Abstract
The central role of beneficial mutations for adaptive processes in natural populations is well established. Thus, there has been a long-standing interest to study the nature of beneficial mutations. Their low frequency, however, has made this class of mutations almost inaccessible for systematic studies. In the absence of experimental data, the distribution of the fitness effects of beneficial mutations was assumed to resemble that of deleterious mutations. For an experimental proof of this assumption, we used a novel marker system to trace adaptive events in an evolving Escherichia coli culture and to determine the selective advantage of those beneficial mutations. Ten parallel cultures were propagated for about 1,000 generations by serial transfer, and 66 adaptive events were identified. From this data set, we estimate the rate of beneficial mutations to be 4 × 10−9 per cell and generation. Consistent with an exponential distribution of the fitness effects, we observed a large fraction of advantageous mutations with a small effect and only few with large effect. The mean selection coefficient of advantageous mutations in our experiment was 0.02.
Advantageous mutations lead to a higher fitness and hence per definition to more offspring of their bearer (1–4). Unfortunately, these beneficial mutations are rare (5–7) and thus difficult to study. Rapidly dividing organisms, such as yeast and bacteria, offer the advantage that beneficial mutations could potentially be monitored in the laboratory. In a growing population many new mutations are introduced, with a large fraction of deleterious mutations (8–10). Depending on the population size, most of the deleterious mutations are purged from the population. Advantageous mutations, however, will spread and increase the overall fitness of the population. To identify the carriers of favorable mutations, an informative genetic marker is required that discriminates between clonal lineages. Until now, no such marker system was available for Escherichia coli or yeast. In this report, we used microsatellites, a highly informative marker, to identify adaptive events in an evolving E. coli culture, to calculate the Malthusian fitness parameter of the beneficial mutations, and to visualize clonal interference among different advantageous mutations.
Materials and Methods
Genetic Marker, Bacterial Strain and Culture Conditions.
The PCR product of the microsatellite locus pnga 255, which contains a (GA)n microsatellite and 80-bp flanking sequence from Arabidopsis thaliana, was cloned into the NruI site of the plasmid pBR322. The common laboratory strain E. coli XL1 blue distributed by Stratagene [recA1 end A1 gyr A96 thi-1 hsdR17 sup E44 relA1 lac (F′ pro AB lacIq ZΔM15 Tn10)] was used for transformation of the cloned dinucleotide microsatellite. An overnight culture was inoculated with a single clone carrying a (GA)30 microsatellite and subsequently used as starter culture for the ten replicate cultures. Hence, all experimental populations were founded by one single ancestral cell and were thus genetically uniform at the beginning of the experiment. Populations were maintained by serial transfer in 5 ml rich media (Lennox L Broth Base, GIBCO/BRL) at 37°C and 250 rpm. Every 12 h, each population was diluted by a factor 1:500, allowing approximately nine generations per transfer. Bacterial density at transfer was ≈5 × 108 cells per ml. Cell numbers were obtained by titration. Samples from each population were periodically (every ≈27th generation) stored at −80°C. About every 54th generation, ampicillin (100 μg/ml) was added to assure maintenance of the pBR322 plasmid in the cultures. No influence on microsatellite allele frequencies by this treatment was noted. The number of generations per growth cycle (g) were estimated as log2 (N12h/N0), where N0 is the number of cells during transfer and N12h is the number of cells after 12 h of growth. Population genetic theory predicts that only a small number of beneficial mutations is not lost by drift. The substitution rate for beneficial mutations per generation (k) can be approximated as k ≅ μN0g2S, where N0 is the cell number after the transfer and S the average selection coefficient of beneficial mutations (11). Hence the beneficial mutation rate can be calculated as
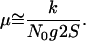 |
Restriction-Analysis and Detection of Adaptive Events.
Plasmid DNA was purified according to standard protocols. Every 90th generation plasmid DNA of each population was digested with HaeIII and separated on a denaturing 7% sequencing gel. DNA fragments and a size standard were transferred to a membrane by capillary blotting. Fragments carrying the microsatellite were detected by hybridization with a 32P-labeled oligonucleotide specific for A. thaliana. Signal intensities were determined by three independent measurements on a phosphorimager (Bio-Rad, GS-525). To determine the experimental error rate, we independently processed 10 samples from the same bacteria culture. The standard deviation was determined for these independent preparations. We observed that the standard deviation depended on the signal intensity. Alleles present at lower frequencies were associated with a higher experimental error. To account for this, we fitted a function that describes the relation between signal intensity and standard deviation: y is the empirically determined standard deviation among independent measurements, which can be expressed as a function of the signal intensity (x) of a given band: y = 0.3065x0.3878.
