passage number and serum starvation, telomerase was determined in two bovine cell lines used in nuclear transfer protocols. The telomerase activity of both the bovine ES-like cells (Fig. 1E) and bovine FFs (Fig. 1F) significantly decreased from early to late cell passage (P < 0.05). Telomerase activity decreased by 31% and 42% from early to late cell passage in ES-like cells (34th passage) and FFs (24th passage), respectively. Similarly, telomerase activity levels of early passage stem cells (eighth passage) and fibroblasts (first passage) significantly decreased by days 5 and 10 of serum starvation conditions (P < 0.05). By 10 days of serum starvation telomerase levels decreased by 44.8% and 51.4% in ES-like cells (Fig. 1E) and FFs (Fig. 1F), respectively, compared with early-passage nontreated cells.
Telomere Length of Early- and Late-Passage Bovine FFs and ES-Like Cells.
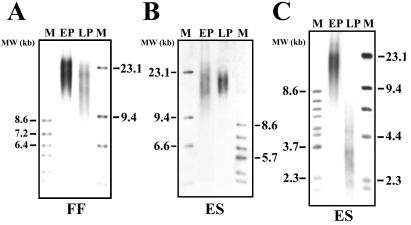
To examine the effect of in vitro cell division on telomere length, the mean TRF length was determined for early- and late-passage bovine FFs and ES-like cells. Mean TRF lengths were determined from two independent samples for each cell line and analyzed twice in separate experiments. The mean TRF length decreased with passage number in FFs (Fig. 2A) and, surprisingly, in telomerase-positive bovine ES-like cells (Fig. 2 B and C). The mean TRF length shortened from 21.17 kb at early passages (population doubling, PD = 4) to 16.45 kb at late PDs (PD = 33) in bovine FFs (Fig. 2A). Analysis of multiple fibroblast samples from different cell passages revealed that the telomeres shortened at a rate of approximately 163 bases with each PD. Telomere length was maintained at around 19 kb from early passage (fourth passage) to late passages (37th passage) in ES-like cell cultures (Fig. 2B). However, after many subcultures (52 passages; approximately 600–650 PDs) ES-like cells displayed extremely short TRF lengths of approximately 3.28 kb (Fig. 2C).
Figure 2.
Chemiluminescent detection of telomere length in bovine cells during in vitro culture. (A–C) TRF analysis demonstrating telomere length in early-passage (EP) and late-passage (LP) FFs and bovine ES-like cells cultured under 5% CO2 in air atmosphere. (A) Telomere length decreased in FF cells from early (4 PD) to late cell passages (33 PD). (B) Telomere length was maintained in the early passages (EP = passage 4; LP = passage 37) of ES-like cell cultures but (C) shortened in ES cells (derived from same starting clone) at very late passages (passage 52; approximately 600–650 population doublings). Molecular weight (MW) markers (lane M) are displayed in kilobases (kb).
Nuclear Reprogramming of Telomerase Activity.
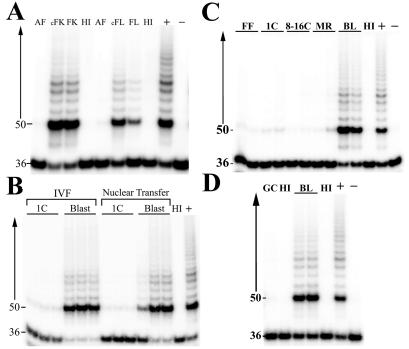
The TRAP assay was used to ascertain whether reprogramming of telomerase expression occurred in cloned embryos. Initial experiments detected the presence of telomerase activity in tissues from a 40-day cloned fetus reconstructed with the use of an adult fibroblast cell line from a 21-year-old bull (Fig. 3A). Protein extracts from kidney and liver samples from the cloned fetus were positive for telomerase activity, which is normally detected in these tissues during fetal gestation (Fig. 3A). Cloned bovine blastocysts, reconstructed with the use of bovine FFs and granulosa cells, exhibited telomerase activity like that of their in vitro-fertilized counterparts (Fig. 3 B and D). A more detailed analysis revealed that high levels of telomerase activity were not detected before the blastocyst stage of cloned embryo development (Fig. 3C).
Figure 3.
