Abstract
Multiple endocrine neoplasia type 1 (MEN1) is an autosomal dominant cancer syndrome, characterized primarily by multiple tumors in the parathyroid glands, endocrine pancreas, and anterior pituitary. Other tumors, including gastrinoma, carcinoid, adrenal cortical tumors, angiofibroma, collagenoma, and lipoma, also occur in some patients. Individuals with MEN1 almost always have loss-of-function mutations in the MEN1 gene on chromosome 11, and endocrine tumors arising in these patients usually show somatic loss of the remaining wild-type allele. To examine the role of MEN1 in tumor formation, a mouse model was generated through homologous recombination of the mouse homolog Men1. Homozygous mice die in utero at embryonic days 11.5–12.5, whereas heterozygous mice develop features remarkably similar to those of the human disorder. As early as 9 months, pancreatic islets show a range of lesions from hyperplasia to insulin-producing islet cell tumors, and parathyroid adenomas are also frequently observed. Larger, more numerous tumors involving pancreatic islets, parathyroids, thyroid, adrenal cortex, and pituitary are seen by 16 months. All of the tumors tested to date show loss of the wild-type Men1 allele, further supporting its role as a tumor suppressor gene.
Multiple endocrine neoplasia type 1 (MEN1) is an autosomal dominant cancer syndrome characterized by multiple tumors of the parathyroid, endocrine pancreas, and the anterior pituitary. Additional tumors have been associated with MEN1, including foregut carcinoid, adrenal cortical tumors, angiofibroma, collagenoma, and lipoma (1, 2). Linkage studies in affected families mapped the MEN1 locus to chromosome 11q13 (3), and the responsible gene, MEN1, was identified by positional cloning in 1997 (4). Germline mutations in MEN1 have been identified in almost all MEN1 kindreds (5–7), and somatic mutations in MEN1 have been reported in sporadic parathyroid adenomas, pituitary tumors, insulinomas, gastrinomas, and lung carcinoids (8–12). Over 70% of germline mutations are nonsense and frameshifts, predicting truncation or absence of the resulting protein. Missense mutations and in-frame deletions account for the remaining 30% of the almost 250 unique mutations identified to date. Hence, MEN1 appears to be a classic tumor suppressor gene with tumors in affected patients showing somatic loss of the wild-type allele.
The MEN1 gene consists of 10 exons (the first of which is untranslated), spanning 7.2 kb of genomic sequence and encoding a protein of 610 amino acids. The protein product, menin, does not reveal homologies to any other known proteins or possess notable motifs from which the putative function of the protein could be deduced. Menin RNA and protein apparently are expressed in all tissues (13), leaving unexplained the basis for endocrine predominance of neoplasia. Menin is found in the nucleus (14) and binds to the AP1 transcription factor JunD (15). This finding suggests a role in transcriptional regulation, although a detailed model for menin tumor suppression activity remains elusive.
In mice, the Men1 gene is localized on chromosome 19 and has exon–intron organization similar to that of the human gene. Men1 demonstrates 97% identity/98% similarity to MEN1 at the amino acid level and is ubiquitously expressed in all tissues and stages of mouse development (16, 17). To pursue an understanding of tumorigenesis upon loss of menin function, a mouse model was generated via homologous recombination. The heterozygous phenotype of menin inactivation in mice is strikingly similar to that of the human disorder MEN1.
Materials and Methods
Men1 Gene Targeting.
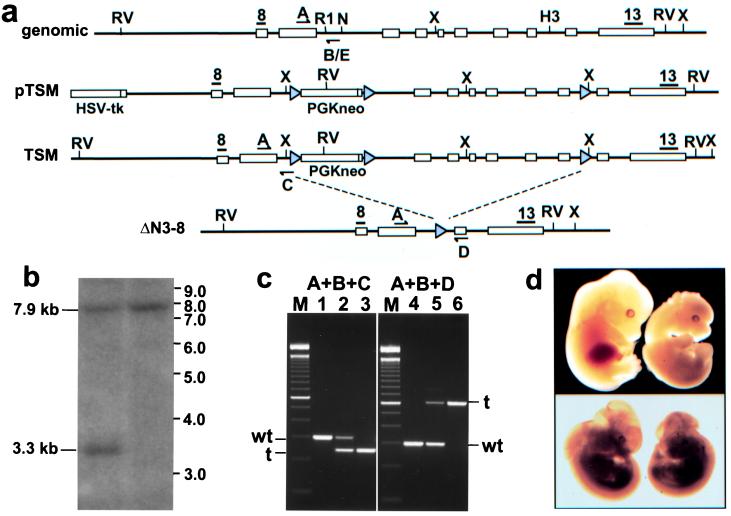
To target the mouse Men1 locus by homologous recombination in embryonic stem (ES) cells, we isolated the 129Sv/cJ7 mouse Men1 gene in a 190-kb bacterial artificial chromosome. This bacterial artificial chromosome was partially digested with Sau3A1, and a pBluescriptIIKS+ subclone containing all 10 exons of Men1 was identified and dubbed 2G4. A 2.1-kb EcoRI genomic fragment containing exons 1 and 2 was isolated from 2G4 and cloned upstream of the floxed phosphoglycerate kinase (PGK)-neomycin cassette in pPNT-loxP2 (a gift of A. Wynshaw-Boris, University of California, San Diego) in the opposite transcriptional orientation. A third loxP site was introduced into intron 8. A 5.1-kb NdeI–BamHI fragment containing exons 3–10 and the newly inserted loxP site in intron 8 was cloned into the pPNT-loxP2 plasmid, which already contained exons 1 and 2, to generate the targeting construct pTSM. A small 134-bp region of the wild-type intron 2 was deleted during cloning and replaced with an 82-bp pPNT-loxP2 vector fragment.
