Abstract
Explicit-solvent molecular dynamics simulations (50 ns, 300 K) of the eight reducing glucose disaccharides (kojibiose, sophorose, nigerose, laminarabiose, maltose, cellobiose, isomaltose, and gentiobiose) have been carried out using the GROMOS 45A4 force field (including a recently reoptimized carbohydrate parameter set), to investigate and compare their conformational preferences, intramolecular hydrogen-bonding patterns, torsional dynamics, and configurational entropies. The calculated average values of the glycosidic torsional angles agree well with available experimental data, providing validation for the force field and simulation methodology employed in this study. These simulations show in particular that: 1) (1→6)-linked disaccharides are characterized by an increased flexibility, the absence of any persistent intramolecular hydrogen bond and a significantly higher configurational entropy (compared to the other disaccharides); 2) cellobiose presents a highly persistent interresidue hydrogen bond and a significantly lower configurational entropy (compared to the other disaccharides); 3) persistent hydrogen bonds are observed for all disaccharides (except (1→6)-linked) and typically involve a hydrogen donor in the reducing residue and an acceptor in the nonreducing one; 4) the probability distributions associated with the glycosidic dihedral angles φ and ψ are essentially unimodal for all disaccharides, and full rotation around these angles occurs at most once or twice for φ (never for ψ) on the 50-ns timescale; and 5) the timescales associated with torsional transitions (except around φ and ψ) range from ∼30 ps (rotation of hydroxyl groups) to the nanosecond range (rotation of the lactol and hydroxymethyl groups, and around the ω-glycosidic dihedral angle in (1→6)-linked disaccharides).
INTRODUCTION
Carbohydrates play a key role in the molecular logic of life. Present in nearly all biological systems on earth, this class of molecules accounts for most of the organic matter in the biosphere, predominantly in the form of cellulose. The complex functions of carbohydrates in biology may be related to the high density of functional (predominantly hydroxyl) groups present in these compounds, in a virtually infinite variety of possible stereoisomers (1). Due to the ability to encode complex stereochemical information, saccharides are considered key elements in a biochemical alphabet that plays a fundamental (yet not completely understood) role in intra- and intercellular recognition processes (2).
Disaccharides are the simplest molecules presenting all the rotational degrees of freedom determining the conformation and flexibility of more complex oligo- and polysaccharides. For this reason, the investigation of the conformational preferences of disaccharides is not only interesting on its own, but also an important first step toward the understanding of the conformation and dynamics of polysaccharides. From the point of view of computer simulation, they also represent the simplest test systems to validate force fields that ultimately aim at the simulation of more complex saccharides.
In the present work, explicit-solvent molecular dynamics (MD) simulations are used to investigate the behavior of disaccharides of D-glucopyranose (Glc) in water. The eight reducing disaccharides (β-anomeric configuration in the reducing residue) presenting all possible glycosidic linkages considered are (Fig. 1): kojibiose (K), sophorose (S), nigerose (N), laminarabiose (L), maltose (M), cellobiose (C), isomaltose (I), and gentiobiose (G).
FIGURE 1.
The eight reducing disaccharides (β anomeric configuration in the reducing residue) formed by the linkage of two D-Glc units: kojibiose (K; Glcα(1→2)Glcβ), sophorose (S; Glcβ(1→2)Glcβ), nigerose (N; Glcα(1→3)Glcβ), laminarabiose (L; Glcβ(1→3)Glcβ), maltose (M; Glcα(1→4)Glcβ), cellobiose (C; Glcβ(1→4)Glcβ), isomaltose (I; Glcα(1→6)Glcβ), and gentiobiose (G; Glcβ(1→6)Glcβ). The conformations around the glycosidic linkage are arbitrary. The standard atom labeling is illustrated for K. The definitions of the dihedral angles φ (O5-C1-O1-Cn′), ψ (C1-O1-Cn′-Cn−1′),  (O5-C5-C6-O6), and ω̃′ (O5′-C5′-C6′-O6′, where O6′ = O1 for a (1→6)-linkage), are illustrated for I. The H-bonds listed in Table 2 are also indicated along with their occurences during the corresponding simulations.
(O5-C5-C6-O6), and ω̃′ (O5′-C5′-C6′-O6′, where O6′ = O1 for a (1→6)-linkage), are illustrated for I. The H-bonds listed in Table 2 are also indicated along with their occurences during the corresponding simulations.
All of these disaccharides are found in honey (3) and most of them have been the scope of numerous experimental studies. Those constituting the building blocks of naturally abundant polysaccharides like amylose (M) or cellulose (C) have received most attention in the literature (4–21), while those carrying a less common glycosidic linkage (e.g., 1→2) have received more limited consideration (4,5,22,23). Most of these disaccharides have been characterized experimentally using x-ray crystallography (24) or nuclear magnetic resonance (NMR) spectroscopy (7,8,14,15,23) and investigated theoretically using MD simulations (9–17,23,27), molecular mechanics (MM) calculations (4–6,22,28,29) (typically MM3 (30,31)) or quantum mechanics calculations (18–21).
The conformation of disaccharides is mainly determined by the populations of rotamers around the glycosidic linkage. Although the pyranose ring exhibits a certain degree of flexibility, the effect of this flexibility on the overall conformation of oligo- and polysaccharides is limited. Therefore, studies on the conformation of disaccharides mainly focus on the glycosidic torsional angles φ (O5-C1-O1-Cn′) and ψ (C1-O1-Cn′-Cn−1′) around a (1→n)-linkage (24) (with n = 2, 3, 4, 6), in addition to the torsional angle  (O5′-C5′-C6′-O6′, where O6′ = O1) in the case of a (1→6)-linkage (see Fig. 1). The conformation of unsubstituted hydroxymethyl groups in either of the two residues is described similarly through the dihedral angles
(O5′-C5′-C6′-O6′, where O6′ = O1) in the case of a (1→6)-linkage (see Fig. 1). The conformation of unsubstituted hydroxymethyl groups in either of the two residues is described similarly through the dihedral angles  (O5′-C5′-C6′-O6′) and
(O5′-C5′-C6′-O6′) and  (O5-C5-C6-O6).
(O5-C5-C6-O6).
This work focuses on the study of the flexibility and dynamics of disaccharides in water by: 1) analyzing the distribution of glycosidic torsional angles; 2) analyzing the occurrence of intramolecular hydrogen bonds; 3) evaluating the dynamics of the various torsional angles; and 4) estimating the configurational entropies of the saccharides. These analyses are based on long (50 ns) MD simulations of the eight disaccharides in explicit water based on the most recent version (45A4) of the GROMOS carbohydrate force field (32). The results contribute to the validation of the force field and provide valuable insight into (thermodynamical and dynamical) properties not directly accessible from experimental measurements.
