Abstract
Visual pigments (rhodopsins) are composed of a chromophore (vitamin A derivative) bound to a protein moiety embedded in the retinal membranes. Animals cannot synthesize the visual chromophore de novo but rely on the uptake of carotenoids, from which vitamin A is formed enzymatically by oxidative cleavage. Despite its importance, the enzyme catalyzing the key step in vitamin A formation resisted molecular analyses until recently, when the successful cloning of a cDNA encoding an enzyme with β,β-carotene-15,15′-dioxygenase activity from Drosophila was reported. To prove its identity with the key enzyme for vitamin A formation in vivo, we analyzed the blind Drosophila mutant ninaB. In two independent ninaB alleles, we found mutations in the gene encoding the β,β-carotene-15,15′-dioxygenase. These mutations lead to a defect in vitamin A formation and are responsible for blindness of these flies.
The vertebrate and invertebrate phototransduction system is a rhodopsin-based G protein-coupled signaling cascade displaying exquisite sensitivity and broad dynamic range. The visual stimulus triggers a change of the ionic permeability of the photoreceptor cell membranes (1, 2). To analyze the molecular mechanisms of the visual process, Drosophila has served for decades as a model system for studies using electrophysiology, photochemistry, genetics, and molecular biology (1, 3, 4). Powerful screening strategies and subsequent genetic dissection led to the characterization of several mutants affected in different parts of the visual process, e.g., in the protein moiety of the visual pigments (ninaE; ref. 5), rhodopsin assembly (ninaA; ref. 6), and signal transduction (norpA, rdgA, arr1; refs. 7–9). The molecular identification of most of the components responsible for the phototransduction system has since been achieved. However, molecular characterization of mutants affecting the formation of the visual chromophores still is missing.
In both the vertebrate and invertebrate visual systems, vitamin A derivatives such as retinal or 3-hydroxyretinal serve as chromophores for the various visual pigments (rhodopsins; refs. 10 and 11). In vertebrates, besides a role in vision, vitamin A and its derivatives exert multiple functions in development, embryogenesis, and cell differentiation (12, 13), whereas in invertebrates vitamin A functions seem to be restricted to the visual system (14). Animals in general are not able to synthesize vitamin A de novo. For the formation of the visual chromophores, they rely on the uptake in the diet of carotenoids with provitamin A activity or on preformed retinoids from animal food sources. The key step in the formation of vitamin A was shown 40 years ago to be the oxidative cleavage of carotenoids by carotenoid dioxygenases (15). Molecular data on this important reaction have been missing since then, and there is a controversy regarding the reaction mechanism (16). A breakthrough in understanding the oxidative cleavage reaction of carotenoids came from earlier work on a carotenoid-cleavage enzyme in the pathway leading to the plant-growth regulator abscisic acid in maize and other plants. In plants, various carotenoid-cleavage products have been described, e.g., saffron in crocus, citraurin and other apocarotenals in citrus fruits, and abscisic acid. The analysis of the maize mutant, vp14, led to the identification of the gene and cDNA encoding a 9-cis-epoxy-carotenoid dioxygenase, the key enzyme in the formation of abscisic acid (17, 18). By sequence similarity to this plant enzyme, a cDNA encoding a putative animal carotene dioxygenase could be isolated from Drosophila melanogaster in our laboratory (19). Heterologous expression in Escherichia coli and subsequent biochemical analyses revealed that the encoded protein, β,β-carotene-15,15′-dioxygenase (β-diox), was able to catalyze the symmetric oxidative cleavage of β-carotene to form two molecules of vitamin A aldehyde (retinal). Soon thereafter, a cDNA encoding its vertebrate homologue was identified (20). Furthermore, in the databases there are cDNAs encoding putative β-carotene dioxygenases from several species, including human. Sequence comparison showed a high overall sequence similarity among the animal enzymes and a significant similarity to some domains of the plant dioxygenase (Fig. 1). This similarity indicates that the animal proteins belong to a large and diverse class of dioxygenases heretofore described only in plants and microorganisms (21). The identification of cDNAs encoding β-diox may provide the key toward understanding vitamin A formation in animals. However, direct evidence that the encoded proteins catalyze vitamin A formation in vivo, and therefore the generation of the visual chromophores is still missing.