pTSM was electroporated into TC-1 mouse ES cells, and correctly targeted clones were identified by Southern blot. ES cell genomic DNA was digested with EcoRV and probed with probe 8 (Fig. 1a) to reveal 3.3-kb target and 7.9-kb wild-type bands, or with probe 13 to obtain 5.3-kb target and 7.9-kb wild-type bands. To verify the presence of the third loxP site, XbaI digests of ES cell genomic DNA were blotted and hybridized with probe 13 to obtain 2.6-kb targeted and 4.2-kb wild-type bands. Correctly targeted clones were injected into C57BL/6J blastocysts to generate male chimeras. Male chimeras were bred to NIH Black Swiss females or 129/SvEvTacFBR females to generate Men1TSM/+ animals, with subsequent mating of these heterozygotes to generate homozygous Men1TSM/TSM animals.
Figure 1.
(a) Targeting strategy for the Men1 knockouts. Restriction sites: RV, EcoRV; X, XbaI; R1, EcoRI; n, NdeI; H3, HindIII. Gray triangles, LoxP sites; black bars, probes 8 and 13; arrows, genotyping primers. (b) Southern blot of ES cell genomic DNA digested with EcoRV and probed with probe 8. Wild-type allele (7.9 kb) and targeted allele (3.3 kb) are indicated. (c) Agarose gel electrophoresis of genotype PCRs. Lanes 1–3 are wild-type, heterozygote, and homozygote TSM mice, respectively. Lanes 4–6 are wild-type, heterozygote, and homozygote ΔN3–8 mice, respectively. (d) Lethality in homozygote embryos. (Upper) Wild-type E12.5 embryo on left, TSM homozygote E12.5 embryo on right. (Lower) Wild-type E11.5 embryo on left, ΔN3–8 E11.5 embryo on right.
Men1ΔN3–8/+ mice were obtained by crossing Men1TSM/+ mice with the ubiquitously expressing EIIa-cre mouse line (18). Heterozygote mice lacking both the PGK-neomycin cassette and exons 3–8 of the Men1 gene were bred to obtain Men1ΔN3–8/ΔN3–8 homozygotes. All mice were maintained in accordance with National Institutes of Health and American Association of Laboratory Animal Care guidelines.
Genotyping.
Wild-type, Men1TSM/+, and Men1TSM/TSM mice were genotyped by multiplex PCR (95°C, 1 min; 95°C, 30 s; 60°C, 30 s; 72°C, 1 min, for 35 cycles; 72°C, 10 min; 4°C hold) with the use of three primers. Primer A (CCCACATCCAGTCCCTCTTCAGCT) is anchored in exon 2, primer B (CCCTCTGGCTATTCAATGGCAGGG) is specific for the 134-bp wild-type sequence deleted when the TSM construct was cloned, and primer C (CGGAGAAAGAGGTAATGAAATGGC) is specific for the inserted 82-bp vector fragment (Fig. 1a for primer locations). Therefore, the combination of primers A and B yields a wild-type 300-bp amplicon, and primers A and C yield a 239-bp targeted amplicon (Fig. 1c, lanes 1–3).
Wild-type, Men1ΔN3–8/+, and Men1ΔN3–8/ΔN3–8 mice were genotyped with a multiplex PCR of primers A, B, and D (CATAAAATCGCAGCAGGTGGGCAA). Combination of primers A and B yields the wild-type amplicon of 300 bp, and combination of primers A and D, which is located in exon 9, yields a 638-bp ΔN3–8-specific amplicon (Fig. 1c, lanes 4–6). Viable mice were genotyped with the use of tail genomic DNA, and embryos were genotyped with the use of tail and/or amniotic sac, depending on the age of embryo.
Cesarean Sections.
Embryos were removed by Cesarean section after CO2 asphyxiation of pregnant females. The age of embryos was determined by considering the date of plug detection as embryonic day 0.5 (E0.5) and confirmed by staging from the development of external structures. Embryos were evaluated at each 1-day time point between E8.5 and E15.5.
Time Study/Autopsy/Blood Chemistry.
Heterozygous Men1TSM/+ mice were placed on a time study, with mice necropsied every 3 months, beginning at 6 months. Mice were anesthetized with a 0.02 ml/g body weight i.p. injection of Avertin (2.5%), and blood was drawn from the retroorbital sinus. Blood serum was evaluated for insulin, total calcium, and parathyroid hormone levels (Anilytics, Gaithersburg, MD). Glucose levels were analyzed with a Glucometer Elite XL (Bayer, Elkhart, IN) on whole blood at the time of collection. Blood was drawn either in late afternoon or after an 8-h fast. Autopsies were performed after CO2 asphyxiation.
Histology.