METHODS
All MD simulations were performed using the GROMOS96 program (33) together with the GROMOS 45A4 force field (including a parameter set recently developed for carbohydrates (32)), and the SPC water model (34). Newton's equations of motion were integrated using the leap-frog algorithm (35) with a 2-fs timestep. The SHAKE algorithm (36) was applied to constrain all bond lengths with a relative geometric tolerance of 10−4. The simulations were carried out in the isothermal-isobaric ensemble (at a temperature of 300 K and a pressure of 1 atm), by separately coupling the temperature of solute and solvent degrees of freedom to a heat bath (37) (relaxation time 0.1 ps) and by coupling the system pressure to a pressure bath via isotropic coordinate scaling (37) (relaxation time 0.5 ps; isothermal compressibility 0.46 × 10−3 · kJ−1 · mol · nm3). Nonbonded interactions were truncated at a distance of 1.4 nm, reevaluated every timestep in the range 0.0–0.8 nm, and evaluated every five timesteps in the range 0.8–1.4 nm (twin-range cutoff scheme (38)). A reaction-field correction (39) was applied to account for the mean effect of neglected interactions beyond 1.4 nm, using a relative dielectric permittivity of 66 for the SPC water model. All simulated systems were composed of one disaccharide molecule surrounded by ∼1000 water molecules (K: 1012; S: 988; N: 972; L: 1109; M: 1002; C: 1048; I: 924; G: 1070) in a truncated-octahedron box simulated under periodic boundary conditions. The initial configurations for the disaccharides S, L, M, C, and G, were taken from the Cambridge Structural Database (CSD) (this database is available from Cambridge Crystallographic Data Centre, http://www.ccdc.cam.ac.uk/products/csd/. For the disaccharides K, N, and I, crystallographic structures were not directly available in the CSD and the structures of similar compounds (methyl-2-O-α-D-mannopyranosyl-β-D-glucopyranoside, methyl-3-O-D-glucopyranosyl-α-D-glucopyranoside and 6-O-α-D-galactopyranosyl-α-D-glucopyranoside, respectively) were used as a basis to model the corresponding disaccharide (using the modeling program XCHemEdit (40)). The CSD reference codes of the structures used in this study are: RESMOR (K), SOPHROS (S), MOGLPR (N), LAMBIO (L), MALTOS (M), CELLOB (C), MELIBM (I), and GENTBS (G)). All simulations were carried out for 50 ns after an equilibration period of 0.1 ns. Atomic coordinates were saved every 0.5 ps for analysis.
Solute (absolute) configurational entropies were estimated for all simulations based on a quasi-harmonic analysis ((41), R. Baron, W. F. van Gunsteren, and P. H. Hünenberger, to be submitted), by calculating the solute all-atom mass-weighted covariance matrix in Cartesian coordinates, after least-square fit superposition (42) of the successive trajectory configurations onto the corresponding initial structure (so as to eliminate overall translational and rotational motions (43)). The quasi-harmonic entropy estimate ( ) was then evaluated as the entropy of a multidimensional quantum-mechanical harmonic oscillator with the same mass-weighted covariance matrix. The six (nearly zero) eigenvalues corresponding to the suppressed rigid-body motion were left out of the analysis. The convergence of the estimated entropies with time was assessed by repeating the analysis for increasingly long time-periods along the simulations (differing in length by 0.5 ns). Entropy corrections for anharmonicities in the quasi-harmonic modes (
) was then evaluated as the entropy of a multidimensional quantum-mechanical harmonic oscillator with the same mass-weighted covariance matrix. The six (nearly zero) eigenvalues corresponding to the suppressed rigid-body motion were left out of the analysis. The convergence of the estimated entropies with time was assessed by repeating the analysis for increasingly long time-periods along the simulations (differing in length by 0.5 ns). Entropy corrections for anharmonicities in the quasi-harmonic modes ( ) and for (supralinear) pairwise correlations among the modes (
) and for (supralinear) pairwise correlations among the modes ( ) were evaluated at the classical level as detailed elsewhere (R. Baron, W. F. van Gunsteren, and P. H. Hünenberger, to be submitted). The anharmonicity correction
) were evaluated at the classical level as detailed elsewhere (R. Baron, W. F. van Gunsteren, and P. H. Hünenberger, to be submitted). The anharmonicity correction  was calculated by summing the corresponding per-mode contributions up to eigenvector 50 (i.e., in the domain of validity of the classical approximation and for the modes where anharmonicity effects are significant. The pairwise (supralinear) correlation correction
was calculated by summing the corresponding per-mode contributions up to eigenvector 50 (i.e., in the domain of validity of the classical approximation and for the modes where anharmonicity effects are significant. The pairwise (supralinear) correlation correction  was calculated by summing the corresponding contributions over all (unique) pairs of modes. Additional information on the underlying theory, assumptions, approximations and practical implementation can be found (R. Baron, W. F. van Gunsteren, and P. H. Hünenberger, to be submitted).
was calculated by summing the corresponding contributions over all (unique) pairs of modes. Additional information on the underlying theory, assumptions, approximations and practical implementation can be found (R. Baron, W. F. van Gunsteren, and P. H. Hünenberger, to be submitted).
RESULTS AND DISCUSSIONS
The average values of the φ- and ψ-dihedral angles, together with the conformer populations around the  -dihedral angle of the reducing residue (i.e., the free hydroxymethyl group in K, S, N, L, M, and C, or the third glycosidic dihedral angle in I and G), are reported in Table 1 for all disaccharides considered. The simulation results are presented along with available experimental (solution NMR (14,23,26) and solid-state x-ray (24)) and theoretical (MM (4,5,28) and MD (10,11,13–15,27)) data from the literature. The corresponding normalized probability distributions for the glycosidic dihedral angles φ, ψ, and
-dihedral angle of the reducing residue (i.e., the free hydroxymethyl group in K, S, N, L, M, and C, or the third glycosidic dihedral angle in I and G), are reported in Table 1 for all disaccharides considered. The simulation results are presented along with available experimental (solution NMR (14,23,26) and solid-state x-ray (24)) and theoretical (MM (4,5,28) and MD (10,11,13–15,27)) data from the literature. The corresponding normalized probability distributions for the glycosidic dihedral angles φ, ψ, and 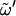 (reducing residue) are also displayed in Fig. 2. In general, the simulation results of this work are in good qualitative agreement with experimental and theoretical data from the literature (Table 1). The best agreement is met for the average values of the φ-glycosidic dihedral angle, the preferences around which are largely dictated by the exo-anomeric effect that causes polar substituents to be oriented away from the ring (24). For α-linked disaccharides (K, N, M, I) this average angle is in the range [70°; 100°] for all entries of Table 1, while for β-linked disaccharides (S, L, C, G) it is typically in the range [−60°; −90°] (except L with MD (27), −58.6°; G with x-ray (24), −58.3°). The corresponding probability distributions from the present simulations (Fig. 2 a) for the β-linked disaccharides and (to a lesser extent) the α-linked disaccharides are indeed very similar for all linkages considered. The variability is more important for the average values of the ψ-dihedral angle (Table 1). Considering all entries of Table 1, disaccharides with a R configuration at Cn′ (K, S, M, C) present values in the range [90°; 120°] (except K with MM (5), 78.8°; S with x-ray (31), 128.5°; C with MM (4), 73.8). Disaccharides with an S configuration at Cn′ (N, L) present values in the range [−150°; −110°] (except N with MM (5), −72.6°; L with x-ray (24), −109.1°, MM (4), −162.3° and MD, −94.5°), and (1→6)-linked disaccharides (I, G) values in the range [160°; 180°] (except G with NMR (14), −129.0° and x-ray (24), −156.3°). These differences are clearly reflected in the corresponding probability distributions from the simulations (Fig. 2 b). The corresponding maxima are found at ∼110° (K, S), −120° (N, L), 100° (M, C), and 180° (I, G). Note also the significantly broader distribution in the case of (1→6)-linked disaccharides (I, G). Finally, the conformational distributions around the dihedral angle