Reprogramming of telomerase activity in cloned bovine fetuses and embryos. Telomerase activity was assessed in protein extracts (0.5 μg) of bovine cell lines and fetuses and in the equivalent of one bovine embryo. (A) Telomerase activity status of bovine adult fibroblasts (AF) and the cloned fetal kidney (cFK) and liver (cFL) tissues produced from the reconstruction of adult fibroblast nuclei with enucleated bovine oocytes. Telomerase activity of fetal kidney (FK) and liver (FL) tissues from a natural mating is shown as positive controls. (B) Comparison of telomerase activity between in vitro-fertilized zygotes (1C) and blastocysts (Blast) with reconstructed embryos (Nuclear Transfer), with FFs as the donor nuclei. (C) Developmental expression of telomerase activity within cloned embryos. Telomerase activity was analyzed in cloned one-cell (1C) and 8- to 16-cell (8–16C) morulae (MR) and blastocysts (BL) reconstructed with nuclei from bovine FFs. (D) Reprogramming of telomerase activity in cloned bovine blastocysts (BL) derived from bovine granulosa cells (GC). Heat-inactivated (HI), positive cell extract control (+), and negative lysis buffer control (−) samples were used in each assay.
Analysis of Telomere Length in Cloned Cattle.
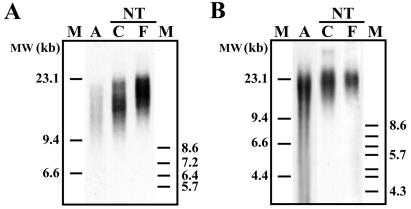
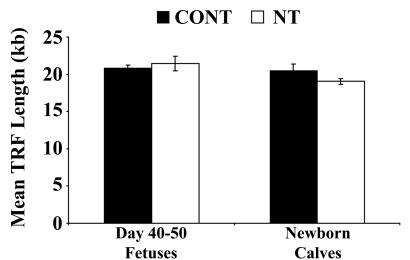
We sought to determine, with the use of TRF analysis, whether the telomere lengths of cloned fetuses and offspring reflect those of progenitor donor nuclei or age-matched control fetuses and animals. Chemiluminescent detection of telomeric DNA in cloned fetal and calf animals revealed that telomere lengths were longer than those measured for their donor cells (Fig. 4). Mean TRF lengths increased from 13.68 kb in progenitor adult fibroblasts from a 9-year-old Charolais female to 17.95 kb and 15.32 kb in cloned FFs and cloned calf fibroblasts, respectively (Fig. 4A). Although frozen aliquots of adult donor cell samples from a 21-year-old Brahman bull were destroyed during a freezer malfunction and the bull had died, and recovered tissue was unsuitable for cell culture (26), DNA recovery from frozen muscle tissue was achieved for TRF analysis. Mean TRF lengths were also shown to increase from 17.85 kb in adult muscle tissue from a 21-year-old Brahman bull to 22.74 kb and 21.33 kb in cloned fetal liver tissue and cloned calf fibroblasts, respectively (Fig. 4B). Telomere length analysis of different tissue samples from multiple cloned calves (<3 weeks old) generated from the same donor cell line displayed distinct variations in mean TRF lengths between cloned animals and between different tissues within each clone as well (Fig. 5). After we compiled and compared the mean TRF length data on all cloned fetal (n = 5) and cloned calf samples (n = 16) with age-matched and tissue-matched controls (n = 22), no significant differences (P > 0.05) in telomere lengths were observed (Fig. 6). Mean TRF lengths were calculated to be approximately 20.90 ± 0.33 kb and 20.54 ± 0.85 kb in control fetal and newborn animals, respectively, and were 21.45 ± 0.97 kb and 19.03 ± 0.39 kb in cloned fetuses and newborn calves, respectively (Fig. 6).
Figure 4.
Rebuilding of telomere length in cloned cattle. Chemiluminescent detection of TRF lengths of RsaI/HindfI-digested DNA from cells and tissues of cloned fetuses and calves compared with their donor cells from a 9-year-old Charolais female (A) and from a 21-year-old Brahman bull (B). Mean TRF lengths were measured in DNA samples from adult (A) donor cells and samples from cloned (NT) fetal (F) and calf (C) animals. Molecular weight (MW) markers (lane M) are displayed in kb.
Figure 5.
