Abstract
Acute myeloid leukemia (AML) is a heterogeneous group of diseases. Normal cytogenetics (CN) constitutes the single largest group, while trisomy 8 (+8) as a sole abnormality is the most frequent trisomy. How trisomy contributes to tumorigenesis is unknown. We used oligonucleotide-based DNA microarrays to study global gene expression in AML+8 patients with +8 as the sole chromosomal abnormality and AML-CN patients. CD34+ cells purified from normal bone marrow (BM) were also analyzed as a representative heterogeneous population of stem and progenitor cells. Expression patterns of AML patients were clearly distinct from those of CD34+ cells of normal individuals. We show that AML+8 blasts overexpress genes on chromosome 8, estimated at 32% on average, suggesting gene-dosage effects underlying AML+8. Systematic analysis by cellular function indicated up-regulation of genes involved in cell adhesion in both groups of AML compared with CD34+ blasts from normal individuals. Perhaps most interestingly, apoptosis-regulating genes were significantly down-regulated in AML+8 compared with AML-CN. We conclude that the clinical and cytogenetic heterogeneity of AML is due to fundamental biological differences.
Normal hematopoiesis is organized in a hierarchical fashion: normal pluripotent stem cells give rise to progeny that progressively lose their capacity for self-renewal as they become committed to certain lineages (1). This process of differentiation and commitment is thought to be controlled at the level of transcription by the interaction of lineage-specific transcription factors (2). When normal stem cell differentiation is blocked, malignant neoplastic proliferation and accumulation of immature hematopoietic stem cells is the result. The excessive accumulation of immature nonlymphatic bone marrow (BM) precursor cells in the marrow could be caused by increased cell proliferation and/or reduced cell death. The latter, characterized by aberrant differentiation, has been suggested as the more important mechanism in the majority of acute myeloid leukemia (AML) cases (3).
Approximately 55% of de novo AMLs show clonal cytogenetic abnormalities; the rest show no cytogenetic changes, which mask any clues to their molecular pathogenesis (4). Normal cytogenetics (CN) constitutes the single largest group in AML. However, several nonrandom chromosomal abnormalities are also frequent, of which trisomy 8 (+8) is the most common numerical aberration (12%) as either a sole abnormality (≈4%) or part of more complex karyotypes (≈8%) (4). AML+8 is generally associated with poor outcome, whereas AML-CN shows varied responses (5, 6). The question as to whether +8 is a chromosomal aberration likely to be critical in initiating or promoting leukemia, or whether +8 occurs after the development of leukemia and without an important contribution, is currently unanswered.
The pathophysiological mechanisms of AML+8 and AML-CN are largely unknown. Two possible molecular scenarios could explain the phenotype of AML+8. First, the AML phenotype may be due to a gene-dosage effect and a direct result of the trisomy with genes on chromosome 8 being overexpressed. This hypothesis is based on the analogous assumption of a gene-dosage effect for constitutional trisomy 21 in Down syndrome, where +21 appears to predispose to an increased risk for hematological malignancies (7, 8). Similarly, constitutional +8 mosaicism has also been postulated to predispose to neoplasms, mainly myelodysplastic syndrome and AML (9). Alternatively, a yet-to-be identified rearrangement of a gene(s) on chromosome 8 could underlie the transformed phenotype, which is similar to that seen in de novo AML associated with +11, where the majority of cases are associated with an intragenic molecular rearrangement of MLL (10).
We hypothesized that whatever the underlying molecular leukemogenic event(s) associated with AML+8 and AML-CN, the molecular changes at the DNA level should be reflected in specific changes at the RNA level. Thus, global expression profiling with DNA microarrays should prove particularly powerful in identifying the downstream genetic programs controlled by the leukemogenic factor(s) in AML+8 and AML-CN. Moreover, a molecular classification scheme based on expression profiles would have the potential to define new subgroups with prognostic and therapeutic significance and to provide a better understanding of the intrinsic disease biology. Several recent studies on the molecular classification of AML/acute lymphocytic leukemia (ALL) (11), diffuse large B-cell lymphoma (12), cutaneous malignant melanoma (13), and breast cancer (14) using similar approaches are particularly encouraging in this respect.
Materials and Methods
BM Sample Collection and Processing.
Upon informed consent, seven normal BM samples were obtained from four male and three female donors. From 40 ml of BM sample, mononuclear cells were isolated immediately by using a Ficoll–Hypaque gradient. After a 2-hr culture to remove adherent growing cells, the purified cells were washed, stained with anti-CD34 antibody, and labeled with a secondary antibody conjugated to magnetic beads (Miltenyi Biotec, Auburn, CA). After selection over a magnetic column, purified cells were immediately snap-frozen in liquid nitrogen. The purity of CD34+ selected cells was determined by flow cytometry. All samples were >97% pure. Twenty pretreatment BM AML samples were obtained from the Cancer and Leukemia Group B (15 samples) and the Ohio State University (5 samples) leukemia cell banks. All 10 AML+8 patients had +8 as a sole cytogenetic abnormality; they had a median age of 70 (range, 30–79) years; 6 were women; and the French American British (FAB) classification breakdown was as follows: one M0, two M1, two M2, two M4, two M5, and one not otherwise specified. The 10 AML-CN patients had a median age of 70 (range, 24–84) years; 3 were women; and the FAB classification breakdown was as follows: one M0, two M1, three M2, one M4, and two M5. Mononuclear cells were isolated from the AML BM samples in a fashion similar to that described above.