(reducing residue) are also displayed in Fig. 2. In general, the simulation results of this work are in good qualitative agreement with experimental and theoretical data from the literature (Table 1). The best agreement is met for the average values of the φ-glycosidic dihedral angle, the preferences around which are largely dictated by the exo-anomeric effect that causes polar substituents to be oriented away from the ring (24). For α-linked disaccharides (K, N, M, I) this average angle is in the range [70°; 100°] for all entries of Table 1, while for β-linked disaccharides (S, L, C, G) it is typically in the range [−60°; −90°] (except L with MD (27), −58.6°; G with x-ray (24), −58.3°). The corresponding probability distributions from the present simulations (Fig. 2 a) for the β-linked disaccharides and (to a lesser extent) the α-linked disaccharides are indeed very similar for all linkages considered. The variability is more important for the average values of the ψ-dihedral angle (Table 1). Considering all entries of Table 1, disaccharides with a R configuration at Cn′ (K, S, M, C) present values in the range [90°; 120°] (except K with MM (5), 78.8°; S with x-ray (31), 128.5°; C with MM (4), 73.8). Disaccharides with an S configuration at Cn′ (N, L) present values in the range [−150°; −110°] (except N with MM (5), −72.6°; L with x-ray (24), −109.1°, MM (4), −162.3° and MD, −94.5°), and (1→6)-linked disaccharides (I, G) values in the range [160°; 180°] (except G with NMR (14), −129.0° and x-ray (24), −156.3°). These differences are clearly reflected in the corresponding probability distributions from the simulations (Fig. 2 b). The corresponding maxima are found at ∼110° (K, S), −120° (N, L), 100° (M, C), and 180° (I, G). Note also the significantly broader distribution in the case of (1→6)-linked disaccharides (I, G). Finally, the conformational distributions around the dihedral angle  of the reducing residue (Table 1 and Fig. 2 c) show comparable populations for the gt (
of the reducing residue (Table 1 and Fig. 2 c) show comparable populations for the gt ( = −60°) and gg (
= −60°) and gg ( = 60°) conformers (the latter being marginally more populated for free hydroxymethyl groups, the opposite being true for hydroxymethyl groups within a (1→6)-linkage), whereas the tg (
= 60°) conformers (the latter being marginally more populated for free hydroxymethyl groups, the opposite being true for hydroxymethyl groups within a (1→6)-linkage), whereas the tg (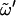 = 180°) conformer is never significantly populated. The analysis of the corresponding distributions for the free hydroxymethyl group in the nonreducing ring of all disaccharides reveals nearly identical features (data not shown). The rotamer populations from the present simulations are in reasonable qualitative agreement with data from NMR measurements (23,26) and other MD simulations (10,11,13,15,27) whenever available (the largest deviations being observed for C). Two-dimensional φ-ψ maps of probability distributions were also generated from the present simulations and found to present very good agreement with corresponding MM3 adiabatic maps for the eight disaccharides (4–6,22,28,29) (data not shown).
= 180°) conformer is never significantly populated. The analysis of the corresponding distributions for the free hydroxymethyl group in the nonreducing ring of all disaccharides reveals nearly identical features (data not shown). The rotamer populations from the present simulations are in reasonable qualitative agreement with data from NMR measurements (23,26) and other MD simulations (10,11,13,15,27) whenever available (the largest deviations being observed for C). Two-dimensional φ-ψ maps of probability distributions were also generated from the present simulations and found to present very good agreement with corresponding MM3 adiabatic maps for the eight disaccharides (4–6,22,28,29) (data not shown).
TABLE 1.
Average values of the φ- and ψ-glycosidic dihedral angles (first line of each entry) and population around the  dihedral angle of the reducing residue (second line of each entry), calculated from the simulations and compared with available experimental (NMR (14,23,26), x-ray (24)), and theoretical (MM (4,5,28), MD (10,11,13–15,27)) results
dihedral angle of the reducing residue (second line of each entry), calculated from the simulations and compared with available experimental (NMR (14,23,26), x-ray (24)), and theoretical (MM (4,5,28), MD (10,11,13–15,27)) results
〈φ〉 [deg] (SD), 〈ψ〉 [deg] (SD), Ptg/Pgg/Pgt for 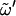 [%] [%]
|
|||||
|---|---|---|---|---|---|
| Sugar | This work | NMR | X-ray | MM | MD |
| K | 95.8 (17.0), 114.7 (15.9) | — | — | 80.6, 78.85 | — |
| 0.0/54.8/45.2 | — | — | — | — | |
| S | −83.8 (44.7), 119.8 (14.1) | — | −81.2, 128.5 (24) | −71.2, 111.74 | — |
| 0.0/59.2/40.8 | 0.0/58.0/42.0 (23) | — | — | — | |
| N | 95.7 (15.6), −119.4 (15.0) | 81.2, −142.6 (14) | 99.9, −135.8 (24) | 85.2, −72.65 | 84.4, −129.6 (14) |
| 0.1/60.9/39.1 | — | — | — | — | |
| L | −72.6 (14.5), −114.2 (13.6) | −85.8, −143.8 (14) | −69.1, −109.1 (24) | −85.0, −162.34 | −58.6, −94.5 (27) |
| 0.0/57.0/43.00 | — | — | — | — | |
| M | 87.2 (17.1), 100.5 (15.5) | 88.5, 95.5 (14) | 96.8, 105.2 (24) | 97.3, 98.35 | 94.7, 106.1 (14) |
| 0.0/65.2/34.7 | — | — | — | 5.0/ 71.0/ 24.0 (13) | |
| C | −77.5 (31.8), 111.4 (10.2) | −87.8, 98.9 (14) | −75.6, 108.2 (24) | −84.0, 73.84 | −75.9, 119.8 (14) |
| 0.0/68.8/31.2 | — | — | — | 12.0/35.0/53.0 (10,11) | |
| I | 82.7 (15.4), 167.5 (31.4) | — | 75.9, −174.2 (24) | 78.7, −173.6 (28) | 80.0, 180.0 (15) |
| 0.0/43.9/56.1 | — | — | — | — | |
| G | −81.6 (29.3), 173.1 (29.4) | −71.4, −129.0 (14) | −58.3, −156.3 (24) | −77.5, 178.4 (28) | −60.8, −177.0 (27) |
| 0.0/46.3/53.8 | 0.0/34.0/66.0 (26) | — | — | 2.0/36.0/62.0 (27) | |
Note that φ = O5-C1-O1-Cn′, ψ = C1-O1-Cn′-Cn−1′, and 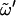 = O5′-C5′-C6′-O6′, with O6′ = O1 for a (1→6)-linkage (I, G); tg,
= O5′-C5′-C6′-O6′, with O6′ = O1 for a (1→6)-linkage (I, G); tg, 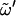 = 180°; gg,
= 180°; gg, 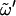 = 60°; and gt,
= 60°; and gt, 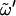 = −60°. Standard deviations (SD) are reported between parentheses. Data from the literature reported in terms of the dihedral angles φH (HC1-C1-O1-Cn′) and ψH (C1-O1-Cn′-HCn′), were transformed to φ- and ψ-values (used in this study) so as to permit a direct comparison. The following transformations were applied (24): φ = φH ± 120°, where the plus symbol applies to the α-linked disaccharides (K, N, M, I), and the minus symbol applies to the β-linked disaccharides (S, L, C, G); and ψ = ψH ± 120°, where the plus symbol applies to the disaccharides with an R-configuration at Cn′ (K, S, M, C) and the minus symbol applies to those with an S configuration at the Cn′ (N, L). The codes used for the disaccharides refer to Fig. 1.