Tissues from autopsied animals were snap-frozen in isopentane over liquid nitrogen, fixed overnight in cold 70% ethanol, or fixed in 10% phosphate-buffered formalin. Frozen tissues were imbedded in Tissue-Tek OCT (Sakura Finetek, Torrance, CA), sectioned at 10 μm, mounted, and stained with Mayer's hematoxylin and eosin (H&E). Tissues fixed in formalin were embedded in paraffin and sectioned at 4–6 μm before staining. Ethanol-fixed tissues were embedded in polyester wax (Galard–Schlesinger Industries, Garden City, NY) and sectioned at 8 μm before staining.
Immunohistochemistry.
Tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, and sectioned at 4–6 μm. For some immunohistochemistry, sections were pretreated in water in a microwave (19). A guinea pig polyclonal antibody to porcine pancreatic insulin (Dako) was used at a 1:500 dilution, and rabbit anti-human somatostatin, glucagon, and gastrin were all used at 1:1,000 dilution (Dako). The Vectastain ABC guinea pig or rabbit kit (Vector Laboratories) was used, with diaminobenzidine as the chromogen. Controls included normal tissues of wild-type mice and normal tissues of Men1TSM/+ mice.
Laser Capture Microdissection and Loss-of-Heterozygosity (LOH) Analyses.
Several mice with histopathological abnormalities similar to those represented in Figs. 2 and 3 were selected for tissue processing by laser capture microdissection. Tissues were either frozen or embedded in polyester wax, sectioned at 8 μm, eosin-stained, and microdissected with the use of a PixCell II laser capture microscope with an infrared diode laser (Arcturus Engineering, Santa Clara, CA) as described (20, 21). DNA was isolated from microdissected tissue (according to the procedure described at http://dir.nichd.nih.gov/lcm/LCMTAP.htm#Sample). To test for LOH, microdissected DNA was subjected to a multiplex PCR. The same PCR primers and conditions used to genotype Men1TSM/+ mice were used for LOH analyses, with one exception: primer B was replaced with primer E (CCCTCTGGCTATTCAATGGCAGGG) to yield a 216-bp wild-type amplicon. With the use of these primer sets, a TSM-specific PCR product of 300 bp was distinguishable from the 216-bp wild-type fragment on a 2% agarose gel. All PCRs were run in duplicate.
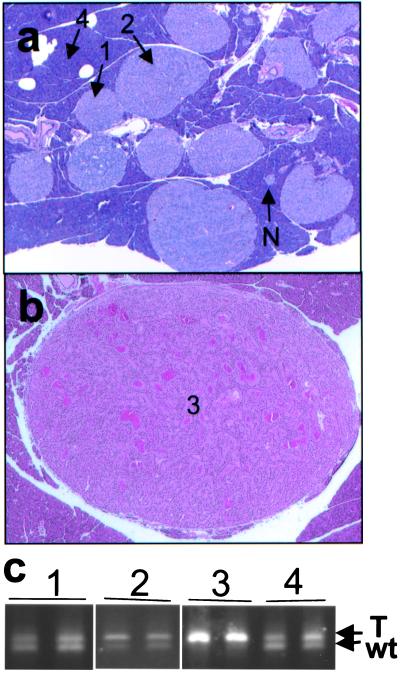
Figure 2.
Pancreatic lesions in Men1TSM/+ mice. (a) H&E-stained section (×15) of pancreas from a 12-month-old mouse, showing normal islets (N), hyperplastic islets (1), hyperplastic islets with dysplasia (2), and acinar pancreas (4). (b) H&E-stained section (×15) of a pancreatic islet cell tumor (3) from a 15-month-old mouse, showing increased vascularization and ribbon-like nuclei formation around capillaries. (c) Agarose gel electrophoresis of PCR from microdissected samples. Numbers correlate with the type of tissue indicated in a and b: 1 is hyperplastic islet, 2 is hyperplastic islet with dysplasia, 3 is pancreatic islet cell tumor, and 4 is acinar tissue. The upper band arises from the TSM allele (T), and the lower band from the wild-type allele (wt).
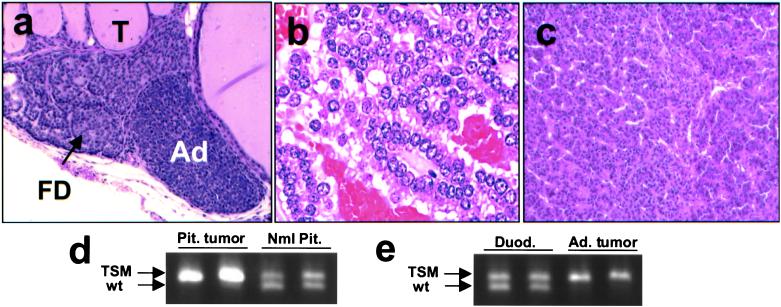
Figure 3.
Other lesions in Men1TSM/+ mice. (a) H&E-stained section (×40) of parathyroid from a 12-month-old mouse with thyroid follicles (T), focal parathyroid dysplasia (FD), and parathyroid adenoma (Ad). (b) H&E-stained section (×120) of pituitary adenoma from a 21-month-old mouse. (c) H&E-stained section (×40) of adrenal cortical tumor from a 15-month-old mouse. (d) Agarose gel electrophoresis, showing loss of the wild-type allele in pituitary adenoma and retention of both alleles in normal pituitary tissue. (e) Agarose gel electrophoresis, showing loss of the wild-type allele in adrenal cortical tumor and retention of both alleles in normal duodenum tissue from the same animal. In LOH the upper band arises from the TSM allele, and the lower band, from the wild-type allele.