Expression Profiling.
Detailed protocols are available at http://cancergenetics.med.ohio-state.edu/microarray and are published as supplemental data on the PNAS web site, www.pnas.org. Briefly, RNA from CD34+ cells was extracted by using the total RNA extraction protocol (Qiagen, Chatsworth, CA). AML BM samples in cell-freezing medium were thawed in 10 vol of RNA Stat-60 (Tel-Test, Houston, TX); 0.2 vol of chloroform was added to the lysed cells, and the aqueous layer containing the RNA was purified with RNeasy columns (Qiagen). The integrity of individual RNA samples was verified by denaturing agarose gel electrophoresis. cRNA target was prepared from a total of 4–8 μg (CD34+ cells) and 8 μg (AML samples) of total cellular RNA, respectively, hybridized to HuGeneFL Affymetrix oligonucleotide arrays, scanned and analyzed according to Affymetrix (Santa Clara, CA) protocols. Expression and the logarithm of expression were further normalized across samples by using a modification of the linear scaling method (11). Analyses were performed primarily by using log expression values, which have reduced skew and desirable variability properties. All primary expression data are available at http://cancergenetics.med.ohio-state.edu/microarrayunit.
Statistical Analysis.
Analyses were performed with S-PLUS 3.4 (Mathsoft, Seattle) and are detailed in the supplemental data. A systematic analysis based on functional categories was also performed. We used the functional terms for all 6,606 genes on the HuGeneFL array compiled by the Whitehead Institute/Massachusetts Institute of Technology Center for Genome Research Molecular Pattern Recognition group using functional annotations from the SWISS-PROT database (http://www.expasy.ch/sprot/) and selected nine categories based on hypothesized involvement in AML. UNIGENE cluster IDs were used to identify genes with putative involvement in the following nine categories (numbers of genes in parentheses): transcription (532), nuclear factors (1,026), DNA-binding (744), apoptosis (68), cell cycle control (74), RNA-binding (171), ATP-binding (551), cell adhesion (172), and signal transduction (1,547). Such a comprehensive approach enables more sensitive investigation of whether an entire group of genes is apparently up- or down-regulated. Genes belonging to these categories were compared by using t tests in the set of AMLs vs. CD34+ and AML+8 vs. AML-CN for a total of 18 comparisons. Among all expressed genes, the t-statistic ranks of those belonging to the functional category were plotted in a histogram. Categories systematically differing in the two groups show a shift to the left or right from the uniform distribution expected under the null hypothesis. A two-sided Wilcoxon rank sum test (15) was used as a statistic to compare the ranks of the genes belonging to a category with the ranks of the remainder of the genes. The standard Wilcoxon P-value approximation was used to select for further study those categories achieving nominal significance with a Bonferroni correction for 18 comparisons (P < 0.0028). However, the genes within each sample are correlated, and proper significance testing requires treating samples as the independent units. Thus, empirical P values were generated by computing the statistic for each of 1,000 permutations in which group membership was randomly assigned to the samples.
Chromosomal Localization of Genes on the HuGeneFL Array.
The genes and expressed sequence tags (ESTs) on the HuGeneFL array (Affymetrix) are indexed by GenBank and The Institute for Genomic Research (TIGR) accession numbers. These accession numbers were used to obtain genomic sequence. The sequences were compared by using blast to the Division of Human Cancer Genetics internal gene index HINT (Human Index of Nonredundant Transcripts; Zhuo, R.K., F.A.W., and B.Y., unpublished work), a set of transcript consensus sequences assembled from UNIGENE EST clusters. Cytogenetic and genetic mapping location was derived from the integration of HINT ESTs with mapping information in UNIGENE and GeneMap '99 (16). Chromosomal assignment was possible for 93% of the HuGeneFL genes. GB4 radiation hybrid map information was available for 45% of the genes and ESTs on the array, G3 information for an additional 20%. The two radiation hybrid maps were integrated for chromosome 8 by interpolating between the 28 anchors having both GB4 and G3 listings in GeneMap '99.
TaqMan Real-Time PCR Assay.
Relative expression levels and differences between AML+8 and AML-CN were validated with the TaqMan 5′ nuclease real-time PCR assay (17). The approach is detailed in the supplemental material. Briefly, a subset of 11 AML samples (6 AML+8 and 5 AML-CN) previously profiled with DNA microarrays was studied. SEQUENCE DETECTOR 1.6 (Applied Biosystems) was used for analysis; the comparative CT method was used to determine the ratio of target and endogenous control according to specifications.
Results
Global Expression Profiling of AML+8, AML-CN, and CD34+ Samples.