Changes in allele frequencies during the experiment (δF) were determined every 90 generations by comparing the signal intensities of equally sized fragments by using the following equation:
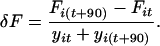 |
Here, Fit is the frequency of allele i at time t (measured in generations), and y is the experimental error as described above. Only δF values larger than 4 were considered significant; thus, values with large experimental errors (y) are not included for further analysis. Persistence time of a selective sweep was determined from the first significant increase in frequency until the allele reaches its maximum frequency.
To verify that the observed changes in allele frequency are caused by frequency changes of the different bacterial genotypes rather than changes in plasmid copy number, we plated individual E. coli cells and typed the microsatellite for each colony separately. The obtained allele frequency distribution corresponded to that obtained from our blotting experiments.
Determination of Fitness Parameter m.
The Malthusian fitness parameter m can be determined from the frequency increase of the carrier of the advantageous mutation (12):
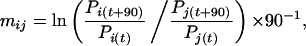 |
where mij is given per generation. Pi is the frequency of the selected lineage at the time point when a statistically significant increase in allele frequency was detected. Pj = 1 − Pi. Time measured in generations is specified by t.
Results and Discussion
Identification of Adaptive Events.
The genetic marker in our experiment consists of a dinucleotide microsatellite cloned into a plasmid vector (pBR322). The high instability of microsatellites rapidly generates different alleles in dividing cells. These alleles at a single microsatellite locus are sufficient to trace clonal lineages, because the Rep protein-mediated copy number control prevents the take up of a second plasmid with the same replication control system (13).
Ten parallel cultures were started from a single cell carrying a cloned microsatellite. The cultures were propagated for about 1,000 generations by serial transfer. Every 90 generations, the allelic spectrum of the microsatellite was monitored in the population. Because of the high mutation rates of cloned microsatellites, all populations had already accumulated substantial variability after 90 generations. Analysis of the bacterial culture in subsequent generations allowed us to follow the shifts in allele frequency at the cloned microsatellite locus. Hence, for each of the ten replicate populations, we surveyed the allelic distribution every 90 generations. Changes in allele frequency could, in principle, have three different causes: (i) new mutations in the microsatellite, (ii) genetic drift, and (iii) selection. The first two evolutionary forces are not directed and affect all alleles to the same extent. In a multiallelic system such as microsatellites, however, directional selection increases only the frequency of the allele associated with the advantageous mutation. Hence, it is possible to trace adaptive events in a growing bacteria population by the significant increase of one microsatellite allele.