= −60°. Standard deviations (SD) are reported between parentheses. Data from the literature reported in terms of the dihedral angles φH (HC1-C1-O1-Cn′) and ψH (C1-O1-Cn′-HCn′), were transformed to φ- and ψ-values (used in this study) so as to permit a direct comparison. The following transformations were applied (24): φ = φH ± 120°, where the plus symbol applies to the α-linked disaccharides (K, N, M, I), and the minus symbol applies to the β-linked disaccharides (S, L, C, G); and ψ = ψH ± 120°, where the plus symbol applies to the disaccharides with an R-configuration at Cn′ (K, S, M, C) and the minus symbol applies to those with an S configuration at the Cn′ (N, L). The codes used for the disaccharides refer to Fig. 1.
FIGURE 2.
Normalized probability distributions of the glycosidic dihedral angles φ (a), ψ (b), and  (c, reducing residue) for the eight disaccharides considered, either α-linked, (K, N, M, I; left panels), or β-linked (S, L, C, G; right panels). The codes used for the disaccharides refer to Fig. 1.
(c, reducing residue) for the eight disaccharides considered, either α-linked, (K, N, M, I; left panels), or β-linked (S, L, C, G; right panels). The codes used for the disaccharides refer to Fig. 1.
The data reported in Table 1 represent an extensive validation of the new carbohydrate parameter set recently developed for the GROMOS (45A4) force field (32) and tested up to now only for a limited set of compounds (32,44,45).
The occurrences of intramolecular hydrogen bonds (H-bonds) during the simulations are reported in Table 2 (only H-bonds with occurrences larger than 5% are indicated) and Fig. 1. Significantly-populated intramolecular H-bonds are observed in all simulations, except for the two (1→6)-linked disaccharides (I, G), and all of them are interresidue H-bonds. The complete absence of intraresidue H-bonds is probably related to the rather restrictive geometrical criterion used here to define the presence of an H-bond, namely a hydrogen-acceptor distance shorter than 0.25 nm and donor-hydrogen-acceptor angle larger than 135°. MD simulations of β-D-glucose in solution show that when the angle criterion is relaxed (135° → 100°), intraresidue H-bonds become observable (data not shown). Interestingly, all but one of the nine persistent H-bonds observed involve a hydrogen donor from the reducing ring and an acceptor from the nonreducing one. The absence of H-bonds in I and G is probably due to the larger spacing between the rings and the increased flexibility caused by an additional bond within the glycosidic linkage. The absence of intramolecular H-bonds in I and the presence of interresidue H-bonds between the hydroxyl groups at positions 2 and 3′ in M were previously suggested by a combined NMR/MD study (15). The H-bonds present in K and S were also observed in the corresponding global minima found in MM3 calculations (22). Finally, the H-bond observed for C was reported in a number of previous experimental and theoretical studies (6,10,11,20,21,46). In these simulations, this specific H-bond is the one with the highest occurrence (nearly 70%). It is known to persist in the polysaccharide chains (cellulose) formed by this disaccharide in the crystalline state (47,48), and to be an important determinant of the physico-chemical and mechanical properties of this material (stability, rigidity, and insolubility in water (49)).
TABLE 2.
Occurrences of intramolecular H-bonds evaluated from the present simulations
| Sugar | Donor H | Acceptor O | Distance [nm] | Occurrence % |
|---|---|---|---|---|
| K | HO3′ | O5 | 0.206 | 14.9 |
| K | HO1′ | O2 | 0.191 | 48.2 |
| S | HO1′ | O5 | 0.194 | 38.7 |
| N | HO4′ | O2 | 0.194 | 49.5 |
| N | HO2′ | O5 | 0.208 | 21.8 |
| L | HO4′ | O5 | 0.192 | 42.1 |
| M | HO2 | O3′ | 0.189 | 9.8 |
| M | HO3′ | O2 | 0.191 | 32.3 |
| C | HO3′ | O5 | 0.184 | 69.7 |
| I | — | — | — | — |
| G | — | — | — | — |
Only H-bonds with occurrences >5% are reported. An H-bond is assumed to be present in a specific configuration if the hydrogen-acceptor distance is shorter than 0.25 nm (the corresponding distance averaged over all configurations is indicated) and the donor-hydrogen-acceptor angle is larger than 135°. These H-bonds are also reported graphically in Fig. 1. The codes used for the disaccharides refer to Fig. 1.
The timescales associated with dihedral-angle transitions occurring around  (nonreducing residue),
(nonreducing residue), 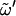 (reducing residue), and the different χn (n = 2,3,4,6; nonreducing residue) and χn′ (n = 1,2,3,4,6; reducing residue) dihedral angles defining the orientation of the hydroxyl groups in the two rings (defined as Cn−1-Cn-On-HOn and Cn−1′-Cn′-On′-HOn′, respectively) are reported in Table 3. These timescales were evaluated by considering transitions between the three dihedral-angle wells centered at staggered conformations. The probability distributions associated with the glycosidic dihedral angles φ and ψ (Fig. 2, a and b) are essentially unimodal for all disaccharides considered (except for minor peaks at φ = 60° in the case of S, C, and G). During the 50-ns simulations, full rotation around these angles occurs at most once or twice for φ (in S, C, and G only) and never for ψ. The timescale associated with the rotation of free hydroxymethyl groups and with transitions around
(reducing residue), and the different χn (n = 2,3,4,6; nonreducing residue) and χn′ (n = 1,2,3,4,6; reducing residue) dihedral angles defining the orientation of the hydroxyl groups in the two rings (defined as Cn−1-Cn-On-HOn and Cn−1′-Cn′-On′-HOn′, respectively) are reported in Table 3. These timescales were evaluated by considering transitions between the three dihedral-angle wells centered at staggered conformations. The probability distributions associated with the glycosidic dihedral angles φ and ψ (Fig. 2, a and b) are essentially unimodal for all disaccharides considered (except for minor peaks at φ = 60° in the case of S, C, and G). During the 50-ns simulations, full rotation around these angles occurs at most once or twice for φ (in S, C, and G only) and never for ψ. The timescale associated with the rotation of free hydroxymethyl groups and with transitions around 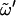 in (1→6)-linked disaccharides is in the range 0.5–1.5 ns. The typical timescales associated with the rotation of hydroxyl groups are 30–40 ps (χ2, χ2′, χ3, χ3′, χ6, χ6′), 80–100 ps (χ4, χ4′), and 0.5–1.2 ns (χ1′, with a dominant trans conformation). However, deviations to larger timescales are observed for specific hydroxyl groups in the reducing residue, typically for those involved in interresidue H-bonds (e.g., χ3′ in C, χ4′ in N and L, and χ1′ in S).
in (1→6)-linked disaccharides is in the range 0.5–1.5 ns. The typical timescales associated with the rotation of hydroxyl groups are 30–40 ps (χ2, χ2′, χ3, χ3′, χ6, χ6′), 80–100 ps (χ4, χ4′), and 0.5–1.2 ns (χ1′, with a dominant trans conformation). However, deviations to larger timescales are observed for specific hydroxyl groups in the reducing residue, typically for those involved in interresidue H-bonds (e.g., χ3′ in C, χ4′ in N and L, and χ1′ in S).
TABLE 3.