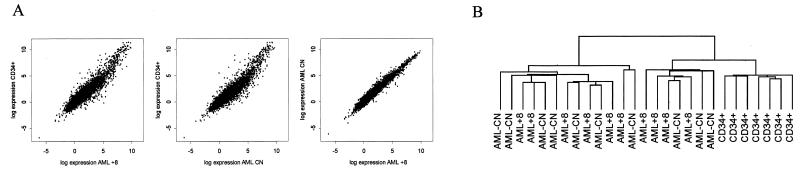
Using the HuGeneFL GeneChip (18), we compared the overall expression patterns for 6,606 unique genes in the three sample groups (Fig. 1A). The averaged expression values for each gene are presented for the three groups after appropriate normalization and scaling (Fig. 1A). Expression values of the two AML groups were highly correlated, and each showed a lower correlation with the CD34+ group. Two-way hierarchical cluster analysis (Fig. 1B and Fig. 5 in the supplemental data) was performed on 1,959 genes passing a variation filter to group genes and samples on the basis of similarity in the pattern with which expression varied over all samples (19). The CD34+ samples clustered into a distinct group; AML+8 and AML-CN samples did not, but were intercalated with each other (Fig. 1B). This clustering is a reflection of the similar expression profiles and scatter plot for the AML+8 and AML-CN samples (Fig. 1A).
Figure 1.
(A) Scatter plots of the log-intensity values for the 6,606 unique genes assayed with the HuGeneFL array. The intensity values were scaled and averaged over each of three groups: AML+8, AML-CN, and CD34+ cells. (B) Dendrogram from two-way hierarchical cluster analysis of 1,959 genes passing a variation filter generated by using the programs cluster and TreeView (19). See also Fig. 5, which is published as supplemental data on the PNAS web site, www.pnas.org.
Classification of AML and CD34+ Samples Based on Their Expression Profiles.
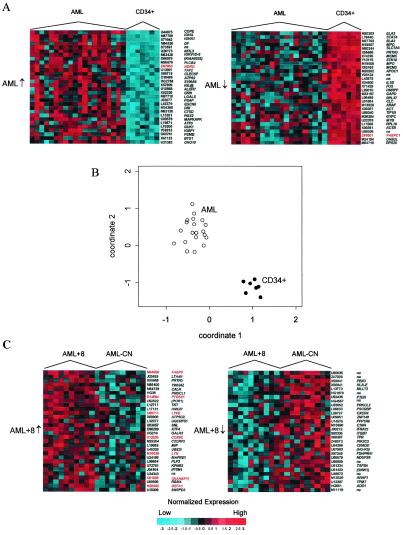
To identify genes that differentiate AML blasts from normal immature progenitor CD34+ cells, we first compared the expression profiles of both AML groups together against the CD34+ samples. Relative to the CD34+ samples, 1,030 genes were up-regulated and 754 genes were down-regulated in AML, respectively. The 30 most significantly down- and up-regulated genes and their corresponding expression profiles are shown in Fig. 2A. On the basis of these 60 dysregulated genes, the AML samples could be clearly distinguished from the CD34+ samples by using simple z-statistic comparisons of the groups (Fig. 2B). However, our selection procedure for the 60 genes could potentially exaggerate this distinction. Thus, we applied a cross-validation procedure to this comparison, confirming that the groups can be reliably distinguished, with 100% of samples correctly reclassified.
Figure 2.
(A) Genes distinguishing AML and CD34+ samples. Expression profiles for the 30 most up-regulated (left, AML↑, from highest to lower) and down-regulated (right, AML↓, from lowest to higher) genes between AML and CD34+ samples. Normalized intensities are presented for each gene as standard deviations of log intensity above the mean (red) and below the mean (blue) across samples. Similarly generated data for genes distinguishing AML+8 and AML-CN are shown in Fig. 6, which is published as supplemental data on the PNAS web site at www.pnas.org. (B) Coordinate plot obtained by averaging the unit normal deviates for genes in A that were down-regulated (coordinate 1) and up-regulated (coordinate 2) in AML vs. CD34+. Cross-validation revealed perfect class prediction based on this simple rule. (C) The corresponding analyses for the most significantly dysregulated genes in AML+8 vs. AML-CN. Genes are identified by their GenBank accession number and symbol. Preliminary symbols are indicated in parentheses. Genes on chromosome 8 are highlighted (red).
Known myeloid markers, such as ELA2 and MPO, were highly expressed in both CD34+ and AML samples, but were relatively more highly expressed in CD34+ than in AML (Fig. 2A). In addition, several known hematopoietic transcription factors showed down-regulation in AML (Fig. 2A): STAT4 (20), FUS (21, 22), and MCM3 and MCM5 (23). Genes that were up-regulated in AML (Fig. 2A) included BTG1, a gene on chromosome 12 associated with the t(8;12) translocation in B-cell chronic lymphatic leukemia (24), and ATF3 (25).