Results
Men1 Gene Targeting.
To target the mouse gene by homologous recombination in ES cells, an intended conditional knockout construct was developed that contained the mouse 129/SvJ Men1 locus in the targeting vector backbone, pPNT-loxP2 (Fig. 1a). A floxed PGK-neomycin cassette was inserted into intron 2, and a third loxP site was introduced into intron 8 of the Men1 gene to generate the TSM construct. Linearized pTSM was electroporated into TC-1 ES cells, and the transfectants were selected in the presence of G418 and 2′-fluoro-2′-deoxy-5-iodo-1-β-d-arabinofuranosyluracil. Southern blot analysis of the resulting clones indicated homologous recombination in 8/234 (3.4%) clones (Fig. 1b and data not shown). Of these, three clones were injected into blastocysts, and male chimeras were obtained that transmitted the targeted allele to their progeny when mated to wild-type NIH Black Swiss or 129/SvEvTacFBR females. Genotypes were assigned with the use of a PCR assay (Fig. 1c).
The Homozygous Phenotype Is Embryonic Lethal.
Matings between heterozygous Men1TSM/+ mice yielded no viable Men1TSM/TSM pups (wild type: 29% or 28/98; Men1TSM/+: 71% or 70/98), suggesting that disruption of the Men1 allele by the PGK-neomycin cassette, even though it is located in an intron, resulted in loss of gene function and embryonic lethality. Homozygous embryos removed at E9.5 (4/21) demonstrated no gross abnormalities in comparison to wild-type (4/21) and heterozygous (13/21) littermates, but by E11.5–12.5, Men1TSM/TSM embryos (4/34) were developmentally delayed and significantly smaller, with 20% of embryos exhibiting defects in cranial and/or facial development (Fig. 1d, Upper) in comparison with wild-type (8/34) or heterozygote (22/34) littermates. The majority of Men1TSM/TSM embryos were resorbed by E14.5.
These data suggest that the TSM allele is functioning effectively as a null, but it was desirable to confirm this conclusion by producing an unequivocal null with an intragenic deletion of Men1. Men1TSM/+ mice were crossed with the ubiquitous cre-expressing line EIIa-cre (18) to remove both the neomycin cassette and Men1 exons 3–8. Heterozygous mice exhibiting the appropriate deletion were dubbed Men1ΔN3–8/+. The Men1ΔN3–8/ΔN3–8 genotype was lethal at E10.5–11.5 (Fig. 1d Lower), slightly earlier than Men1TSM/TSM.
Pancreatic Islet Tumors in Heterozygotes.
The incidence of pancreatic islet cell tumors in MEN1 patients, who are heterozygous for a loss-of-function mutation in one MEN1 allele, varies widely from 30% to 80% in different clinical series (22). To detect pancreatic islet tumor development in this model, Men1TSM/+ mice were necropsied at 3-month intervals, beginning at 6 months. No significant abnormalities were observed at 6 months of age. Four of 10 Men1TSM/+ mice had developed large, hyperplastic pancreatic islets and pancreatic islet cell tumors at 9 months of age (Fig. 2 a and b). Larger (1–8-mm diameter), more numerous tumors were visible upon gross analysis by 16 months of age. Capsular invasion was found in one 22-month-old animal. These abnormalities developed initially as medium to large hyperplastic islets (i.e., composed of an excessive number of apparently normal islet cells), into large islets with focal atypia or dysplasia (defined as abnormal cell proliferation), and finally into pancreatic islet tumors. Pancreatic islet tumors were found in 20/71 (28%) Men1TSM/+ mice by 22 months of age or younger. Islet cell adenomas of this type were not found in wild-type, untargeted littermates (n = 2) and were rarely seen in aged mice on the B6, 129 background (2/48, 4.2%; J.M.W., unpublished data). Other minor pancreatic lesions, including duct inflammation, acinar cysts, fatty replacement, focal acinar dysplasia, and focal acinar necrosis, were occasionally detected in Men1TSM/+ mice.
Heterozygous Men1ΔN3–8/+ mice (5/6, or 83%) showed a progression similar to that in Men1TSM/+ from normal pancreatic islets to hyperplastic islets to pancreatic islet tumor formation, with grossly visible pancreatic tumors detected after 9 months of age. Although numbers of Men1ΔN3–8/+ animals are currently small, this finding suggests that the pancreatic islet cell neoplasia may begin at a slightly earlier age in these mice as compared with Men1TSM/+ mice and could define a slight difference in the phenotypes of these two mouse lines.
Elevated serum insulin levels (3.5–39 ng/ml) were noted in all Men1TSM/+ mice with large, hyperplastic islets; hyperplastic islets with atypia; and/or pancreatic islet cell tumors, as compared with the wild-type range of 0.04–2.4 ng/ml. Fasted or unfasted sample collection had no impact on serum insulin levels. The degree of insulin elevation correlated with islet phenotype severity. Slightly elevated insulin levels (2.4–15 ng/ml) indicated mild atypical hyperplasia and possibly the development of a single, small tumor, whereas more elevated insulin levels (15–39 ng/ml) appeared in mice with a combination of islet hyperplasia, islet atypia/dysplasia, and multiple or large tumors. No mice had blood glucose levels below 40 mg/dl or died of suspected hypoglycemia.