Timescale (in nanoseconds) associated with transitions occurring around the dihedral angles 
 and the different χn and χn′
and the different χn and χn′
| Sugar | 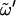 |
 |
χ2 | χ3 | χ4 | χ6 | χ1′ | χ2′ | χ3′ | χ4′ | χ6′ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| K | 0.82 | 0.83 | 0.04 | 0.03 | 0.08 | 0.04 | 0.86 | — | 0.04 | 0.11 | 0.03 |
| S | 0.88 | 0.70 | 0.04 | 0.03 | 0.08 | 0.04 | 2.94 | — | 0.04 | 0.09 | 0.03 |
| N | 0.89 | 0.89 | 0.03 | 0.03 | 0.08 | 0.03 | 0.82 | 0.03 | — | 0.16 | 0.03 |
| L | 0.83 | 0.64 | 0.04 | 0.03 | 0.09 | 0.04 | 0.68 | 0.03 | — | 0.25 | 0.03 |
| M | 1.14 | 0.68 | 0.03 | 0.03 | 0.08 | 0.04 | 1.22 | 0.03 | 0.04 | — | 0.04 |
| C | 1.43 | 0.53 | 0.04 | 0.03 | 0.09 | 0.04 | 1.22 | 0.03 | 0.17 | — | 0.04 |
| I | 1.35 | 0.86 | 0.03 | 0.03 | 0.09 | 0.03 | 0.88 | 0.03 | 0.03 | 0.11 | — |
| G | 1.00 | 0.61 | 0.04 | 0.03 | 0.10 | 0.03 | 0.55 | 0.02 | 0.03 | 0.09 | — |
Note that  = O5-C5-C6-O6 and
= O5-C5-C6-O6 and  = O5′-C5′-C6′-O6′, with O6′ = O1 for a (1→6)-linkage (I, G); χn = Cn−1-Cn-On-HOn (n = 2,3,4,6; nonreducing residue) and χn′ = Cn−1′-Cn′-On′-HOn′ (n = 1,2,3,4,6; reducing residue). The timescales represent the average time spent in the different dihedral-angle wells centered at staggered conformations (intervals [−120°; 0°], [0°; 120°], and [120°; −120°]). Changes of well occurring for <5 ps (allowed excursion time) are not counted as transitions. The codes used for the disaccharides refer to Fig. 1.
= O5′-C5′-C6′-O6′, with O6′ = O1 for a (1→6)-linkage (I, G); χn = Cn−1-Cn-On-HOn (n = 2,3,4,6; nonreducing residue) and χn′ = Cn−1′-Cn′-On′-HOn′ (n = 1,2,3,4,6; reducing residue). The timescales represent the average time spent in the different dihedral-angle wells centered at staggered conformations (intervals [−120°; 0°], [0°; 120°], and [120°; −120°]). Changes of well occurring for <5 ps (allowed excursion time) are not counted as transitions. The codes used for the disaccharides refer to Fig. 1.
The convergence properties of the estimated configurational (quasi-harmonic) entropy  as a function of the sampling time are illustrated in Fig. 3 a for the eight disaccharides. These entropy values are directly comparable because all molecules contain an identical number of atoms (31 atoms). The curves show a stepwise buildup in the first 15–20 ns of the simulations, which suggests that simulations of at least this length are required for a disaccharide in water to sample a significant fraction of its accessible configurational space. After this time-period, the curves are characterized by long plateaus and convergence appears to be essentially reached after ∼30–40 ns. The numerical error on the final
as a function of the sampling time are illustrated in Fig. 3 a for the eight disaccharides. These entropy values are directly comparable because all molecules contain an identical number of atoms (31 atoms). The curves show a stepwise buildup in the first 15–20 ns of the simulations, which suggests that simulations of at least this length are required for a disaccharide in water to sample a significant fraction of its accessible configurational space. After this time-period, the curves are characterized by long plateaus and convergence appears to be essentially reached after ∼30–40 ns. The numerical error on the final  estimates (evaluated based on block averaging of five 10-ns subensembles of structures) is ∼7 J × K−1 × mol−1 (average value over all disaccharides, with a maximum of 16 J × K−1 × mol−1 for I). The contributions
estimates (evaluated based on block averaging of five 10-ns subensembles of structures) is ∼7 J × K−1 × mol−1 (average value over all disaccharides, with a maximum of 16 J × K−1 × mol−1 for I). The contributions  of the successive quasi-harmonic modes to the total configurational entropy
of the successive quasi-harmonic modes to the total configurational entropy 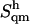 (evaluated from the full 50-ns simulations) are displayed in Fig. 3 b as a function of the eigenvector index m (arranged in order of increasing frequency) for all disaccharides considered. For all systems, the single-mode contribution from the first eigenvector (which dominantly accounts for the relative rotation of the two rings) is markedly higher than that from all the following eigenvectors. The single-mode contributions also show a noticeable decrease after eigenvector 20. The corresponding cumulative estimates of the entropy upon summing the successive per-mode contributions are also displayed in Fig. 3 b. A significant number of low-frequency modes must be included to obtain an accurate estimate of the total quasi-harmonic entropy, in agreement with previous observations in the context of the reversible-folding of β-peptides in methanol (R. Baron, W. F. van Gunsteren, and P. H. Hünenberger, to be submitted). In the present case, the inclusion of ∼60 or 80 modes is required to account for 90 or 99% of the total entropy, respectively.
(evaluated from the full 50-ns simulations) are displayed in Fig. 3 b as a function of the eigenvector index m (arranged in order of increasing frequency) for all disaccharides considered. For all systems, the single-mode contribution from the first eigenvector (which dominantly accounts for the relative rotation of the two rings) is markedly higher than that from all the following eigenvectors. The single-mode contributions also show a noticeable decrease after eigenvector 20. The corresponding cumulative estimates of the entropy upon summing the successive per-mode contributions are also displayed in Fig. 3 b. A significant number of low-frequency modes must be included to obtain an accurate estimate of the total quasi-harmonic entropy, in agreement with previous observations in the context of the reversible-folding of β-peptides in methanol (R. Baron, W. F. van Gunsteren, and P. H. Hünenberger, to be submitted). In the present case, the inclusion of ∼60 or 80 modes is required to account for 90 or 99% of the total entropy, respectively.
FIGURE 3.
Build-up curves of the quasi-harmonic entropy  for the eight disaccharides considered as a function of the sampling time (a, the inset focuses on the last 20 ns of the main graph), and corresponding entropy contributions
for the eight disaccharides considered as a function of the sampling time (a, the inset focuses on the last 20 ns of the main graph), and corresponding entropy contributions  per mode (based on the full 50-ns simulations) displayed as a function of the eigenvector index m (b, left scale and decreasing curves) together with the corresponding cumulative sum (b, right scale and increasing curves). The last six eigenvectors with (nearly) vanishing eigenvalues are omitted from the analysis. The codes used for the disaccharides refer to Fig. 1.
per mode (based on the full 50-ns simulations) displayed as a function of the eigenvector index m (b, left scale and decreasing curves) together with the corresponding cumulative sum (b, right scale and increasing curves). The last six eigenvectors with (nearly) vanishing eigenvalues are omitted from the analysis. The codes used for the disaccharides refer to Fig. 1.