To identify differences specific to either group of AML, we carried out similar comparisons for each AML group against the CD34+ samples. When compared separately to CD34+, the 60 genes showing the most significant differential expression in AML+8 (Fig. 6A in the supplemental data) and AML-CN (Fig. 6B in the supplemental data) were quite different. The AML+8 vs. CD34+ and AML-CN vs. CD34+ comparisons identified seven genes concordantly up-regulated in AML+8 and AML-CN relative to CD34+: DF, COPEB, NDRG1, TAF2H, NFIL3, ATP6S1, and PLCB2. Among the 23 discordant genes that were up-regulated only in AML+8, LYN, TCEB1, and PTK2B are located on chromosome 8 (Fig. 6A). Genes up-regulated only in AML-CN included DAD1 and MLC1 (Fig. 6B).
Nine genes were concordantly down-regulated between the two AML groups relative to the CD34+ group: STAT4, ELA2, MPO, SLC7A5, MCM5, PRG2, APOC1, GAPD, and MCM3 (Fig. 6). However, STAT4 ranked higher in the AML+8 vs. CD34+ than the AML-CN vs. CD34+ comparison. Genes down-regulated only in AML+8 relative to CD34+ (Fig. 6B) included the protooncogene MYC, which is located in chromosome 8q and known for its role as a transcriptional inducer in Burkitt's lymphoma (26, 27), and RALGDS, which maps to the BCR-ABL breakpoint region in 9q34 (28) and acts as an effector molecule for the Ras protein family (29). Interestingly, five different ribosomal proteins were down-regulated in the AML-CN group only (Fig. 6B).
Comparison of Expression Profiles of AML+8 and AML-CN Suggests a Gene-Dosage Effect.
As in the AML vs. CD34+ comparison, we attempted to differentiate AML+8 and AML-CN and compared the overall expression profiles of the two against each other. Class prediction approaches (11) were applied to consider the ability of expression profiling to clearly distinguish the groups. Despite examination using several approaches, careful cross-validation of the prediction rules revealed that the classification precision was not substantially higher than expected by chance. Such unstructured classification procedures involve thousands of genes and exact a large penalty for multiple comparisons. Thus, the result was not entirely unexpected, as the overall gene expression values were very similar in the two groups. More structured comparisons driven by clear hypotheses, such as comparing only those genes on chromosome 8, and comparisons involving genes of prespecified functional categories, can have greater resolving power.
While automatically derived prediction rules did not reliably classify AML+8 and AML-CN samples, the most highly dysregulated genes in the two groups have roles in cell growth regulation (Fig. 2C). Interestingly, of 29 genes most significantly up-regulated in AML+8 (Fig. 2C) for which a chromosomal assignment could be made, 7 map to chromosome 8: FABP5, PTDSS1, LY6E, COX6C, LYN, SIAHBP1, and DEFA1. In contrast, none of the 25 mapped (5 were unmapped) genes up-regulated in AML-CN (Fig. 2C) map to chromosome 8.
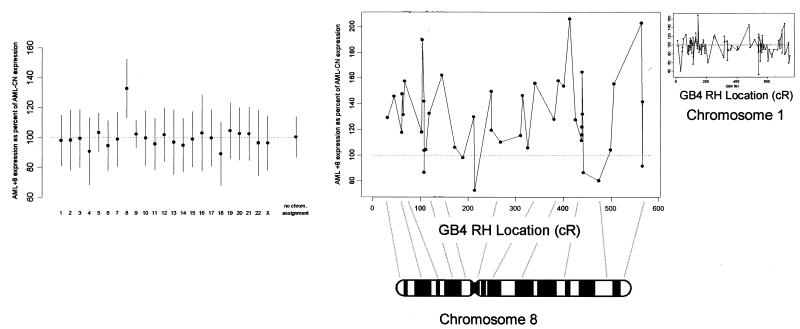
To test our hypothesis of a gene-dosage effect in the origin of AML+8, we identified all mapped genes from the 6,606 unique genes on the HuGeneFL array. Two hundred and thirteen genes mapped to chromosome 8, whereas 495 could not be mapped. Fig. 3 Left shows estimates of the overall AML+8/AML-CN expression ratios by chromosome, using 1,813 genes that were expressed in a majority of the 20 samples. Genes on chromosome 8 showed a clear increase in expression in the AML+8 samples (estimated at 32%, 95% confidence interval 13–52%; P < 0.005) compared with the other 22 chromosomes. An additional procedure was performed using all 213 genes on chromosome 8, in which 1,000 random permutations of group assignment (+8 vs. CN) were applied to the data. None of the values from this permutation distribution was as strong as the statistic computed by using the observed data (empirical P value < 0.001).
Figure 3.
(Left) AML+8 samples exhibit higher expression for genes on chromosome 8 than do AML-CN samples. The average expression levels for genes by chromosome in AML+8 relative to AML-CN are shown. (Right) The increased expression of genes on chromosome 8 as a function of chromosomal location. Of the 169 genes with a specific localization on the integrated radiation hybrid map of chromosome 8, 42 were expressed in a majority of AML samples. For these, the intensities in the AML+8 group are expressed as a percentage of the intensities in the AML-CN group. For comparison, the results for chromosome 1, which shows no gene dosage effect, are presented in the Inset.