Other Tumors in Men1TSM/+ Mice.
One of the most common features of MEN1 patients is primary hyperparathyroidism, which occurs in over 95% of MEN1 patients over age 40 as a result of parathyroid adenoma or hyperplasia (1, 23–27). Consistent with this human phenotype, Men1TSM/+ mice develop focal dysplasia throughout the course of the time study, with 6/25 (24%; 4/14 male, 2/11 female) animals developing parathyroid adenoma as early as 9 months of age (Fig. 3a). One carcinoma was detected. These tumors are rare in control mice (1/47, 2%; J.M.W., unpublished data). Total serum calcium levels were unremarkable.
Pituitary tumors, adrenal cortical tumors, and gastric neuroendocrine tumors are also present in MEN1 patients, with the documented incidence ranging from 15% to 90% for pituitary tumors (1, 24) and up to 40% for adrenal cortical tumors (28). Seven of 27 (26%; 2/15 male, 5/12 female) Men1TSM/+ mice analyzed had a large pituitary adenoma by 16 months of age (Fig. 3b). Adrenal cortical carcinomas (Fig. 3c) were detected in 4/27 Men1TSM/+ mice (20%; 4/15 male, 0/12 female), bilateral pheochromocytoma was detected in 2/27 mice (7.4%; 0/15 male, 2/12 female), lung adenocarcinoma was detected in 6/27 (22%, 5/15 male, 1/12 female), and one gastric neuroendocrine tumor was detected.
Some thyroid abnormalities were also detected in these mice, including cysts and follicular adenoma. In human MEN1 patients, thyroid tumors have been reported to occur in as many as 25% of cases (2, 24). However, because of the high prevalence of thyroid abnormalities in the general population and lack of somatic MEN1 mutations in sporadic thyroid tumors (29), these data are typically discounted as incidental and not significant. Additional tumors not known to be associated with the human phenotype were occasionally found in Men1TSM/+ mice, including prostate carcinoma (n = 1) and ovarian adenoma (n = 1).
Immunohistochemistry.
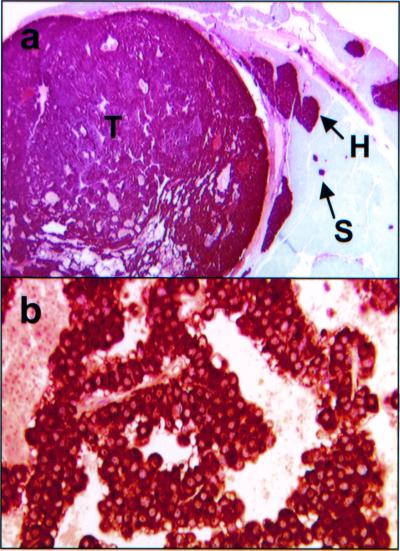
Immunohistochemical data showed that the hyperplastic pancreatic islets and pancreatic islet tumors produced insulin, although at a slightly reduced level compared with wild type (Fig. 4a). Hyperplastic islets and pancreatic islet cell tumors did not express glucagon, somatostatin, or gastrin, with the exception of one case, which expressed insulin and glucagon (data not shown).
Figure 4.
Immunohistochemistry of Men1TSM/+ mouse lesions. (a) Insulin immunohistochemical staining of a pancreatic section from a 20-month-old mouse. This section (×10) shows small islets (S), hyperplastic islets (H), and tumor (T), all containing insulin at appreciable levels. (b) Prolactin immunohistochemical staining of a pituitary adenoma section (×120) from a 21-month-old mouse.
All pituitary tumors detected in Men1TSM/+ mice secrete prolactin, but not ACTH, indicating that these tumors arose from the pars distalis, mimicking the human phenotype (Fig. 4b).
LOH Analysis Shows That the Wild-Type Allele Is Lost in Tumors.
Pancreatic islets showing a range of lesions from hyperplasia to atypia/dysplasia to tumors were microdissected and subjected to LOH analyses (Fig. 2c). Controls included microdissected duodenal or normal acinar tissue from the same mouse. The 216-bp wild-type and 300-bp TSM-specific fragments coamplified in normal tissues and large, hyperplastic islets. In contrast, only the 300-bp TSM-specific fragment amplified in pancreatic tumor tissue, indicating loss of the wild-type allele. LOH analyses of atypical/dysplastic islets typically revealed amplification of both alleles, although in some instances there was reduction in the intensity of the wild-type allele.
Adrenal cortical and pituitary tumors similar to those seen in Fig. 3 were also subjected to laser capture microdissection/LOH. Both the wild-type and TSM alleles were amplified in normal pituitary and duodenal tissues, but only the TSM allele was detected in pituitary and adrenal cortical tumors (Fig. 3 d and e).