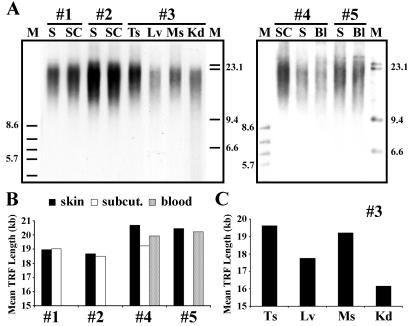
Variation of telomere lengths between cloned calves and among different tissues within each cloned animal. (A) TRF analysis of digested DNA from various tissue samples of multiple clone calves (#1–#5; <3 weeks old) derived from the same bovine FF cell line. Samples are skin (S), s.c. (SC), testis (Ts), muscle (Ms), kidney (Kd), and blood (Bl). Molecular weight markers (M) are in kilobases. (B) Densitometric analysis of TRF profiles revealing mean TRF lengths of different samples (skin, s.c., and blood) from multiple cloned offspring (#1, #2, #4, #5). (C) Mean TRF lengths in testis (Ts) (mean TRF length = 19.63 kb), liver (Lv) (mean TRF length = 17.76 kb), muscle (Ms) (mean TRF length = 19.23 kb), and kidney (Kd) (mean TRF length = 16.16) tissues from clone #3.
Figure 6.
Comparison of telomere lengths between cloned cattle and age-matched controls. Densitometric analysis of TRF lengths generated from RsaI/HindfI digestion of genomic DNA from cloned (NT) fetuses and newborn calves compared with age-matched control (CONT) cattle.
Discussion
The results presented in this study clearly show that telomere loss occurs in cultured bovine fibroblasts and ES-like cells; however, cloned cattle, reconstructed from cultured adult and fetal somatic cells, display telomere lengths similar to those of age-matched control animals. The rebuilding of telomeric sequences from shorter telomere lengths observed in the donor cells could be due to the presence of telomerase activity, detected as early as the first week of postcloning embryonic development. Reprogramming of telomerase activity was observed as early as the blastocyst stage in nuclear transfer bovine embryos that were reconstructed with the use of various donor nuclei that display low/nondetectable levels of telomerase. This appearance of telomerase activity in cloned embryos is delayed compared with fertilization-derived bovine embryos, in which relatively high levels of telomerase activity are observed after activation of the embryonic genome at the 8–16-cell stage (24). Other studies have examined structural and functional reprogramming events in reconstructed embryos, including nucleolar and mitochondrial morphology (31), nuclear swelling (32), and the exchange of somatic histone H1 subtypes (25). These studies have demonstrated that nuclear reprogramming takes place over several cell cycles and may be delayed or incomplete in the transferred nucleus (32, 33). Recently, a differential display analysis that compared cDNA profiles between nuclear-transferred bovine blastocysts with in vivo- and in vitro-derived blastocysts demonstrated that most but not all of the mRNAs are reprogrammed in cloned embryos (34). Specific epigenetic DNA modifications are probably required for proper transcriptional activation of the embryonic genome. The lack of complete genetic reprogramming of gene expression and chromatin structure may lead to the developmental failures and abnormalities observed in cloned embryos, fetuses, and offspring (3, 4).
The telomere loss observed in donor somatic cells has been attributed to the age of the donor animal and culture propagation of the cells in vitro (21–23). In this study, telomere attrition was observed in bovine FFs and surprisingly within telomerase-positive bovine ES-like cells after many cell passages. Although the low levels of telomerase activity can account for the shorter telomeres observed in late passage bovine fibroblasts, the shortening of telomere length observed in late-passage ES-like cells may be due to the observed decline in telomerase activity. Other reports have demonstrated decreased telomerase expression in dividing stem cells in culture, with an associated shortening of telomere lengths (35, 36). These studies suggest that the observed telomerase activity in candidate bovine stem cells is not sufficient to prevent telomere shortening. Single-stranded telomeric DNA damage, caused by oxygen free radicals, has been shown to accumulate in cells after prolonged periods of culture and confluency, leading to an increased rate of telomeric shortening in fibroblasts (37). This oxidative stress-induced telomeric damage has also been observed in telomerase-positive cells grown in culture (38). The reduced telomerase activity levels observed in late-passage bovine ES-like cells may repair telomeric damage but do not prevent the telomere shortening produced by the “end replication problem.” Early-passage cells cultured under serum starvation conditions may also be susceptible to increased telomeric damage/shortening because of reduced telomerase levels due to cell cycle exit into the quiescent (Go) state (39).