To determine whether a specific region on chromosome 8 was concordantly dysregulated in AML+8, we plotted the expression profile of 41 genes that were expressed in the majority of samples and could be localized on chromosome 8 (Fig. 3 Right). No broad-scale regional effects on chromosome 8 were apparent.
Analysis of Expression Profiles Based on Gene Function.
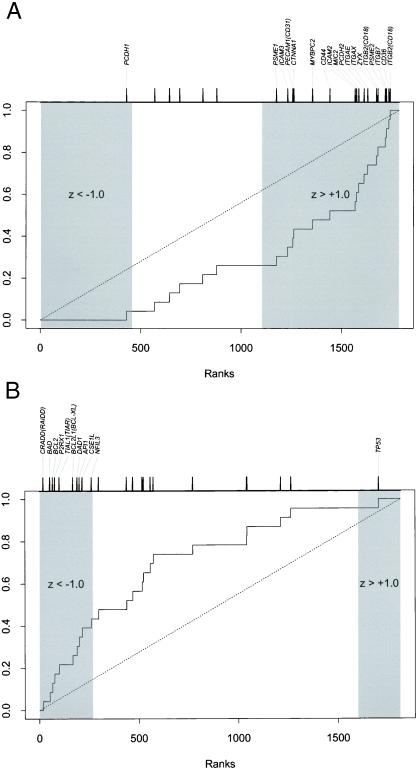
To investigate in more detail the expression profiles of the AML and CD34+ samples we analyzed the genes according to their function(s). We used the functional terms for all 6,606 genes on the HuGeneFL array. Functional annotation was available for 80% of the genes. Most genes have multiple annotations. Our analysis focused on genes involved in nine critical cellular pathways (see Materials and Methods) that have been implicated in leukemogenesis and would likely be associated with the molecular pathology of AML. All genes were examined in a comparison of AML vs. CD34+ (1,784 genes expressed in a majority of samples) and AML+8 vs. AML-CN (1,813 genes expressed in a majority of samples) for a total of 18 comparisons. For each gene, a z score was calculated for the difference in the two comparison groups, and genes were ranked according to the score. The overall involvement of a functional group would be observable as a shift in the ranks of genes in the functional group compared with the remaining genes. We used a rank-based nonparametric procedure as an initial screen for potentially significant results. Three of the 18 comparisons met this criterion: RNA-binding was lower in AML than in CD34+, cell adhesion was higher in AML than in CD34+, and apoptosis was lower in AML+8 than in AML-CN. We subjected each of these comparisons to more rigorous testing using 1,000 random permutations of group membership. The RNA-binding result (data not shown) was still suggestive of greater expression in CD34+ samples, but no longer significant (P = 0.102). The other two comparisons remained significant. Genes involved in cell adhesion were up-regulated in AML relative to CD34+ (P = 0.010; Fig. 4A): genes included several integrins [e.g., ITGB7 and ITGB2 (CD18)] and various members of the immunoglobulin superfamily (e.g., ICAM2 and ICAM3). The most striking result was obtained for apoptosis genes, which were down-regulated in AML+8 vs. AML-CN (P < 0.001; Fig. 4B): for example, CRADD, an apoptosis inducer (30). The second highest level of down-regulation was observed for the Bcl-2 antagonist, BAD (31). In contrast, expression levels of the tumor suppressor and apoptosis inducer TP53 (32) were increased in AML+8 (Fig. 4B).
Figure 4.
Systematic functional analysis of AML samples based on SWISS-PROT database functional annotations for expressed genes. z statistics were calculated for each gene to describe the expression difference in that gene across sample groups. The plots show the cumulative distribution for the ranks of the genes in the functional category relative to all expressed genes. The tick marks at the top of the plots show the ranks of the individual genes; only those showing significant dysregulation (z scores below −1 or above +1) are indicated. (A) Twenty-three expressed genes involved in cell adhesion showed a shift to the right indicating up-regulation of genes in the AML compared with CD34+ samples (P = 0.010). (B) Twenty-three genes involved in apoptosis showed a shift to the left indicating down-regulation in AML+8 compared with AML-CN (P < 0.001).
Validation of Oligonucleotide-Array Gene Expression Results.
We used the TaqMan assay (17) to validate the expression differences for 11 genes of interest. Relative expression levels initially determined with the HuGeneFL array were correlated with TaqMan results for a subset of samples (6 AML+8 and 5 AML-CN samples). We chose FABP5, ATF4, MIF, and SIAHBP1, which were up-regulated in AML+8, and PBX3, MLLT2, PTEN, and CRADD, which were up-regulated in AML-CN (Fig. 2C), in addition to MPO, FUS, and CD34. Fig. 7 in the supplemental data shows a comparison of the GeneChip and TaqMan data for MLLT2 and FABP5, the two most extreme genes in the AML+8 vs. AML-CN comparison. Relative mRNA expression levels of MLLT2 and FABP5 are plotted. The correlation of the z-score comparisons (AML+8 vs. AML-CN) between the two assays for all 11 genes validated was high (r = 0.88), indicating substantial agreement between the two assays for the identification of dysregulated genes (Fig. 8 in the supplemental data).