Discussion
We have assessed the function of Men1 in tumor development with the use of homologous recombination in mice. The original intention with the TSM construct was to produce a conditional knockout that would function as a wild-type allele but could subsequently be inactivated by tissue-specific, cre-mediated recombination. However, the embryonic lethal phenotype of the homozygote indicates that insertion of the PGK-neomycin cassette in intron 2 inactivates the gene. This hypothesis was confirmed by generating an unequivocal null, the ΔN3–8 allele, and noting a similar embryonic lethal phenotype. The slightly earlier lethality in the Men1ΔN3–8/ΔN3–8 homozygotes suggests that the TSM allele may have some residual function. This conclusion is also suggested by the appearance of pancreatic islet tumors in the Men1ΔN3–8/+ mice a few weeks earlier than in Men1TSM/+, although the differences are modest.
The tumor incidence and pattern of tissue distribution in the Men1TSM/+ mice correlate closely with the human MEN1 phenotype. The pancreatic lesions show a morphologic, neoplastic progression from normal islets to large islets, to large islets with dysplasia, and finally to pancreatic islet cell tumors. PCR analysis of DNA from laser capture microdissected tissue from these various stages of islet transformation reveals that the wild-type Men1 allele is retained until tumor formation. We hypothesize that the gradual enlargement of pancreatic islets is initially nonclonal and occurs as a consequence of a dosage effect resulting from reduced amounts of menin protein (50%). Presumably, the subsequent transition from hyperplastic islets with dysplasia to islet cell tumor is the result of loss of the wild-type Men1 allele. The other endocrine-related tumors found in Men1 heterozygote mice, including pituitary adenoma, parathyroid adenoma, and adrenal cortical tumors, further substantiate the faithfulness of this model in reproducing the MEN1 phenotype. However, several differences were noted. No gastrinomas were detected in any of these animals. A few of the rarer tumors seen in the mouse model (prostate, ovarian) are not thought to be a feature of the human disorder. Because of the small number observed, however, these tumors could be spontaneous and unrelated in the mouse. Whereas parathyroid hyperplasia and adenoma formation were frequent features of the mouse model, elevated serum calcium was not observed.
Murine tumor suppressor knockout models (including Rb, Apc, Nf1, Nf2, Ink4a, Brca1, Brca2, Wt-1, and Vhl) provide a wide range of variability with respect to the human syndromes they were designed to emulate. Rb, Apc, Nf1, Nf2, Brca1, Brca2, Wt-1, and Vhl knockout models result in homozygote lethality at varying ages (30–38), which demonstrates that these genes play an important role in embryonic development, whereas p53 and Ink4a homozygotes are viable (although 20% of p53 homozygotes exhibit exencephaly; refs. 34, 39, and 40). p53, Apc, and Nf1 heterozygotes develop tumors similar to those in their human counterparts and have proved to be useful resources in the study of the disease pathogenesis (31, 32, 41, 42). However, other heterozygote tumor suppressor knockout models, such as Rb, do not always develop tumors associated with the human disease being studied. Human patients with a null allele at RB-1 develop retinoblastoma and osteosarcoma, but heterozygous mice deleted in Rb develop pituitary adenoma (43–45). In other cases, such as the breast cancer gene homologs Brca1 and 2 (35, 36), the kidney cancer homolog Wt-1 (37), and the renal cell carcinoma/pancreatic cancer homolog Vhl (38), the heterozygous mice do not develop any tumors. A prior endocrine tumor model with homozygous inactivation of two cdk inhibitors in mice showed overlap of MEN1 and MEN2 phenotypes (46, 47). In contrast, our data indicate that germ-line disruption of Men1 in heterozygous mice produces a phenotype that is remarkably similar to that observed in human MEN1 kindreds.
Specifically, this model will be quite useful for examining the transitions to hyperplastic islets and pancreatic islet tumors. Is loss of the wild-type Men1 allele both necessary and sufficient for tumor formation, or are other clonal genetic events required? Detection of additional genes that are activated or repressed in the early stages of these transitions will be instrumental in understanding the role of menin in this endocrine tumor syndrome.
The presence of the three loxP sites in the TSM allele also allows the generation of mouse lines with conditional targeting of Men1. We recently have developed lines in which the PGK-neomycin cassette has been deleted, leaving a loxP site in intron 2 and another in intron 8. Recent data (J.S.C., unpublished observations) indicate that homozygotes for this allele are viable, indicating that Men1 can be homozygously inactivated in a tissue-specific manner.
Ultimately, the availability of a mouse model for MEN1 should be an asset for the testing of possible therapeutic approaches, which are still often unsatisfactory for this inherited disorder and its sporadic counterparts.
Acknowledgments
We thank Amy Chen, Theresa Hernandez, Gene Elliot, Jun Cheng, and Shelley Hoogstraten-Miller for excellent technical assistance; Anthony Wynshaw-Boris, Pamela Schwartzberg, and David Bodine for helpful discussions; Donna Butcher for immunohistochemistry; and John Gillespie for laser capture microdissection.