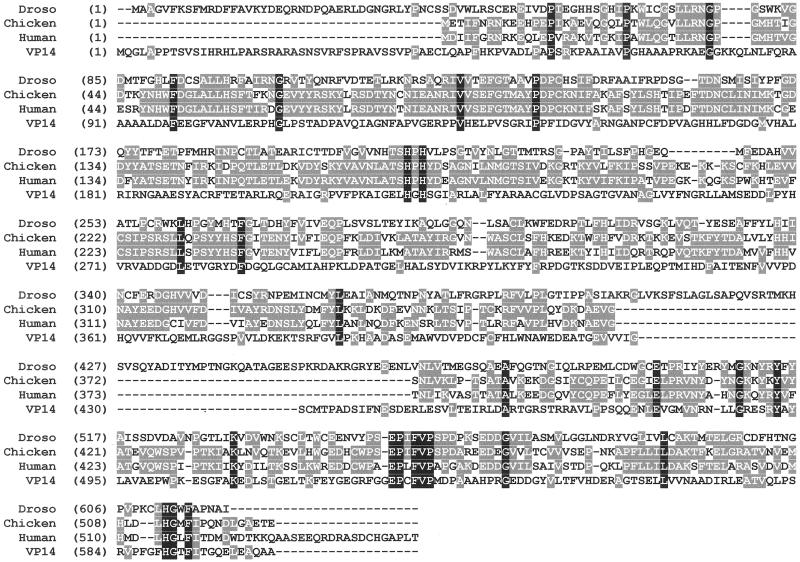
Figure 1.
Linear alignment of the deduced amino acid sequences of the β-diox from Drosophila compared with its vertebrate homologues from chicken (20) and human (GenBank accession no. AF294900) and to the plant epoxy-carotenoid cleaving enzyme (VP14) from maize (17). Identity of the amino acids in all four amino acid sequences is indicated in black, and identities in at least two amino acid sequences are indicated in dark gray.
Therefore, we searched for Drosophila mutants affecting vision. Among the various mutants, two mutants, ninaB and ninaD, have been described (22) for which the corresponding genes have not yet been identified. Both mutants exhibit a strongly reduced visual sensitivity and display an abnormal electroretinogram (nina: neither inactivation nor afterpotential). The mutant phenotypes can be rescued by supplementing the diet with retinal, the direct precursor of the fly's visual chromophore 3-hydroxy-retinal. However, the ninaD phenotype can be rescued also by feeding high doses of β-carotene, and therefore it has been concluded that a function involved in carotenoid uptake, transport, or storage is affected by this mutation (23). In contrast, the ninaB phenotype can be rescued exclusively by feeding of retinal but not by high dosages of β-carotene (22). This finding led us to infer that a mutation in the recently characterized β-diox gene might cause this phenotype. This possibility was strengthened by the fact that the ninaB mutation has been mapped to the position 87E-F (22) in the Drosophila genome,coinciding with the physical location of the β-diox gene.
Here we show by analyzing two independent ninaB alleles that β-diox indeed is encoded by this gene. Molecular and biochemical analyses of the β-diox function in both ninaBP315 and ninaB360d Drosophila flies revealed defects leading to a loss of the ability to form the visual chromophore, thus identifying the ninaB gene to encode the key enzyme for vitamin A formation in vivo.
Materials and Methods
Flies Stocks and Growth Conditions.
Homozygous ninaBP315 and ninaB360d flies were reared on standard corn medium from a heterozygous w;;w ninaBP315/TM3,Sb,Ser stock and a heterozygous w;;w ninaB360d/TM3,Sb,Ser stock, respectively. For control experiments, heterozygous ninaBP315 flies or wtB (Berlin) flies were used. For vitamin A deprivation, the flies were raised on carotenoid-depleted white corn (DWC) medium (24). β-Carotene or all-trans retinal were supplemented in ethanol to the appropriate medium.
Cloning of the β-diox from ninaB Flies.
For cloning of β-diox from wt, ninaBP315, and ninaB360d Drosophila melanogaster, we isolated total RNA from heads of adult flies obtained by hand dissection. Reverse transcription (RT) was performed by using the primer 5′-CTAAATGGCATTGGGTGCAAACC-3′ and superscript reverse transcriptase (Life Technologies, Gaithersburg, MD). Subsequently, PCR was performed with the primer 5′-GACGCCGGTGTCTTCAAGAG-3′ and the primer used for the RT assay and the Expand PCR system (Roche Molecular Biochemicals) and Pwo-Polymerase (PEQ-Lab, Erlangen, Germany), respectively. The PCR products were cloned directly in the vectors pTOPO-BAD and pCR2.1-Topo (Invitrogen), respectively. For cloning of the genomic DNA, a PCR was performed with the same set of primers by using genomic DNA as a template. The PCR products were cloned directly in the vector pCR2.1-TOPO (Invitrogen). To analyze the resultant plasmids and to confirm their structure, both strands were sequenced completely.
Site-Directed Mutagenesis.