Discussion
We used global expression profiling with DNA microarrays to systematically characterize gene expression in BM samples of AML patients with AML+8 as the sole abnormality or AML-CN, as well as CD34+ cells of normal individuals. The pathophysiological mechanisms underlying both groups of AML are largely unknown. However, it is believed that normal hematopoietic differentiation, which is regulated predominantly at the transcriptional level, is blocked. We therefore reasoned that the leukemogenic events at the DNA level are reflected in specific gene expression changes and profiles at the RNA level.
Expression patterns of AML patients were clearly distinct from those of CD34+ cells of normal individuals. In contrast, overall expression patterns of AML+8 and AML-CN were more alike. While AML samples could be clearly distinguished from CD34+ samples on the basis of their expression profiles, class prediction techniques indicated that the identification of molecular patterns to distinguish the two AML groups is more difficult than with different leukemias (AML vs. ALL) (11) or specific lymphomas (12). Even so, many of the genes dysregulated in the AML+8 vs. AML-CN comparison have a role in cell growth regulation and may thus be useful in generating hypotheses for future study.
AML+8 was clearly associated with an overexpression of genes on chromosome 8. FABP5 and PTDSS1 were recently identified in a search for leukemia candidate genes in a mouse model (33). LY6E has been identified as a hematopoietic stem cell marker in mice (34) and is induced by retinoic acid in acute promyelocytic leukemia (35). Moreover, HMGIY (36), which is required for interferon induction of LY6E, is also up-regulated in AML+8. Up-regulation of LYN and its use as an AML class predictor (11) as well as its inhibition with antisense oligonucleotides in leukemic cell growth (37) have previously been reported. These observations are consistent with a gene-dosage effect for genes on chromosome 8 associated with leukemogenesis in AML+8. To our knowledge, this is the first study to establish globally increased expression on a chromosome caused by trisomy. Although we cannot exclude the rearrangement of a gene(s) in chromosome 8 as an additional underlying cause, we found no evidence for specific regions on chromosome 8 showing significantly higher expression.
Analysis by cellular functions indicated significant up-regulation of genes involved in cell adhesion for both groups of AML compared together against CD34+ samples. There is increasing evidence (38) for an interaction between AML cells and the BM microenvironment that is mediated by cell adhesion molecules such as the β integrins or CD44. Moreover, integrin-mediated signal transduction has been implicated in many cellular functions, including cell proliferation, cell cycle progression, and cell survival (39, 40).
The most significant differences were observed between AML+8 and AML-CN samples for apoptotic genes. Several genes involved in controlled cell death appeared to be specifically down-regulated in AML+8 patients, suggesting that the two groups of AML are fundamentally different regarding apoptosis. In this context it is worth noting that AML+8 patients are known to be incurable with cytarabine-based chemotherapy alone (5) and that cytarabine has been shown to induce apoptosis (41, 42). Moreover, resistance to chemotherapeutic drugs has been linked to increased inhibition of normal apoptosis (43, 44). Specifically, it has been suggested that high levels of Bcl-2 are responsible for resistance to chemotherapy by protecting CD34+ AML blasts from induced apoptosis (30). Thus, the high level of down-regulation we observed for BAD, which encodes a Bcl-2 antagonist that dimerizes with Bcl-xL to reverse its death repressor activity (31), is of particular interest. Down-regulation of the apoptosis inducers CRADD and BAD in AML+8 is consistent with the idea of lowered apoptosis. Interestingly, expression levels of the apoptosis inducer TP53 (32) were increased in AML+8, whereas BCL2 expression, which is repressed by p53, appeared to be down-regulated. The increase in TP53 expression at the mRNA level could be a physiological response to the decreased transcript levels of other apoptosis inducers, including CRADD and BAD. Because TP53 expression is also subject to translational regulation (45), p53 protein and mRNA expression levels may not necessarily correspond. The same may apply to any of the other genes shown to be dysregulated at the mRNA level. Whereas AML+8 samples showed down-regulation of pro-apoptotic genes, AML-CN samples showed up-regulation of DAD1. Mutant DAD1 inhibits apoptosis (46) and induces T cell hyperproliferation when overexpressed in vivo in mice (47). Opposite trends in the dysregulation of expression levels for key apoptosis regulators, including TP53 as well as p53 transcriptional targets, between the two AML groups suggest that AML+8 and AML-CN escape apoptosis by using distinct pathways.
No significant differences in gene expression were observed for other leukemogenic candidate functions: transcriptional regulation, cell cycle control, or signal transduction. The exclusion of specific functional classes as a whole, however, does not necessarily preclude the involvement of specific genes already implicated in leukemogenesis. For example, several hematopoietic transcription factors such as STAT4, which is involved in IL-12 signal transduction through the JAK/STAT pathway (20), or FUS, which encodes an RNA-binding protein with roles in transcription, DNA repair, and recombination (21) and is translocated with ERG1 in myeloid leukemias (22), are down-regulated in AML. Interestingly, five of the genes identified as AML genes in our AML vs. CD34+ comparison have previously been noted as AML vs. ALL predictor genes (11). Three genes were more highly expressed in AML: DF, LGALS3, and CTSD, a metastatic marker in solid tumors (48). Among the genes down-regulated in AML compared with CD34+, MYB and MCM3 were also down-regulated in the AML vs. ALL comparison (11). Thus, these AML-specific expression changes are likely biologically important in the pathophysiology of AML.