Abbreviations
- MEN1
multiple endocrine neoplasia type 1
- ES
embryonic stem
- LOH
loss of heterozygosity
- H&E
hematoxylin and eosin
- E
embryonic day
- PGK
phosphoglycerate kinase
References
- 1.Trump D, Farren B, Wooding C, Pang J T, Besser G M, Buchanan K D, Edwards C R, Heath D A, Jackson C E, Jansen S, et al. Q J Med. 1996;89:653–669. doi: 10.1093/qjmed/89.9.653. [DOI] [PubMed] [Google Scholar]
- 2.Marx S. In: Genetic Basis of Human Cancer. Vogelstein B, Kinzler K, editors. New York: McGraw–Hill; 1998. pp. 489–506. [Google Scholar]
- 3.Larsson C, Skogseid B, Oberg K, Nakamura Y, Nordenskjold M. Nature (London) 1988;332:85–87. doi: 10.1038/332085a0. [DOI] [PubMed] [Google Scholar]
- 4.Chandrasekharappa S C, Guru S C, Manickam P, Olufemi S E, Collins F S, Emmert-Buck M R, Debelenko L V, Zhuang Z, Lubensky I A, Liotta L A, et al. Science. 1997;276:404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 5.Agarwal S K, Kester M B, Debelenko L V, Heppner C, Emmert-Buck M R, Skarulis M C, Doppman J L, Kim Y S, Lubensky I A, Zhuang Z, et al. Hum Mol Genet. 1997;6:1169–1175. doi: 10.1093/hmg/6.7.1169. [DOI] [PubMed] [Google Scholar]
- 6.Bassett J H, Forbes S A, Pannett A A, Lloyd S E, Christie P T, Wooding C, Harding B, Besser G M, Edwards C R, Monson J P, et al. Am J Hum Genet. 1998;62:232–244. doi: 10.1086/301729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poncin J, Abs R, Velkeniers B, Bonduelle M, Abramowicz M, Legros J J, Verloes A, Meurisse M, Van Gaal L, Verellen C, et al. Hum Mutat. 1999;13:54–60. doi: 10.1002/(SICI)1098-1004(1999)13:1<54::AID-HUMU6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 8.Debelenko L V, Brambilla E, Agarwal S K, Swalwell J I, Kester M B, Lubensky I A, Zhuang Z, Guru S C, Manickam P, Olufemi S E, et al. Hum Mol Genet. 1997;6:2285–2290. doi: 10.1093/hmg/6.13.2285. [DOI] [PubMed] [Google Scholar]
- 9.Heppner C, Kester M B, Agarwal S K, Debelenko L V, Emmert-Buck M R, Guru S C, Manickam P, Olufemi S E, Skarulis M C, Doppman J L, et al. Nat Genet. 1997;16:375–378. doi: 10.1038/ng0897-375. [DOI] [PubMed] [Google Scholar]
- 10.Zhuang Z, Vortmeyer A O, Pack S, Huang S, Pham T A, Wang C, Park W S, Agarwal S K, Debelenko L V, Kester M, et al. Cancer Res. 1997;57:4682–4686. [PubMed] [Google Scholar]
- 11.Zhuang Z, Ezzat S Z, Vortmeyer A O, Weil R, Oldfield E H, Park W S, Pack S, Huang S, Agarwal S K, Guru S C, et al. Cancer Res. 1997;57:5446–5451. [PubMed] [Google Scholar]
- 12.Farnebo F, Teh B T, Kytola S, Svensson A, Phelan C, Sandelin K, Thompson N W, Hoog A, Weber G, Farnebo L O, et al. J Clin Endocrinol Metab. 1998;83:2627–2630. doi: 10.1210/jcem.83.8.4846. [DOI] [PubMed] [Google Scholar]
- 13.Wautot V, Khodaei S, Frappart L, Buisson N, Baro E, Lenoir G M, Calender A, Zhang C X, Weber G. Int J Cancer. 2000;85:877–881. doi: 10.1002/(sici)1097-0215(20000315)85:6<877::aid-ijc23>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 14.Guru S C, Goldsmith P K, Burns A L, Marx S J, Spiegel A M, Collins F S, Chandrasekharappa S C. Proc Natl Acad Sci USA. 1998;95:1630–1634. doi: 10.1073/pnas.95.4.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agarwal S K, Guru S C, Heppner C, Erdos M R, Collins R M, Park S Y, Saggar S, Chandrasekharappa S C, Collins F S, Spiegel A M, et al. Cell. 1999;96:143–152. doi: 10.1016/s0092-8674(00)80967-8. [DOI] [PubMed] [Google Scholar]
- 16.Stewart C, Parente F, Piehl F, Farnebo F, Quincey D, Silins G, Bergman L, Carle G F, Lemmens I, Grimmond S, et al. Oncogene. 1998;17:2485–2493. doi: 10.1038/sj.onc.1202164. [DOI] [PubMed] [Google Scholar]
- 17.Guru S C, Crabtree J S, Brown K D, Dunn K J, Manickam P, Prasad N B, Wangsa D, Burns A L, Spiegel A M, Marx S J, et al. Mamm Genome. 1999;10:592–596. doi: 10.1007/s003359901051. [DOI] [PubMed] [Google Scholar]
- 18.Lakso M, Pichel J G, Gorman J R, Sauer B, Okamoto Y, Lee E, Alt F W, Westphal H. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frost A R, Sparks D, Grizzle W E. Appl Immunohistochem Mol Morphol. 2000;8:236–243. doi: 10.1097/00129039-200009000-00011. [DOI] [PubMed] [Google Scholar]
- 20.Bonner R F, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta L A. Science. 1997;278:1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 21.Emmert-Buck M R, Bonner R F, Smith P D, Chuaqui R F, Zhuang Z, Goldstein S R, Weiss R A, Liotta L A. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 22.Pannett A A, Thakker R V. Endocr Relat Cancer. 1999;6:449–473. doi: 10.1677/erc.0.0060449. [DOI] [PubMed] [Google Scholar]