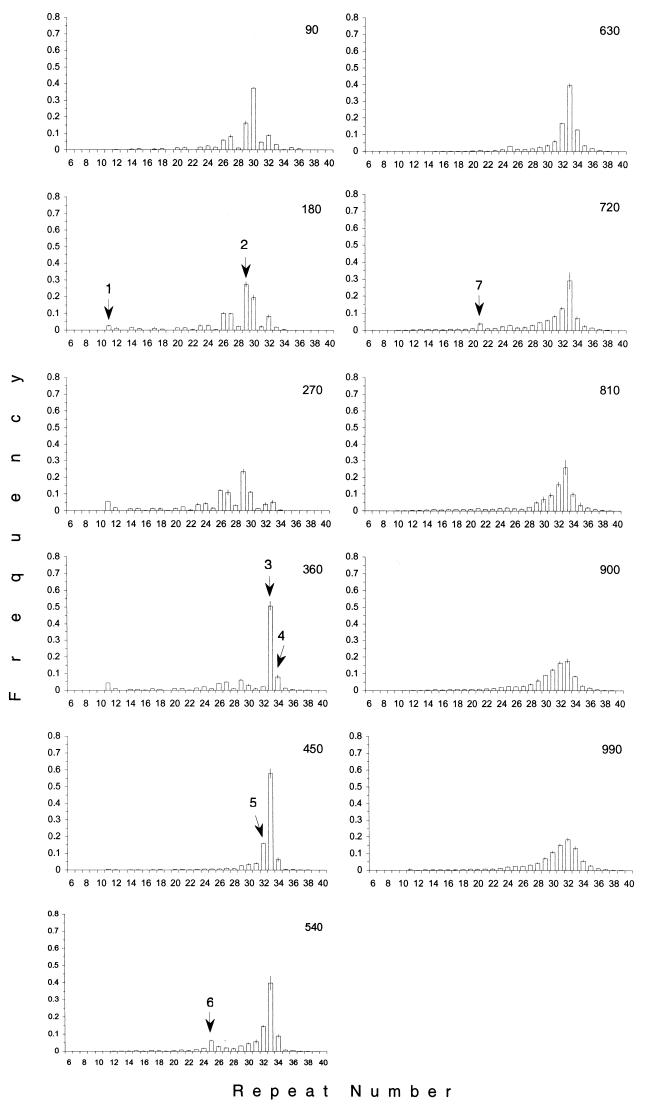
Fig. 1 shows the typical temporal fluctuations in allele frequencies of one bacterial culture. Visual inspection already indicates that the allele distribution is not constant over time. By contrast, some systematic changes can be recognized. A striking shift in allele distribution becomes apparent after generation 270, where a single allele with 33 repeats has markedly increased its frequency during 90 generations (Fig. 1). This pattern is consistent with an advantageous mutation in the cells carrying the allele with 33 repeats. For an objective criterion to identify selective sweeps in growing E. coli cultures, we determined the change in frequency of each allele relative to the previous measurement 90 generations ago. After accounting for measurement errors, we identified 66 significant increases of a single allele in the 10 replicate cultures (see supplementary Figs. 4–12, which are published as supplemental data on the PNAS web site, www.pnas.org). Hence, at least 66 advantageous mutations have occurred over approximately 10,000 generations. The beneficial mutation rate was estimated by the equation μ ≅ k/N0g2S (see Materials and Methods). Given an approximate substitution rate for beneficial mutations (k) of 6.7 × 10−3 per generation, a population size after transfer (N0) of 5 × 106, nine generations per growth cycle (g), and a mean selection coefficient (S) of 0.02 (see below), we obtained 4 × 10−9 per cell generation as an estimate for the beneficial mutation rate.
Figure 1.
Allele frequencies for one of the ten replicate populations. During the entire experiment, every 90th generation, a “snapshot” of the allele distribution in the evolving population was taken. The number in the upper right corner of each diagram indicates the number of generations; error bars show standard deviations of three independent measurements. Each column indicates the frequency of the corresponding allele in the bacterial culture at a given time. The shown frequencies are based on approximately 5 × 108 cells. Arrows indicate the alleles, which significantly increase in frequency. All significant increases are consecutively numbered.
Selective Sweeps Are Reproducible.
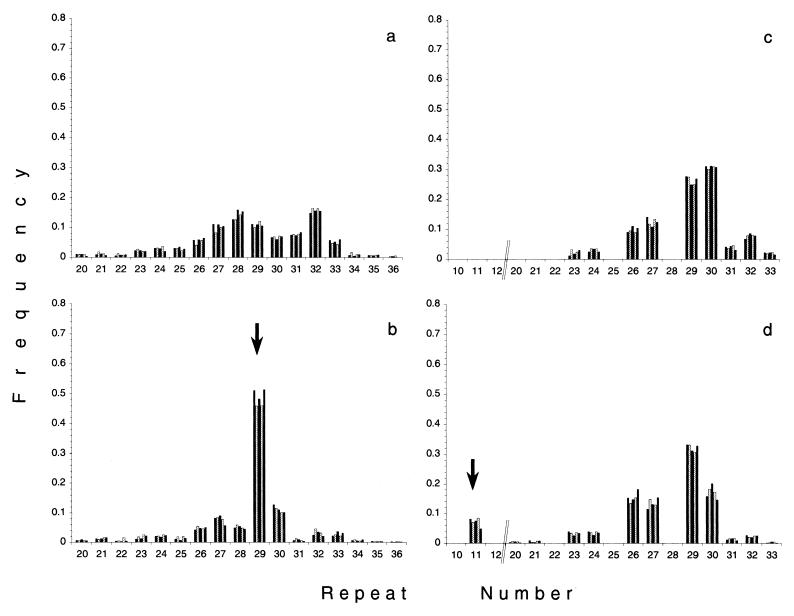
For an experimental test of our criterion to identify beneficial mutations, we repeated the spread of beneficial mutations. Replicas were inoculated with cells that had been frozen at least 90 generations before a statistically significant adaptive event could be identified. Four different populations were replicated in five parallel cultures each and propagated for 180 generations. The four different populations were selected to include different types of alleles with a significant increase in frequency: alleles that reached a high frequency (>0.4) as well as alleles that did not reach a high frequency (<0.1).