In conclusion, we identified a number of genes whose expression is correlated with the leukemic phenotype. The significant increase in and dysregulation of cell adhesion and apoptotic genes is consistent with the biological and clinical characteristics of these leukemias. Moreover, several of the genes have been implicated in other studies as diagnostic in AML/ALL (11). The identification of abnormalities in these basic cellular functions provides insight into the downstream effects of the molecular events responsible for the initiation of leukemia. The observed differences in the dysregulation of specific functional subsets of genes between the different subclasses of AML (+8 vs. CN) should stimulate further functional studies of the individual components of the pathways implicated.
Supplementary Material
Acknowledgments
We thank J. Palatini, E. Hertlein, R. Gama, X. Gao, H. Yu, J. VanDeusen, D. Bucci, and K. Archer for technical assistance, and P. Tamayo, N. Siemers, and E. Lander of the Whitehead Institute/Massachusetts Institute of Technology Center for Genome Research for making available the functional annotation of the HuGeneFL GeneChip array. This study was supported in part by an Ohio State University Comprehensive Cancer Center James Cancer Hospital and Solove Research Institute institutional seed grant (R.K.), the Leukemia Clinical Research Foundation (C.D.B.), and National Institutes of Health Grants GM58934 (F.A.W.), CA77658 (C.D.B.), CA31946 (C.D.B., M.A.C.), and CA09338 (M.A.C.) and P30 CA16058 from the National Cancer Institute.
Abbreviations
- AML
acute myeloid leukemia
- CN
normal cytogenetics
- +8
trisomy 8
- BM
bone marrow
- ALL
acute lymphocytic leukemia
Footnotes
K.V., F.A.W., and R.K. contributed equally to this work.
References
- 1.Orkin S H. Curr Opin Cell Biol. 1995;7:870–877. doi: 10.1016/0955-0674(95)80072-7. [DOI] [PubMed] [Google Scholar]
- 2.Tenen D, Hromas R, Licht J, Zhang D. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 3.Wickremasinghe R, Hoffbrand A. Blood. 1999;93:3587–3600. [PubMed] [Google Scholar]
- 4.Heim S, Mitelman F. Cancer Cytogenetics. New York: Wiley; 1995. [Google Scholar]
- 5.Byrd J C, Lawrence D, Arthur D C, Pettenati M J, Tantravahi R, Qumsiyeh M, Stamberg J, Davey F R, Schiffer C A, Bloomfield C D. Clin Cancer Res. 1998;4:1235–1241. [PubMed] [Google Scholar]
- 6.Mrózek K, Heinonen K, de la Chapelle A, Bloomfield C D. Semin Oncol. 1997;24:17–31. [PubMed] [Google Scholar]
- 7.Pritchard M A, Kola I. J Neural Transm Suppl. 1999;57:293–303. [PubMed] [Google Scholar]
- 8.Taub J W, Huang X, Matherly L H, Stout M L, Buck S A, Massey G V, Becton D L, Chang M N, Weinstein H J, Ravindranath Y. Blood. 1999;94:1393–1400. [PubMed] [Google Scholar]
- 9.Seghezzi L, Maserati E, Minelli A, Dellavecchia C, Addis P, Locatelli F, Angioni A, Balloni P, Miano C, Cavalli P, et al. Genes Chromosomes Cancer. 1996;17:94–101. doi: 10.1002/(SICI)1098-2264(199610)17:2<94::AID-GCC4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 10.Caligiuri M A, Strout M P, Lawrence D, Arthur D C, Baer M R, Yu F, Knuutila S, Mrozek K, Oberkircher A R, Marcucci G, et al. Cancer Res. 1998;58:55–59. [PubMed] [Google Scholar]
- 11.Golub T R, Slonim D K, Tamayo P, Huard C, Gaasenbeek M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 12.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 13.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hemndrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, et al. Nature (London) 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 14.Perou C M, Sorlie T, Eisen M B, van de Rijn M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johsen H, Akslen L A, et al. Nature (London) 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 15.Ott R L. An Introduction to Statistical Methods and Data Analysis. Belmont, CA: Duxbury; 1993. [Google Scholar]
- 16.Deloukas P, Schuler G D, Gyapay G, Beasley E M, Soderlund C, Rodriguez-Tome P, Hui L, Matise T C, McKusick K B, Beckmann J S, et al. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- 17.Heid C A, Stevens J, Livak K J, Williams P M. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 18.Lipshutz R J, Fodor S P, Gingeras T R, Lockhart D J. Nat Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 19.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K S, Frank D A, Ritz J. Blood. 2000;95:3183–3190. [PubMed] [Google Scholar]