The quasi-harmonic entropies 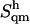 for all disaccharides considered (calculated based on the full 50-ns simulations and including the contributions from all quasi-harmonic modes), together with the associated corrections
for all disaccharides considered (calculated based on the full 50-ns simulations and including the contributions from all quasi-harmonic modes), together with the associated corrections  for anharmonicity in the individual modes and
for anharmonicity in the individual modes and  for (supralinear) pairwise correlations among the different modes, are reported in Table 4. The corrections
for (supralinear) pairwise correlations among the different modes, are reported in Table 4. The corrections  for anharmonicity are negative and relatively small (at most 1.7% of
for anharmonicity are negative and relatively small (at most 1.7% of 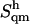 ), which agrees with previous results in the context of small solute molecules (50,51) (R. Baron, W. F. van Gunsteren, and P. H. Hünenberger, to be submitted) but does not necessarily apply to larger macromolecules. These corrections are of similar magnitudes for all compounds. The corrections
), which agrees with previous results in the context of small solute molecules (50,51) (R. Baron, W. F. van Gunsteren, and P. H. Hünenberger, to be submitted) but does not necessarily apply to larger macromolecules. These corrections are of similar magnitudes for all compounds. The corrections 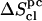 for pairwise (supralinear) correlations are also negative, but of significantly larger magnitudes (19–26% of
for pairwise (supralinear) correlations are also negative, but of significantly larger magnitudes (19–26% of  ), which also agrees with previous results in the context of the reversible folding of β-peptides in methanol (R. Baron, W. F. van Gunsteren, and P. H. Hünenberger, unpublished). This correction is far from negligible, and omitting it would lead to a significant overestimation of the configurational entropy. Higher-order (beyond pairwise) correlations are expected to further decrease the entropy, but are increasingly difficult to estimate (requirement for more extensive sampling, large computational and memory costs involved in their evaluation). Both anharmonicity and correlation effects are likely to become even more important when considering longer polysaccharide chains.
), which also agrees with previous results in the context of the reversible folding of β-peptides in methanol (R. Baron, W. F. van Gunsteren, and P. H. Hünenberger, unpublished). This correction is far from negligible, and omitting it would lead to a significant overestimation of the configurational entropy. Higher-order (beyond pairwise) correlations are expected to further decrease the entropy, but are increasingly difficult to estimate (requirement for more extensive sampling, large computational and memory costs involved in their evaluation). Both anharmonicity and correlation effects are likely to become even more important when considering longer polysaccharide chains.
TABLE 4.
Estimated configurational entropies and corresponding correction terms for the eight disaccharides considered
| Sugar |  |
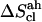 |
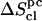 |
Sctd | −TΔSctd |
|---|---|---|---|---|---|
| K | 1638 | −26 (1.6) | −381 (23) | 1231 | −18.30 |
| S | 1642 | −25 (1.5) | −383 (23) | 1234 | −19.20 |
| N | 1632 | −26 (1.6) | −390 (24) | 1216 | −13.80 |
| L | 1654 | −22 (1.3) | −384 (23) | 1248 | −23.40 |
| M | 1624 | −25 (1.5) | −398 (24) | 1201 | −9.30 |
| C | 1622 | −28 (1.7) | −424 (26) | 1170 | 0.00 |
| I | 1683 | −20 (1.2) | −364 (22) | 1299 | −38.70 |
| G | 1700 | −17 (1.0) | −328 (19) | 1355 | −55.50 |
The quasi-harmonic configurational entropies 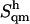 (evaluated using the quantum-mechanical formula for the entropy of a harmonic oscillator) are reported together with their (classically derived) corrections for mode anharmonicities (
(evaluated using the quantum-mechanical formula for the entropy of a harmonic oscillator) are reported together with their (classically derived) corrections for mode anharmonicities (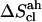 ) and (supralinear) pairwise mode correlations (
) and (supralinear) pairwise mode correlations (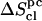 ), leading to the corrected values
), leading to the corrected values 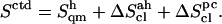 The quantity −TΔSctd, where T is the temperature (300 K) and ΔSctd the difference in Sctd between C and the specific disaccharide, is also reported as an indication of the corresponding free-energy contribution (relative to C). The codes used for the disaccharides refer to Fig. 1. Relative values of the entropy corrections (in percent with respect to
The quantity −TΔSctd, where T is the temperature (300 K) and ΔSctd the difference in Sctd between C and the specific disaccharide, is also reported as an indication of the corresponding free-energy contribution (relative to C). The codes used for the disaccharides refer to Fig. 1. Relative values of the entropy corrections (in percent with respect to 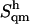 ) are also indicated between parentheses. All entropy values are given in J × K−1 × mol−1 while −TΔSctd is given in kJ × mol−1.
) are also indicated between parentheses. All entropy values are given in J × K−1 × mol−1 while −TΔSctd is given in kJ × mol−1.
The corrected entropy values 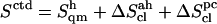 for the (1→6)-linked disaccharides (I, G) are significantly larger than the corresponding values for the other disaccharides. This observation is intuitively justified by the presence of an additional dihedral angle within the glycosidic linkage, and agrees with the results of a chromatographic study comparing estimated conformational entropies for M, C, I, and G in aqueous solution (52). The disaccharide C has a significantly lower entropy compared to all other disaccharides (in large part due to a more negative correlation correction term
for the (1→6)-linked disaccharides (I, G) are significantly larger than the corresponding values for the other disaccharides. This observation is intuitively justified by the presence of an additional dihedral angle within the glycosidic linkage, and agrees with the results of a chromatographic study comparing estimated conformational entropies for M, C, I, and G in aqueous solution (52). The disaccharide C has a significantly lower entropy compared to all other disaccharides (in large part due to a more negative correlation correction term 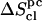 ). This observation is probably related to the presence of a highly-persistent HO3′–O5 H-bond in this sugar (Table 2). This peculiarity of the disaccharide building block of cellulose in terms of conformational entropy may also play a role in determining the specific physico-chemical properties of this polysaccharide. The corrected entropy values for all other disaccharides (K, S, N, L, and M) fall in a narrower range of ∼50 J × K−1 × mol−1. All β-linked disaccharides have a higher entropy than the corresponding α-linked disaccharides, with the exception of the M/C pair. This observation is in qualitative agreement with analyses of the relative flexibilities of disaccharides based on QM and MM potential energy maps (4–11,14,18–23,28,29). These studies suggest that di-equatorial linkages (as found in the β-linked disaccharides) are intrinsically more flexible then axial-equatorial ones (as found in α-linked disaccharides) (4–6,18–22,28,29). The above-mentioned chromatographic study of M, C, I, and G (52) also supports this conclusion. The trends in configurational entropies suggested by the present simulation study are G≫I≫L>S>K>N>M≫C (Table 4).
). This observation is probably related to the presence of a highly-persistent HO3′–O5 H-bond in this sugar (Table 2). This peculiarity of the disaccharide building block of cellulose in terms of conformational entropy may also play a role in determining the specific physico-chemical properties of this polysaccharide. The corrected entropy values for all other disaccharides (K, S, N, L, and M) fall in a narrower range of ∼50 J × K−1 × mol−1. All β-linked disaccharides have a higher entropy than the corresponding α-linked disaccharides, with the exception of the M/C pair. This observation is in qualitative agreement with analyses of the relative flexibilities of disaccharides based on QM and MM potential energy maps (4–11,14,18–23,28,29). These studies suggest that di-equatorial linkages (as found in the β-linked disaccharides) are intrinsically more flexible then axial-equatorial ones (as found in α-linked disaccharides) (4–6,18–22,28,29). The above-mentioned chromatographic study of M, C, I, and G (52) also supports this conclusion. The trends in configurational entropies suggested by the present simulation study are G≫I≫L>S>K>N>M≫C (Table 4).