Site-directed mutagenesis was performed by PCR with a set of primers spanning the entire coding region: up, 5′- GACGCCGGTGTCTTCAAGAG-3′ and down, 5′-CTAAATGGCATTGGGTGCAAACC-3′. A set of reverse-oriented overlapping primers for the introduction of the desired base-pair alteration was used: for the Glu-to-Lys exchange at position 280 up, 5′-CTACTTTGTGATTGTGAAGCAGCC-3′ and down, 5′-ACGGACAACGGCTGCTTCACAATC-3′; for the Met-to-Leu exchange at position 471 up, 5′-GTCAATCTGGTTACCCTGGAGGGC-3′ and down, 5′-GCTTGACTGCCCTCCAGGGTAACC-3′; and for the Glu-to-Ala exchange at position 477 up, 5′-GGGCAGTCAAGCGGCGGCGTTTCA-3′ and down, 5′-CCCTGAAACGCCGCCGCTTGACTG-3′. For the Lys-to-Glu reversion at position 280 in the ninaBP315 cDNA, the primers used were: up, 5′-CTACTTTGTGATTGTGGAGCAGCC-3′ and down, 5′-ACGGACAACGGCTGCTCCACAATC-3′. The PCR products were cloned in the expression vector pTOPO-BAD (Invitrogen), and the mutations in the plasmid constructs were verified by sequence analyses.
Estimation of the ninaE mRNA Levels by Semiquantitative RT-PCR Analyses.
Total RNA was isolated from 10 flies reared on the appropriate media. Three different amounts of total RNA (100, 33, and 11 ng) were subjected to RT-PCR analyses as has been described (25). For ninaE, the following set of intron-spanning primers was used: up, 5′-TCTGTATTTCGAGACCTGG-3′ and down, 5′-TTCTCGGCATCCTCAGAGGAG-3′. Due to stochastic deviations between different sets of experiments, only samples from the same set of experiments were compared.
Extraction of β-Carotene and Retinoids from E. coli and HPLC Analysis.
The E. coli strains were grown under a red safe light in 50-ml flasks in LB until the cultures had reached an OD600 of 1.0. Expression of β-diox was induced by the addition of L-arabinose (0.2% wt/vol) for 6 or 16 h. Then the bacteria were harvested by centrifugation. The pellets were extracted and HPLC analyses were carried out as has been described (19).
Visualization of the Deep Pseudopupil.
For viewing the deep pseudopupil with orthodromic (direct-frontal) illumination at 573 nm in the compound eye of living white-eyed Drosophila, the eyes were adapted by saturating illumination at 450 nm, leading to a photo equilibrium with 20% rhodopsin (R480) and 80% metarhodopsin (M570). Monochromatic light was generated by a xenon lamp (Schölly, Freiburg, Germany) combined with an optical fiber with a filter wheel with interference line filters (Schott, Mainz, Germany). Photographs were taken under a binocular microscope (MZ12, Leica, Deerfield, IL) equipped with a VarioCam monochrome CCD video camera (PCO Computer Optics, Kelheim, Germany) with a variable time exposure, Peltier CCD cooling, and digital interface.
Results and Discussion
Cloning of the β-diox from ninaB Flies and Biochemical Characterization of the Encoded Protein.
We established homozygous ninaBP315 and ninaB360d flies from TM3,Sb,Ser balanced stocks and isolated genomic DNA and total RNA from homozygous ninaBP315 and ninaB360d flies, heterozygotes, and wt flies (22). Because a putative mutation was expected to affect the β-diox function at several levels, we tested whether β-diox mRNA is still present in the ninaB flies first. For this purpose, we performed RT-PCR analyses by using a pair of primers spanning the entire coding region of β-diox. In both ninaB strains, the β-diox mRNA levels were similar to the wild-type control, and the amplified cDNAs had the same size. These results eliminated the possibility of a mutation affecting the transcriptional regulation of the β-diox gene. We next analyzed the ability of the encoded proteins to catalyze vitamin A formation. For this purpose, we cloned β-diox cDNAs from ninaBP315 and ninaB360d homozygous flies and wild-type flies into the expression vector pTOPO-BAD. To test directly for enzymatic activity of the encoded proteins, we transformed the resultant plasmid constructs into the genetic background of an E. coli strain able to synthesize and accumulate β-carotene. With the aid of this test system, it is possible to assess β-diox enzymatic activity by a color change of the bacteria (19). This color change caused by the cleavage of β-carotene (yellow) to retinoids (colorless) became visible in the E. coli strain expressing the wild-type β-diox. In contrast, the E. coli strains expressing the β-diox encoded by the ninaB alleles remained yellow and therefore failed to catalyze retinoid formation (Fig. 2A). For more detailed analyses, overnight cultures were grown and HPLC analyses of lipids were performed as described (19). In the E. coli strain expressing β-diox isolated from wt flies, significant amounts of retinal as well as the corresponding retinols could be detected, whereas in the E. coli strain expressing β-diox isolated from the ninaB mutants, no traces of retinoids were detectable (Fig. 2B). Thus, the cDNAs isolated from ninaB homozygous flies did not encode functional β-diox proteins.