- 23.Marx S J, Vinik A I, Santen R J, Floyd J C, Jr, Mills J L, Green J D. Medicine (Baltimore) 1986;65:226–241. [PubMed] [Google Scholar]
- 24.Thakker R. In: Endocrinology. De Groot L J, Besser G, Jameson J, Loriaux D, Marshall J, Odell W, Potts J, Rubenstein A, editors. Philadelphia: Saunders; 1995. pp. 2815–2831. [Google Scholar]
- 25.Bensen L, Ljunghall S, Akerstrom G, Oberg K. Am J Med. 1987;82:731–737. doi: 10.1016/0002-9343(87)90008-8. [DOI] [PubMed] [Google Scholar]
- 26.Friedman E, Sakaguchi K, Bale A E, Falchetti A, Streeten E, Zimering M B, Weinstein L S, McBride W O, Nakamura Y, Brandi M L, et al. N Engl J Med. 1989;321:213–218. doi: 10.1056/NEJM198907273210402. [DOI] [PubMed] [Google Scholar]
- 27.Arnold A, Brown M F, Urena P, Gaz R D, Sarfati E, Drueke T B. J Clin Invest. 1995;95:2047–2053. doi: 10.1172/JCI117890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skogseid B, Larsson C, Lindgren P G, Kvanta E, Rastad J, Theodorsson E, Wide L, Wilander E, Oberg K. J Clin Endocrinol Metab. 1992;75:76–81. doi: 10.1210/jcem.75.1.1352309. [DOI] [PubMed] [Google Scholar]
- 29.Nord B, Larsson C, Wong F K, Wallin G, Teh B T, Zedenius J. Genes Chromosomes Cancer. 1999;26:35–39. doi: 10.1002/(sici)1098-2264(199909)26:1<35::aid-gcc5>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 30.Moser A R, Shoemaker A R, Connelly C S, Clipson L, Gould K A, Luongo C, Dove W F, Siggers P H, Gardner R L. Dev Dyn. 1995;203:422–433. doi: 10.1002/aja.1002030405. [DOI] [PubMed] [Google Scholar]
- 31.Cichowski K, Shih T S, Jacks T. Semin Cancer Biol. 1996;7:291–298. doi: 10.1006/scbi.1996.0037. [DOI] [PubMed] [Google Scholar]
- 32.Jacks T, Shih T S, Schmitt E M, Bronson R T, Bernards A, Weinberg R A. Nat Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 33.McClatchey A I, Saotome I, Ramesh V, Gusella J F, Jacks T. Genes Dev. 1997;11:1253–1265. doi: 10.1101/gad.11.10.1253. [DOI] [PubMed] [Google Scholar]
- 34.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig T, Chapman D L, Papaioannou V E, Efstratiadis A. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 36.Hakem R, de la Pompa J L, Sirard C, Mo R, Woo M, Hakem A, Wakeham A, Potter J, Reitmair A, Billia F, et al. Cell. 1996;85:1009–1023. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 37.Kreidberg J A, Sariola H, Loring J M, Maeda M, Pelletier J, Housman D, Jaenisch R. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 38.Gnarra J R, Ward J M, Porter F D, Wagner J R, Devor D E, Grinberg A, Emmert-Buck M R, Westphal H, Klausner R D, Linehan W M. Proc Natl Acad Sci USA. 1997;94:9102–9107. doi: 10.1073/pnas.94.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armstrong J F, Kaufman M H, Harrison D J, Clarke A R. Curr Biol. 1995;5:931–936. doi: 10.1016/s0960-9822(95)00183-7. [DOI] [PubMed] [Google Scholar]
- 40.Sah V P, Attardi L D, Mulligan G J, Williams B O, Bronson R T, Jacks T. Nat Genet. 1995;10:175–180. doi: 10.1038/ng0695-175. [DOI] [PubMed] [Google Scholar]
- 41.Donehower L A, Godley L A, Aldaz C M, Pyle R, Shi Y P, Pinkel D, Gray J, Bradley A, Medina D, Varmus H E. Genes Dev. 1995;9:882–895. doi: 10.1101/gad.9.7.882. [DOI] [PubMed] [Google Scholar]
- 42.Luongo C, Moser A R, Gledhill S, Dove W F. Cancer Res. 1994;54:5947–5952. [PubMed] [Google Scholar]
- 43.Jacks T, Fazeli A, Schmitt E M, Bronson R T, Goodell M A, Weinberg R A. Nature (London) 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 44.Lee E Y, Chang C Y, Hu N, Wang Y C, Lai C C, Herrup K, Lee W H, Bradley A. Nature (London) 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 45.Clarke A R, Maandag E R, van Roon M, van der Lugt N M, van der Valk M, Hooper M L, Berns A, te Riele H. Nature (London) 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 46.Rane S G, Dubus P, Mettus R V, Galbreath E J, Boden G, Reddy E P, Barbacid M. Nat Genet. 1999;22:44–52. doi: 10.1038/8751. [DOI] [PubMed] [Google Scholar]
- 47.Franklin D S, Godfrey V L, O'Brien D A, Deng C, Xiong Y. Mol Cell Biol. 2000;20:6147–6158. doi: 10.1128/mcb.20.16.6147-6158.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]