The outcome of five replicates for one population is given in Fig. 2 a and b. Fig. 2a displays the allele distribution in the replicate cultures, before the spread of the beneficial mutation could be detected (but the advantageous mutation was already present at low frequencies). Fig. 2b indicates that the outcome of the beneficial mutation was deterministic in all five parallel cultures. It should be noted that all replicas are very similar despite that they had already been independently cultivated for ≈90 generations. Fig. 2 c and d shows the replication of the spread of a beneficial allele, which did not reach high frequency. Fig. 2d clearly indicates that this class of changes is also reproducible and therefore driven by selection. In 1 of 20 replicate cultures, 1 single replica showed a different pattern. A novel advantageous mutation arose during the propagation of this replicate, and the frequency of a former inconspicuous allele increased significantly. In summary, these experiments demonstrated that our method reliably identifies beneficial mutations with low and high fitness effects in growing E. coli cultures.
Figure 2.
Experimental reproduction of selective sweeps, which were identified by a statistically significant change in allele frequency. (a and b) The replication of a significant increase in allele frequency, which reaches a high frequency. (c and d) The replication of a significant increase in allele frequency, which does not reach a high frequency. For each replication experiment, five replicate cultures were inoculated from a frozen stock (representing generation 378 in the original culture for the high frequency event, 108 for the low frequency event respectively). Allele frequency distributions are shown after 18 generations (a and c) and 90 generations (b and d) of independent propagation of the replicate cultures. Replicas were analyzed and tested for significant allele frequency shifts. In all replicate cultures, significant change in allele frequency was observed for the allele carrying 29 repeats (a and b) and for the 11-repeat allele (c and d). Arrows indicate the alleles, that significantly increase in frequency.
Distribution of Adaptive Events.
Previously, it has been shown that the frequency of beneficial mutations decreases with time (14). To test whether a similar pattern can be detected in our experiment, we determined the temporal distribution of adaptive events. In particular, the occurrence and persistence time of selective sweeps was analyzed in all ten replicate cultures. However, no temporal pattern could be identified, indicating that 1,000 generations of serial propagation were not enough to identify a temporal pattern in the distribution of adaptive events. Each of the ten experimental populations had its own specific pattern, and no decrease in the frequency of adaptive events could be observed in the latter half of the experiment. This observation is consistent with previous results that most beneficial mutations are occurring during the first 2,000 generations (14). Similarly, we did not detect a correlation between the time a selective sweep occurred and the associated selection coefficient.
Clonal Interference.
In the absence of recombination, two advantageous mutations that have arisen in two cells cannot be combined into a single, superior genotype. Hence, their carriers will compete with each other, a phenomenon called clonal interference (15–17). Whereas theory predicts clonal interference, in particular for large populations and high mutation rates (17), so far only indirect proofs exist (18–21). Our marker system, however, permits the direct observation of clonal interference in a growing E. coli population.
One example for clonal interference can be seen in Fig. 1. The allele with 11 repeats rises significantly in frequency from generation 90 to 270 (it should be noted that this spread was reproduced in five replica cultures), but, when an advantageous mutation occurs in one cell associated with a 33-repeat microsatellite allele, the spread of the allele with 11 repeats is prevented and it is eventually lost from the population. Overall, clonal interference is common in our experiment: often more than a single beneficial mutation can be detected in a given time interval. As a consequence of clonal interference, concurrent selective sweeps have different persistence times. The outcome of clonal interference, however, depends on the selection coefficient of the advantageous mutations.
Distribution of Selection Coefficients.