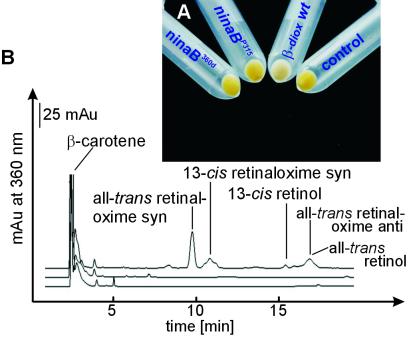
Figure 2.
Biochemical analyses of the β-diox encoded by the ninaBP315 and ninaB360d alleles compared with the wt allele. (A) The picture shows the color shift of the β-carotene-producing and -accumulating E. coli strain from yellow to white caused by the formation of retinoids from β-carotene by the β-diox enzymatic activity. Although the E. coli strain expressing the β-diox encoded by the wt allele becomes white, the E. coli expressing β-diox encoded by the ninaBP315 and ninaB360d alleles remain yellow. A control E. coli strain transformed with the expression vector (pBAD-TOPO) alone remains yellow. (B) HPLC analyses of the products formed in the different E. coli strains. In the E. coli strain expressing β-diox encoded by the wt allele, significant amounts of retinoids are found (upper trace), whereas in the E. coli strain expressing β-diox encoded by the ninaBP315 (middle trace) or ninaB360d (lower trace) alleles, no retinoids become detectable.
A Single Lys-to-Glu Substitution in the ninaBP315 Allele and a Nonsense Mutation in the ninaB360d Allele Abolish the β-diox Enzymatic Activity.
To analyze the molecular basis for this defect, we performed sequence analyses of cDNAs isolated from ninaBP315 and ninaB360d flies by three independent RT-PCRs and compared the sequence with cDNAs isolated from both wt and heterozygous flies.
The sequence analysis of the ninaB360d allele revealed a nonsense mutation, TCG to TAG, at position 41 of the deduced amino acid sequence, whereas in the ninaBP315 allele, three base-pair exchanges compared with the wt allele were found (Fig. 3). In ninaBP315, all three mutations lead to an alteration of the deduced amino acid sequence, but do not interfere with the ORF. To determine which of these three mutations is responsible for the loss of the β-diox enzymatic activity, we introduced each of these mutations separately into the wt allele by site-directed mutagenesis. The resultant expression plasmids harboring the three different mutant cDNAs were transformed into the β-carotene-producing E. coli strain and tested for β-diox enzymatic activity of the encoded mutant protein. The cDNA mutated at position 838 leading to a Glu-to-Lys exchange in the deduced amino acid sequence resulted in a loss of the β-diox enzymatic activity, whereas the mutations at the positions 1,411 (Met-to-Leu) and 1,430 (Glu-to-Ala) did not affect the β-diox enzymatic activity significantly as shown by the formation of retinoids in the E. coli test system (Fig. 4). This result was verified by converting the base-pair exchange at position 838 in the ninaBP315 cDNA to the wt sequence by site-directed mutagenesis, resulting in a restoration of the β-diox enzymatic activity in the E. coli test system (Fig. 4).
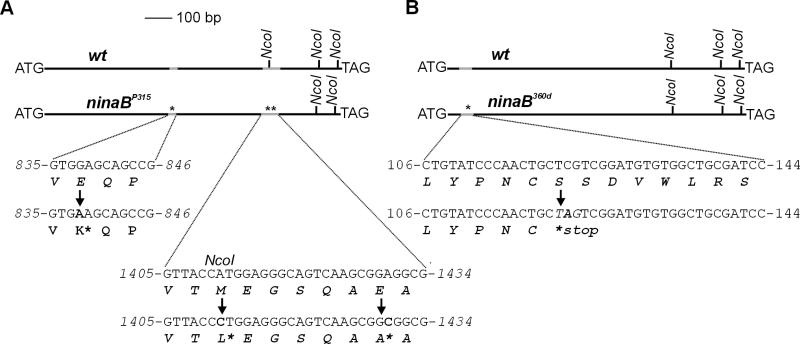
Figure 3.