In our study, telomere rebuilding was observed in multiple tissue and cell samples from cloned bovine fetuses and newborn calves derived from cultured fetal and adult cell lines. The mean TRF lengths of nuclear transfer bovine fetuses and offspring were not significantly different from those of age-matched control samples. However, we did find animal-to-animal variation and tissue-to-tissue variation between and within newborn cloned calves, respectively. Previous studies have shown significant telomere length differences among individuals, and the telomere synchrony observed in fetal tissues is lost during postnatal life (7, 40). The telomere length variations among and within newborn calves could be due to donor cell selection, tissue/animal-specific differences in telomere rebuilding, and/or the different proliferative rates of different tissues (40). Our results and those of Lanza et al. (23) are in contrast to observations on cloned sheep, which were shown to have shortened telomeres (21, 22). Despite having shorter mean TRF lengths, nuclear-transfer sheep are healthy, fertile, and typical for sheep of their breeds (21, 22). The birth of cloned offspring and their development to adulthood strongly suggests that proliferative capacity of late-passage tissue culture cells (23) and cells from very old adult animals (1, 26) can be restored to a considerable degree by nuclear transplantation (41). However, it has not been determined whether telomere length accurately reflects the physiological age of an animal. Mice deficient in telomerase activity only show a disrupted phenotype after 4–5 generations (20, 42), and the sequential cloning of mice by transfer of adult cumulus cell nuclei shows no adverse effects (43). It must be noted, however, that mice telomeres are substantially longer than those of other species, and telomerase activity is present in most of their adult somatic cells (7).
We did not observe any significantly elongation of telomere length in any of the cells and tissues examined from cloned cattle beyond that of control animals, as Lanza et al. (23) recently reported for nucleated blood cells of cloned calves. The discrepancies in telomere length reported for cloned animals could be due to differences in donor cell type, nuclear transfer procedure, and species (23), but also suggest that other regulatory mechanisms other than just the presence of telomerase activity may be involved in determining telomere length. Each vertebrate species has a set characteristic maximum telomere length (7, 44, 45), and many immortalized and cancer cell lines where telomerase has been reactivated show extremely high levels of telomerase activity that only maintains short telomeres (17). Telomere-specific binding proteins such as telomeric repeat binding factor-1, telomeric repeat binding factor-2, tankyrase, and telomeric repeat binding factor-1 interacting nuclear protein-2 have recently been implicated as mediators of telomere length (46–48). These proteins may directly inhibit/facilitate the binding of telomerase to telomeric DNA or provide structural changes within the telomere that prevent/promote telomerase binding. Interestingly, telomeric repeat binding factor-2 is up-regulated in senescent human fibroblasts (49). Complex telomere remodeling and telomerase regulation during nuclear reprogramming may promote telomere restoration and possibly telomere length extension in cloned offspring. Alternatively, because most of the cloned calves studied by Lanza et al. (23) experienced pulmonary hypertension, respiratory distress and fever before 4 months of age, an activated immune response might have triggered an up-regulation of telomerase activity and subsequent lengthening of the telomeres in nucleated blood cells, as demonstrated previously (50).
In summary, we have observed the reprogramming of telomerase activity and the restoration of telomere length in cloned cattle derived from the nuclear transfer of cultured and aged somatic cells. The telomere rebuilding observed in cloned cattle may be attributed to the nuclear reprogramming of telomerase activity that was detected at the blastocyst stage of cloned embryo development. Detection of telomerase reexpression could be used as a marker to assess the extent and timing of nuclear reprogramming in reconstructed embryos. These results demonstrate that cloned offspring repair genomic modifications acquired during the donor nuclei's in vivo and in vitro period before nuclear transfer, suggesting that along with telomere shortening, nuclear reprogramming outside of the germ line may repair other forms of DNA alterations, such as DNA damage. Telomere research on cloned mammals may determine the mechanism and timing of telomere restoration and any physiological effects of somatic cell nuclear transfer on the aging process.
Acknowledgments
We thank Liz St. John and Ed Reyes for their technical assistance. We thank Dr. Wafa Slimane and Dr. Jim Petrik for critical reading of the manuscript and Dr. Charles Long for the production of cloned cattle from the 9-year-old Charolais female. This work was funded by the Natural Science and Engineering Research Council of Canada, the Barbara Graham Memorial Trust, and the Texas Co-ordinating Board of Higher Education, Advanced Technology Program.