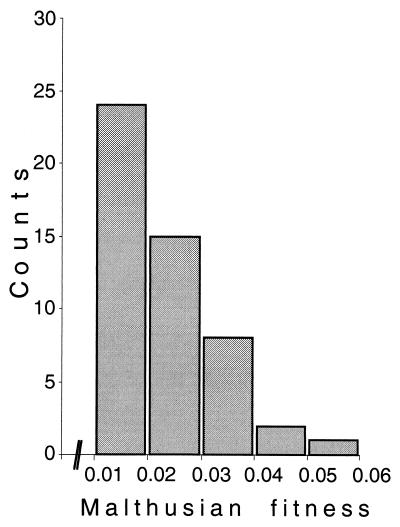
To infer the selective advantage of clones sweeping through a bacterial culture, we determined the Malthusian fitness parameter (12) of all 66 identified selective sweeps. In contrast to previous studies (11, 14), we did not measure the selective advantage by competition experiments with non-evolved bacteria, but determined fitness relative to the remaining culture. The obtained Malthusian fitness parameters range from 0.006 to 0.059. As suggested by population genetic theory (16), most advantageous mutations had a small effect (Fig. 3). Only a small fraction had large selection coefficients. The simplest model that captures this feature is the exponential distribution. Whether the beneficial mutations in our experiment really follow an exponential distribution or are better described by a gamma distribution requires a larger number of adaptive events. Nevertheless, it has to be noted that our calculations probably underestimate the true selection coefficients for two reasons. First, new mutations at the microsatellite locus in those cells bearing the advantageous mutation will affect estimates of the selection coefficients. Whereas new microsatellite mutations will not impair the identification of adaptive events, the determined selection coefficient could be underestimated. Second, clonal interference will cause a less rapid spread of beneficial mutations, leading to an underestimate of the actual selection coefficient as measured in the absence of the competing beneficial mutation. Despite these limitations, our results are most likely one of the best estimates for the distribution of fitness effects of beneficial mutations.
Figure 3.
Distribution of selection coefficients combined over 1,000 generations and 10 experimental cultures. Selection coefficients smaller than 0.01 were not included, because most of them could not be discriminated from experimental errors. It should be noted that all selection coefficients were determined without accounting for clonal interference.
Previous experiments used minimal medium, which represents a novel and restricted environment to cultivate E. coli cells (11). In such an environment, the entire metabolism has to be modified to be autotrophic. Many operons are expected to change their regulation to constitutively produce many compounds absent in the environment. In contrast, our experiments were carried out in rich LB media, as recommended by the supplier of the E. coli strain. Even thought this medium should not impose any restrictions on the availability of resources, we observed a high rate of adaptive events, suggesting that the cells still have a considerable potential for improvement. Interestingly, the estimated rate of advantageous mutations is similar to a previously reported one that accounted for clonal interference (17). Hence, counter to intuition, culture conditions seem not to influence the beneficial mutation rate. For stringent comparisons, it would be required to use the same method of mutation rate measurements for both culture conditions. It should be noted, however, that our estimated beneficial mutation rates are conservative for two reasons. First, the conservative threshold used to identify adaptive events inevitably excludes those events with a very small selection coefficient. Second, because the deleterious mutation rate in E. coli is estimated to be in the order of 10−4 (22), we can assume that favorable mutations do not appear in deleterious mutation-free genomes. Thus, those cells carrying a new beneficial mutation but suffering a negative net selection coefficient cannot be identified (23).
Perspectives.
One of the primary goals of evolutionary biology is to understand which genetic changes are driving adaptive evolution. In contrast to insightful experiments with bacteriophages (24, 25), for E. coli, the current state of sequencing technology precludes sequencing the whole genome of several E. coli lineages to identify those mutations. Some preliminary data suggest that base substitutions are infrequent, but jumping of transposable elements may be an important evolutionary force (20). An alternative approach would be to monitor the changes in expression level of the whole organism. Comparison of non-evolved and evolved cells could, in principle, be used to identify those changes. Using DNA chip technology, a recent study in yeast, however, found several hundred genes with a significantly altered expression in the evolved cells (26). The high number of affected genes could have two reasons: first, gene cascades are affected and, second, the analyzed population carried more than a single cell type. Recently, it has been shown that more than a single cell lineage could be maintained during the course of experimental evolution (27). Using our marker technology, it would be possible to discriminate between cell lineages and thus to obtain a purer signal than in the previous study. The expression pattern could be compared directly between the carrier of the advantageous mutation and other cells in the population. Furthermore, the availability of a genetic marker system will provide further insight into the shape of the adaptive landscape.