CONCLUSIONS
The present article reports a comparative study of the eight reducing glucose-based disaccharides using explicit-solvent MD simulations. The properties of all possible glycosidic linkages, i.e., α(1→2), β(1→2), α(1→3), β(1→3), α(1→4), β(1→4), α(1→6), and β(1→6) have thus been investigated in the context of the disaccharides K, S, N, L, M, C, I, and G, respectively (with the β-anomer at the reducing residue). The conformational preferences associated with the glycosidic dihedral angles φ, ψ, and  (the latter for I and G) are found to agree well with available experimental and theoretical data. This agreement provides additional validation for the new GROMOS 45A4 force-field parameter set recently developed for carbohydrates (32). The H-bond analysis reveals the presence of significantly populated interresidue H-bonds (nearly always involving a hydrogen donor in the reducing residue) in all disaccharides except I and G. The characteristic timescales associated with transitions in the various dihedral angles (except φ and ψ) range from ∼30 ps (rotation of hydroxyl groups) to the nanosecond range (rotation of the lactol and hydroxymethyl groups, and around the ω-glycosidic dihedral angle in (1→6)-linked disaccharides). The probability distributions associated with the φ- and ψ-glycosidic dihedral angles are essentially unimodal, and full rotation around theses angles is observed at most once or twice for φ (never for ψ) on the 50-ns timescale. Finally, a quasi-harmonic entropy analysis shows that simulations of at least 30–40 ns are required to adequately sample the conformational space accessible to solvated disaccharides. The corresponding final entropy estimates (after corrections for anharmonicities and pairwise mode correlation) evidence a significantly higher entropy for (1→6)-linked disaccharides (I and G) and a significantly lower entropy for C compared to the five other disaccharides investigated. In addition, for a given linkage, the entropy is generally higher for the β-form compared to the α-form (except for the M/C pair).
(the latter for I and G) are found to agree well with available experimental and theoretical data. This agreement provides additional validation for the new GROMOS 45A4 force-field parameter set recently developed for carbohydrates (32). The H-bond analysis reveals the presence of significantly populated interresidue H-bonds (nearly always involving a hydrogen donor in the reducing residue) in all disaccharides except I and G. The characteristic timescales associated with transitions in the various dihedral angles (except φ and ψ) range from ∼30 ps (rotation of hydroxyl groups) to the nanosecond range (rotation of the lactol and hydroxymethyl groups, and around the ω-glycosidic dihedral angle in (1→6)-linked disaccharides). The probability distributions associated with the φ- and ψ-glycosidic dihedral angles are essentially unimodal, and full rotation around theses angles is observed at most once or twice for φ (never for ψ) on the 50-ns timescale. Finally, a quasi-harmonic entropy analysis shows that simulations of at least 30–40 ns are required to adequately sample the conformational space accessible to solvated disaccharides. The corresponding final entropy estimates (after corrections for anharmonicities and pairwise mode correlation) evidence a significantly higher entropy for (1→6)-linked disaccharides (I and G) and a significantly lower entropy for C compared to the five other disaccharides investigated. In addition, for a given linkage, the entropy is generally higher for the β-form compared to the α-form (except for the M/C pair).
Acknowledgments
The authors thank Roberto D. Lins for insightful discussions.
Financial support by the Swiss National Foundation grant No. 21-105397 is gratefully acknowledged. R.B. is grateful for financial support from the National Center of Competence in Research, Structural Biology, of the Swiss National Foundation.
References
- 1.Lehmann, J. 1998. Carbohydrates Structure and Biology. Georg Thieme Verlag, Stuttgart, Germany.
- 2.Gabius, H. J., H. C. Siebert, S. Andre, J. J. Barbero, and H. Rüdiger. 2004. Chemical biology of the sugar code. ChemBioChem. 5:740–764. [DOI] [PubMed] [Google Scholar]
- 3.Sanz, M. L., J. Sanz, and I. Martinez-Castro. 2004. Gas chromatographic mass spectrometric method for the qualitative and quantitative determination of disaccharides and trisaccharides in honey. J. Chromatogr. A. 1059:143–148. [DOI] [PubMed] [Google Scholar]
- 4.Dowd, M. K., A. D. French, and P. J. Reilly. 1992. Conformational analysis of the anomeric forms of sophorose, laminarabiose, and cellobiose using MM3. Carbohydr. Res. 233:15–34. [DOI] [PubMed] [Google Scholar]
- 5.Dowd, M. K., J. Zeng, A. D. French, and P. J. Reilly. 1992. Conformational analysis of the anomeric forms of kojibiose, nigerose, and maltose using MM3. Carbohydr. Res. 230:223–244. [DOI] [PubMed] [Google Scholar]
- 6.Stortz, C. A., and A. S. Cerezo. 2003. Depicting the MM3 potential energy surfaces of trisaccharides by single contour maps: application to β-cellotriose and α-maltotriose. Carbohydr. Res. 338:95–107. [DOI] [PubMed] [Google Scholar]
- 7.Lipkind, G. M., A. S. Shashkov, and N. K. Kochetkov. 1985. Nuclear Overhauser effect and conformational states of cellobiose in aqueous solution. Carbohydr. Res. 141:191–197. [Google Scholar]
- 8.Shashkov, A. S., G. M. Lipkind, and N. K. Kochetkov. 1986. Nuclear Overhauser effects for methyl β-maltoside and the conformational states of maltose in aqueous solution. Carbohydr. Res. 147:175–182. [Google Scholar]
- 9.Ha, S. N., L. J. Madsen, and J. W. Brady. 1988. Conformational analysis and molecular dynamics simulations of maltose. Biopolymers. 27:1927–1952. [DOI] [PubMed] [Google Scholar]
- 10.Hardy, B. J., and A. Sarko. 1993. Conformational analysis and molecular dynamics simulation of cellobiose and larger cello-oligomers. J. Comput. Chem. 14:831–847. [Google Scholar]
- 11.Hardy, B. J., and A. Sarko. 1993. Molecular-dynamics simulation of cellobiose in water. J. Comput. Chem. 14:848–857. [Google Scholar]
- 12.Brady, J. W., and R. K. Schmidt. 1993. The role of hydrogen-bonding in carbohydrates—molecular dynamics simulations of maltose in aqueous solution. J. Phys. Chem. 97:958–966. [Google Scholar]
- 13.Ott, K. H., and B. Meyer. 1996. Molecular dynamics simulations of maltose in water. Carbohydr. Res. 281:11–34. [Google Scholar]
- 14.Cheetham, N. W. H., P. Dasgupta, and G. E. Ball. 2003. NMR and modelling studies of disaccharide conformation. Carbohydr. Res. 338:955–962. [DOI] [PubMed] [Google Scholar]
- 15.Best, R. B., G. E. Jackson, and K. J. Naidoo. 2001. Molecular dynamics and NMR study of the α(1 → 4) and α(1 → 6) glycosidic linkages: maltose and isomaltose. J. Phys. Chem. B. 105:4742–4751. [Google Scholar]
- 16.Umemura, M., S. Hayashi, T. Nakagawa, S. Yamanaka, H. Urakawa, and K. Kajiwara. 2003. Structure of water molecules in aqueous maltose and cellobiose solutions using molecular dynamics simulation. I. Statics. J. Mol. Struct. THEOCHEM. 624:129–144. [Google Scholar]
- 17.Umemura, M., S. Hayashi, T. Nakagawa, H. Urakawa, and K. Kajiwara. 2003. Structure of water molecules in aqueous maltose and cellobiose solutions using molecular dynamics simulation. II. Dynamics. J. Mol. Struct. THEOCHEM. 636:215–228. [Google Scholar]
- 18.French, A. D., A. M. Kelterer, G. P. Johnson, and M. K. Dowd. 2000. B3LYP/6–31G*, RHF/6–31G* and MM3 heats of formation of disaccharide analogs. J. Mol. Struct. 556:303–313. [Google Scholar]
- 19.French, A. D., A. M. Kelterer, G. P. Johnson, M. K. Dowd, and C. J. Cramer. 2001. HF/6–31G*energy surfaces for disaccharide analogs. J. Comput. Chem. 22:65–78. [Google Scholar]
- 20.Strati, G. L., J. L. Willett, and F. A. Momany. 2002. Ab initio computational study of P-cellobiose conformers using B3LYP/6–311++G. Carbohydr. Res. 337:1833–1849. [DOI] [PubMed] [Google Scholar]
- 21.Strati, G. L., J. L. Willett, and F. A. Momany. 2002. A DFT/ab initio study of hydrogen bonding and conformational preference in model cellobiose analogs using B3LYP/6–311++G. Carbohydr. Res. 337:1851–1859. [DOI] [PubMed] [Google Scholar]
- 22.Stortz, C. A., and A. S. Cerezo. 2003. MM3 potential energy surfaces of the two linked glucosyl trisaccharides α-kojitriose and β-sophorotriose. Carbohydr. Res. 338:1679–1689. [DOI] [PubMed] [Google Scholar]
- 23.Andre, I., K. Mazeau, F. R. Taravel, and I. Tvaroska. 1995. NMR and molecular modeling of sophorose and sophorotriose in solution. New J. Chem. 19:331–339. [Google Scholar]
- 24.Rao, V. S. R., P. K. Qasba, P. V. Balaji, and R. Chandrasekaran. 1998. Conformation of Carbohydrates. Harwood Academic Publishers, Amsterdam, The Netherlands. 93–102.