Abbreviations
- TRAP
telomeric repeat amplification protocol
- TRF
terminal restriction fragment
- ES
embryonic stem
- FFs
fetal fibroblasts
- PD
population doubling
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031559298.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031559298
References
- 1.Kubota C, Yamakuch H, Todoroki J, Mizoshita K, Tabara N, Barber M, Yang X. Proc Natl Acad Sci USA. 2000;97:990–995. doi: 10.1073/pnas.97.3.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cibelli J B, Stice S L, Golueke P J, Kane J J, Jerry J, Blackwell C, Ponce de Leon F A, Robl J M. Science. 1998;280:1256–1258. doi: 10.1126/science.280.5367.1256. [DOI] [PubMed] [Google Scholar]
- 3.Kikyo N, Wolffe A P. J Cell Sci. 2000;113:11–20. doi: 10.1242/jcs.113.1.11. [DOI] [PubMed] [Google Scholar]
- 4.Gurdon J B, Colman A. Nature (London) 1999;402:743–746. doi: 10.1038/45429. [DOI] [PubMed] [Google Scholar]
- 5.Harley C B, Flutcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 6.Allsopp R C, Chang E, Kashefi-Aazam M, Rogaev E I, Piatyszek M A, Shay J W, Harley C B. Exp Cell Res. 1995;220:194–200. doi: 10.1006/excr.1995.1306. [DOI] [PubMed] [Google Scholar]
- 7.Prowse K R, Greider C W. Proc Natl Acad Sci USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackburn E H. Nature (London) 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 9.Olovnikov A M. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 10.Levy M Z, Allsopp R C, Futcher A B, Greider C W, Harley C B. J Mol Biol. 1992;225:951–960. doi: 10.1016/0022-2836(92)90096-3. [DOI] [PubMed] [Google Scholar]
- 11.Harley C B, Futcher A B, Greider C W. Nature (London) 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 12.Coviello-McLaughlin G M, Prowse K R. Nucleic Acids Res. 1997;25:3051–3058. doi: 10.1093/nar/25.15.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaziri H, Benchimol S. Exp Gerontol. 1996;31:295–301. doi: 10.1016/0531-5565(95)02025-x. [DOI] [PubMed] [Google Scholar]
- 14.Greider C W, Blackburn E H. Cell. 1985;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 15.Harley C B. Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 16.Wright W E, Piatyszek M A, Rainey W E, Byrd W, Shay J W. Dev Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Shay J W, Bacchetti S. Eur J Cancer. 1997;3:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 18.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C-P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama J-I, Tahara H, Tahara E, Saito M, Ito K, Nakamura H, Nakanishi T, Tahara E, Ide T, Ishikawa F. Nat Genet. 1998;18:65–68. doi: 10.1038/ng0198-65. [DOI] [PubMed] [Google Scholar]
- 20.Lee H-W, Blasco M A, Gottlieb G J, Horner J W, II, Greider C W, DePinho R A. Nature (London) 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 21.Shiels P G, Kind A J, Campbell K H, Waddington D, Wilmut I, Colman A, Schnieke A E. Nature (London) 1999;399:316–117. doi: 10.1038/20580. [DOI] [PubMed] [Google Scholar]
- 22.Shiels P G, Kind A J, Campbell K H S, Wilmut I, Waddington D, Colman A, Schnieke A E. Cloning. 1999;1:119–125. doi: 10.1089/15204559950020003. [DOI] [PubMed] [Google Scholar]
- 23.Lanza R P, Cibelli J B, Blackwell C, Cristofalo V J, Francis M K, Baerlocher G M, Mak J, Schertzer M, Chavez E A, Sawyer N, et al. Science. 2000;288:665–669. doi: 10.1126/science.288.5466.665. [DOI] [PubMed] [Google Scholar]
- 24.Betts D H, King W A. Dev Genet. 1999;25:397–403. doi: 10.1002/(SICI)1520-6408(1999)25:4<397::AID-DVG13>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 25.Bordignon V, Clarke H J, Smith L C. Biol Reprod. 1999;61:22–30. doi: 10.1095/biolreprod61.1.22. [DOI] [PubMed] [Google Scholar]