Supplementary Material
Acknowledgments
We are grateful to R. Bürger, S. F. Elena, B. Harr, and D. Hartl, and three anonymous reviewers, for helpful comments and discussions. Many thanks to M. Puchinger for help with the phosphorimager analyses. Special thanks to all members of the Schlötterer lab for continued interest and many helpful discussions. This work is supported by Fonds zur Förderung der Wissenschaftlichen Forschung grants to C.S.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fisher R A. Proc R Soc Edinb. 1922;42:321–341. [Google Scholar]
- 2.Fisher R A. Proc R Soc Edingb. 1930;50:204–219. [Google Scholar]
- 3.Haldane J B S. Proc Camb Philos Soc. 1927;23:838–844. [Google Scholar]
- 4.Wright S. Genetics. 1931. 97–159. [Google Scholar]
- 5.Dobzhansky T. Genetics of Evolutionary Process. New York: Columbia Univ. Press; 1970. [Google Scholar]
- 6.Kimura M. Genet Res. 1967;9:25–34. [Google Scholar]
- 7.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 8.Drake J W. Nature (London) 1969;221:1132. doi: 10.1038/2211132a0. [DOI] [PubMed] [Google Scholar]
- 9.Drake J W. Annu Rev Genet. 1991;25:125–146. doi: 10.1146/annurev.ge.25.120191.001013. [DOI] [PubMed] [Google Scholar]
- 10.Drake J W, Charlesworth B, Charlesworth D, Crow J F. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenski R E, Rose M R, Simpson S C, Tadler S C. Am Nat. 1991;138:1315–1341. [Google Scholar]
- 12.Hartl D L, Clark A G. Principles of Population Genetics. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 13.Rasooly A, Rasooly R S. Trends Microbiol. 1997;5:440–446. doi: 10.1016/S0966-842X(97)01143-8. [DOI] [PubMed] [Google Scholar]
- 14.Lenski R E, Travisano M. Proc Natl Acad Sci USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller H J. Am Nat. 1932;66:118–138. [Google Scholar]
- 16.Fisher R A. The Genetical Theory of Natural Selection. Oxford: Oxford Univ. Press; 1930. [Google Scholar]
- 17.Gerrish P J, Lenski R E. Genetica. 1998;102–103:127–144. [PubMed] [Google Scholar]
- 18.Miralles R, Gerrish P J, Moya A, Elena S F. Science. 1999;285:1745–1747. doi: 10.1126/science.285.5434.1745. [DOI] [PubMed] [Google Scholar]
- 19.Miralles R, Moya A, Elena S F. J Virol. 2000;74:3566–3571. doi: 10.1128/jvi.74.8.3566-3571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papadopoulos D, Schneider D, Meier-Eiss J, Arber W, Lenski R E, Blot M. Proc Natl Acad Sci USA. 1999;96:3807–3812. doi: 10.1073/pnas.96.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arjan J A, Visser M, Zeyl C W, Gerrish P J, Blanchard J L, Lenski R E. Science. 1999;283:404–406. doi: 10.1126/science.283.5400.404. [DOI] [PubMed] [Google Scholar]
- 22.Kibota T T, Lynch M. Nature (London) 1996;381:694–696. doi: 10.1038/381694a0. [DOI] [PubMed] [Google Scholar]
- 23.Orr H A. Genetics. 2000;155:961–968. doi: 10.1093/genetics/155.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bull J J, Badgett M R, Wichman H A. Mol Biol Evol. 2000;17:942–950. doi: 10.1093/oxfordjournals.molbev.a026375. [DOI] [PubMed] [Google Scholar]
- 25.Wichman H A, Badgett M R, Scott L A, Boulianne C M, Bull J J. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- 26.Ferea T L, Botstein D, Brown P O, Rosenzweig R F. Proc Natl Acad Sci USA. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rainey P B, Buckling A, Kassen R, Travisano M. Trends Ecol Evol. 2000;15:243–247. doi: 10.1016/s0169-5347(00)01871-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.