- 21.Hicks G G, Singh N, Nashabi A, Mai S, Bozek G, Klewes L, Arapovic D, White E K, Koury M J, Oltz E M, et al. Nat Genet. 2000;24:175–179. doi: 10.1038/72842. [DOI] [PubMed] [Google Scholar]
- 22.Ichikawa H, Shimizu K, Hayashi Y, Ohki M. Cancer Res. 1994;54:2865–2868. [PubMed] [Google Scholar]
- 23.Tye B K. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 24.Rimokh R, Rouault J P, Wahbi K, Gadoux M, Lafage M, Archimbaud E, Charrin C, Gentilhomme O, Germain D, Samarut J, et al. Genes Chromosomes Cancer. 1991;3:24–36. doi: 10.1002/gcc.2870030106. [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann J, Erdmann D, Lalande I, Grossenbacher R, Noorani M, Furst P. Oncogene. 2000;19:2913–2920. doi: 10.1038/sj.onc.1203606. [DOI] [PubMed] [Google Scholar]
- 26.Maguire R T, Robins T S, Thorgeirsson S S, Heilman C A. Proc Natl Acad Sci USA. 1983;80:1947–1950. doi: 10.1073/pnas.80.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishikura K, ar-Rushdi A, Erikson J, Watt R, Rovera G, Croce C M. Proc Natl Acad Sci USA. 1983;80:4822–4826. doi: 10.1073/pnas.80.15.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuler G D, Boguski M S, Stewart E A, Stein L D, Gyapay G, Rice K, White R E, Rodriguez-Tome P, Aggarwal A, Bajorek E, et al. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 29.Hofer F, Fields S, Schneider C, Martin G S. Proc Natl Acad Sci USA. 1994;91:11089–11093. doi: 10.1073/pnas.91.23.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmad M, Srinivasula S M, Wang L, Talanian R V, Litwack G, Fernandes-Alnemri T, Alnemri E S. Cancer Res. 1997;57:615–619. [PubMed] [Google Scholar]
- 31.Yang E, Zha J, Jockel J, Boise L H, Thompson C B, Korsmeyer S J. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 32.Sheikh M S, Fornace A J., Jr J Cell Physiol. 2000;182:171–181. doi: 10.1002/(SICI)1097-4652(200002)182:2<171::AID-JCP5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Shen H, Himmel K L, Dupuy A J, Largaespada D A, Nakamura T, Shaughnessy J D, Jr, Jenkins N A, Copeland N G. Nat Genet. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- 34.Spangrude G J, Brooks D M. Blood. 1993;82:3327–3332. [PubMed] [Google Scholar]
- 35.Mao M, Yu M, Tong J H, Ye J, Zhu J, Huang Q H, Fu G, Yu L, Zhao S Y, Waxman S, et al. Proc Natl Acad Sci USA. 1996;93:5910–5914. doi: 10.1073/pnas.93.12.5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khodadoust M M, Khan K D, Park E H, Bothwell A L. Blood. 1998;92:2399–2409. [PubMed] [Google Scholar]
- 37.Roginskaya V, Zuo S, Caudell E, Nambudiri G, Kraker A J, Corey S J. Leukemia. 1999;13:855–861. doi: 10.1038/sj.leu.2401429. [DOI] [PubMed] [Google Scholar]
- 38.Bradstock K F, Gottlieb D J. Leuk Lymphoma. 1995;18:1–16. doi: 10.3109/10428199509064917. [DOI] [PubMed] [Google Scholar]
- 39.Giancotti F G, Ruoslahti E. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 40.Hynes R O. Trends Cell Biol. 1999;9:M33–M37. [PubMed] [Google Scholar]
- 41.Friesen C, Fulda S, Debatin K M. Cell Death Differ. 1999;6:471–480. doi: 10.1038/sj.cdd.4400512. [DOI] [PubMed] [Google Scholar]
- 42.Mesner P W, Jr, Budihardjo I I, Kaufmann S H. Adv Pharmacol. 1997;41:461–499. doi: 10.1016/s1054-3589(08)61069-8. [DOI] [PubMed] [Google Scholar]
- 43.Hassan H T, Zander A. Acta Haematol. 1996;95:257–262. doi: 10.1159/000203893. [DOI] [PubMed] [Google Scholar]
- 44.Buchner T, Hiddemann W, Wormann B, Zuhlsdorf M, Rottmann R, Innig G, Maschmeier G, Ludwig W D, Sauerland M C, Heinecke A. Semin Oncol. 1997;24:124–131. [PubMed] [Google Scholar]
- 45.Fu L, Minden M D, Benchimol S. EMBO J. 1996;15:4392–4401. [PMC free article] [PubMed] [Google Scholar]
- 46.Nakashima T, Sekiguchi T, Kuraoka A, Fukushima K, Shibata Y, Komiyama S, Nishimoto T. Mol Cell Biol. 1993;13:6367–6374. doi: 10.1128/mcb.13.10.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong N A, Kabra N H, Hsieh S N, Cado D, Winoto A. J Immunol. 1999;163:1888–1893. [PubMed] [Google Scholar]
- 48.Rochefort H, Liaudet-Coopman E. APMIS. 1999;107:86–95. doi: 10.1111/j.1699-0463.1999.tb01530.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.