- 26.Hill J R, Winger Q A, Long C R, Looney C R, Thompson J A, Westhusin M E. Biol Reprod. 2000;62:1135–1140. doi: 10.1095/biolreprod62.5.1135. [DOI] [PubMed] [Google Scholar]
- 27.Sokal R, Rohlf F J. Biometry. 2nd Ed. San Francisco: Freeman; 1981. pp. 245–261. [Google Scholar]
- 28.Burger A M, Bibby M C, Double J A. Br J Cancer. 1997;75:516–522. doi: 10.1038/bjc.1997.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomson J A, Itskovitz-Eldor J, Shapiro S S, Waknitz M A, Swiergiel J J, Marshall V S, Jones J M. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 30.Wilmut I, Campbell K H S. Science. 1998;281:1611. doi: 10.1126/science.281.5383.1611b. (lett.). [DOI] [PubMed] [Google Scholar]
- 31.King W A, Shepherd D L, Plante L, Lavoir M C, Looney C R, Barnes F L. Mol Reprod Dev. 1996;44:499–506. doi: 10.1002/(SICI)1098-2795(199608)44:4<499::AID-MRD10>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 32.Stice S L, Robl J M. Mol Reprod Dev. 1990;25:272–280. doi: 10.1002/mrd.1080250309. [DOI] [PubMed] [Google Scholar]
- 33.Lavoir M C, Kelk D, Rumph N, Barnes F, Betteridge K J, King W A. Biol Reprod. 1997;57:204–213. doi: 10.1095/biolreprod57.1.204. [DOI] [PubMed] [Google Scholar]
- 34.De Sousa P A, Winger Q, Hill J R, Jones K, Watson A J, Westhusin M E. Cloning. 1999;1:63–69. doi: 10.1089/15204559950020102. [DOI] [PubMed] [Google Scholar]
- 35.Yui J, Chiu C P, Landsdorp P M. Blood. 1998;91:3255–3262. [PubMed] [Google Scholar]
- 36.Golubovskaya V M, Filatov L V, Behe C I, Presnell S C, Hooth M J, Smith G J, Kaufmann W K. Mol Carcinog. 1999;24:209–217. doi: 10.1002/(sici)1098-2744(199903)24:3<209::aid-mc7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 37.Sitte N, Saretzki G, von Zglinicki T. Free Radical Biol Med. 1998;24:885–893. doi: 10.1016/s0891-5849(97)00363-8. [DOI] [PubMed] [Google Scholar]
- 38.Saretzki G, Sitte N, Merkel U, Wurm R E, von Zglinicki T. Oncogene. 1999;18:5148–5158. doi: 10.1038/sj.onc.1202898. [DOI] [PubMed] [Google Scholar]
- 39.Holt S E, Wright W E, Shay J W. Mol Cell Biol. 1996;16:2932–2939. doi: 10.1128/mcb.16.6.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Youngren K, Jeanclos E, Aviv H, Kimura M, Stock J, Hanna M, Skurnick J, Bardeguez A, Aviv A. Hum Genet. 1998;102:640–643. doi: 10.1007/s004390050755. [DOI] [PubMed] [Google Scholar]
- 41.Wilmut I, Clark J, Harley C B. Nat Biotechnol. 2000;18:599–600. doi: 10.1038/76430. [DOI] [PubMed] [Google Scholar]
- 42.Herrera E, Samper E, Martin-Caballero J, Flores J M, Lee H W, Blasco M A. EMBO J. 1999;18:2950–2960. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakayama T, Perry A C, Zuccotti M, Johnson K R, Yanagimachi R. Nature (London) 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 44.Kakuo S, Asaoka K, Ide T. Biochem Biophys Res Commun. 1999;263:308–314. doi: 10.1006/bbrc.1999.1385. [DOI] [PubMed] [Google Scholar]
- 45.Kozik A, Bradbury E M, Zalensky A O. Biol Reprod. 2000;62:340–346. doi: 10.1095/biolreprod62.2.340. [DOI] [PubMed] [Google Scholar]
- 46.van Steensel B, de Lange T. Nature (London) 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 47.van Steensel B, Smogorzewska A, de Lange T. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 48.Kim S-H, Kaminker P, Campisi J. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Figueroa R, Lindenmaier H, Hergenhahn M, Vang Neilson K, Boukamp P. Cancer Res. 2000;60:2770–2774. [PubMed] [Google Scholar]
- 50.Weng N-P, Granger L, Hodes R J. Proc Natl Acad Sci USA. 1997;94:10827–10832. doi: 10.1073/pnas.94.20.10827. [DOI] [PMC free article] [PubMed] [Google Scholar]