- 25.Lipkind, G. M., A. S. Shashkov, S. S. Mamyan, and N. K. Kochetkov. 1988. The nuclear Overhauser effect and structural factors determining the conformations of disaccharide glycosides. Carbohydr. Res. 181:1–12. [Google Scholar]
- 26.Poppe, L. 1993. Modeling carbohydrate conformations from NMR data—maximum-entropy rotameric distribution about the C5–C6 bond in gentiobiose. J. Am. Chem. Soc. 115:8421–8426. [Google Scholar]
- 27.Kony, D., W. Damm, S. Stoll, and P. H. Hünenberger. 2004. Explicit-solvent molecular dynamics simulations of the β (1→3)- and β (1→6)-linked disaccharides β-laminarabiose and β-gentiobiose in water. J. Phys. Chem. B. 108:5815–5826. [Google Scholar]
- 28.Dowd, M. K., A. D. French, and P. J. Reilly. 1994. Relaxed-residue conformational mapping of the three linkage bonds of isomaltose and gentiobiose with MM3 (92). Biopolymers. 34:625–638. [DOI] [PubMed] [Google Scholar]
- 29.Stortz, C. A., and A. S. Cerezo. 2002. Potential energy surfaces of α-(1→3)-linked disaccharides calculated with the MM3 force field. J. Carbohydr. Chem. 21:355–371. [Google Scholar]
- 30.Allinger, N. L., Y. H. Yuh, and J.-H. Lii. 1989. Molecular mechanics—the MM3 force-field for hydrocarbons. 1. J. Am. Chem. Soc. 111:8551–8566. [Google Scholar]
- 31.Allinger, N. L., M. Rahman, and J.-H. Lii. 1990. A molecular mechanics force field (MM3) for alcohols and ethers. J. Am. Chem. Soc. 112:8293–8307. [Google Scholar]
- 32.Lins, R. D., and P. H. Hünenberger. 2005. A new GROMOS force field for hexopyranose-based carbohydrates. J. Comput. Chem. 26:1400–1412. [DOI] [PubMed] [Google Scholar]
- 33.van Gunsteren, W. F., S. R. Billeter, A. A. Eising, P. H. Hünenberger, and P. Krüger. 1996. Biomolecular Simulation: The GROMOS96 Manual and User Guide. A. E. Mark, W. R. P. Scott, and I. G. Tironi, editors. Hochschulverlag an der ETH Zürich/BIOMOS, Zürich/Groningen, Switzerland.
- 34.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, and J. Hermans. 1981. Intermolecular Forces. B. Pullman, editor. Reidel, Dordrecht, The Netherlands. 331–342.
- 35.Hockney, R. W. 1970. The potential calculation and some applications. Methods Comput. Phys. 9:136–211. [Google Scholar]
- 36.Ryckaert, J. P., G. Ciccotti, and H. J. C. Berendsen. 1977. Numerical integration of Cartesian equations of motion of a system with constraints—molecular dynamics of n-alkanes. J. Comp. Phys. 23:327–341. [Google Scholar]
- 37.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 38.van Gunsteren, W. F., and H. J. C. Berendsen. 1990. Computer simulation of molecular dynamics—methodology, applications, and perspectives in chemistry. Angew. Chem. Int. Ed. Engl. 29:992–1023. [Google Scholar]
- 39.Tironi, I. G., R. Sperb, P. E. Smith, and W. F. van Gunsteren. 1995. A generalized reaction field method for molecular dynamics simulations. J. Chem. Phys. 102:5451–5459. [Google Scholar]
- 40.XchemEdit, Version 0.93. Http://www.cemcomco.com/ChemEdit_Distribution1074.html.
- 41.Karplus, M., and J. Kushick. 1981. Method for estimating the configurational entropy of macromolecules. Macromolecules. 14:325–332. [Google Scholar]
- 42.McLachlan, A. D. 1979. Gene duplications in the structural evolution of chymotrypsin. J. Mol. Biol. 128:49–79. [DOI] [PubMed] [Google Scholar]
- 43.Schäfer, H., A. E. Mark, and W. F. van Gunsteren. 2000. Absolute entropies from molecular dynamics simulation trajectories. J. Chem. Phys. 113:7809–7817. [Google Scholar]
- 44.Yu, H., M. Amann, T. Hansson, J. Kohler, G. Wich, and W. F. van Gunsteren. 2004. Effect of methylation on the stability and solvation free energy of amylose and cellulose fragments: a molecular dynamics study. Carbohydr. Res. 339:1697–1709. [DOI] [PubMed] [Google Scholar]
- 45.Pereira, C. S., R. D. Lins, I. Chandrasekar, L. C. G. Freitas, and P. H. Hünenberger. 2004. Interaction of the disaccharide trehalose with a phospholipid bilayer: a molecular dynamics study. Biophys. J. 86:2273–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leeflang, B. R., J. F. G. Vliegenthart, L. M. J. Kroonbatenburg, B. P. Vaneijck, and J. Kroon. 1992. A H-1-NMR and MD study of intramolecular hydrogen-bonds in methyl β-cellobioside. Carbohydr. Res. 230:41–61. [DOI] [PubMed] [Google Scholar]
- 47.Gardner, K. H., and J. Blackwell. 1974. Structure of native cellulose. Biopolymers. 13:1975–2001. [Google Scholar]
- 48.Kolpak, F. J., and J. Blackwell. 1976. Determination of structure of cellulose II. Macromolecules. 9:273–278. [DOI] [PubMed] [Google Scholar]
- 49.Kadla, J. F., and R. D. Gilbert. 2000. Cellulose structure: a review. Cellulose Chem. Technol. 34:197–216. [Google Scholar]
- 50.Ichiye, T., and M. Karplus. 1987. Anisotropy and anharmonicity of atomic fluctuations in proteins—analysis of a molecular dynamics simulation. Proteins. 2:236–259. [DOI] [PubMed] [Google Scholar]
- 51.Perahia, D., R. M. Levy, and M. Karplus. 1990. Motions of an α-helical polypeptide—comparison of molecular and harmonic dynamics. Biopolymers. 29:645–677. [Google Scholar]
- 52.Striegel, A. M. 2003. Anomeric configuration, glycosidic linkage, and the solution conformational entropy of O-linked disaccharides. J. Am. Chem. Soc. 125:4146–4148. [DOI] [PubMed] [Google Scholar]