Molecular analyses of the ninaB mutations. For sequence analyses β-diox cDNAs were cloned from homozygous ninaBP315, homozygous ninaB360d, heterozygous, and wt flies by RT-PCR with primers spanning the whole coding region. The alterations found in the ninaB alleles compared with the wt allele are indicated by arrows. (A) The sequence analyses of the cDNA isolated from ninaBP315 revealed three base-pair exchanges at positions 838, 1,411, and 1,430 with respect to the deduced start ATG compared with the cDNA isolated from wt flies (AJ276682). All three alterations led to an alteration of the deduced amino acid sequence. (B) In the ninaB360d, a point mutation at position 122 (C to A) was found, resulting in a nonsense codon in the deduced amino acid sequence.
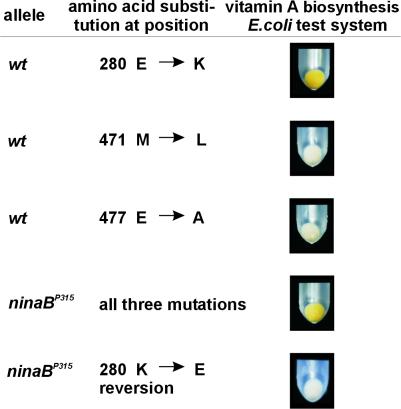
Figure 4.
A single Glu-to-Lys substitution leads to the loss of the β-diox enzymatic activity. The three different mutations found in the β-diox cDNA encoded by the ninaBP315 allele were introduced separately in the wild-type cDNA by site-directed mutagenesis, and the resultant plasmids were tested for enzymatic activity in the E. coli test system. Only the mutation at position 838 leading to a Glu-to-Lys exchange in the deduced amino acid sequence results in a loss of β-diox enzymatic activity, whereas the mutations at positions 1,411 and 1,430 do not affect the enzymatic activity of β-diox. By converting the mutation at position 838 in the ninaBP315 cDNA, the enzymatic activity of the encoded protein could be restored.
In summary, in both ninaB alleles, amino acid sequence alterations could be detected and shown to lead to a loss of function of the encoded β-diox protein. In the ninaB360d allele, a nonsense mutation at position 41 interrupts the ORF, whereas in the ninaBP315 allele, the missense mutation at position 838 allele leading to a Glu-to-Lys exchange in the encoded β-diox protein is responsible for the loss of its enzymatic activity.
The Rhodopsin Content Is Drastically Reduced in ninaB Flies.
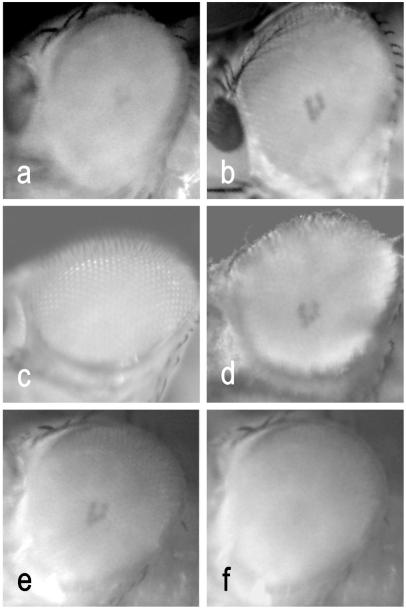
We expected that the mutations found in the ninaB alleles abolished the ability to form the visual chromophore from carotenoids in vivo. To visualize this phenotype directly in the compound eyes of the living fly, we used the appearance of the deep pseudopupil as a measure for the visual pigment content (26–28). For this purpose, we used orthodromic illumination with 573-nm light, which is close to the metarhodopsin absorption maximum (M570) (29). For viewing the deep pseudopupil at 573 nm, the photo equilibrium between rhodopsin (P480) and metarhodopsin (M570) was shifted maximally to the M570 state with saturating illumination at 450 nm. As shown in Fig. 5, in heterozygous flies, a distinct pseudopupil became visible. In homozygous ninaBP315 flies, only a weak shadow was detectable, whereas in ninaB360d flies, a pseudopupil could not be detected. To verify that this phenotype is caused by vitamin A deficiency, we raised ninaB flies on corn medium supplemented with all-trans retinal. This led to a recovery of the deep pseudopupil because of a restoration of the visual pigments. The weak remainder of visual pigments detectable in nonsupplemented ninaBP315 flies has been shown indirectly by electrophysiological measurements previously (22). It may be caused by a residual enzymatic activity of the ninaBP315 mutant protein in vivo. Sequence comparison uncovered no other gene with significant similarity to β-diox in the entire Drosophila genome. Thus, in Drosophila, the visual chromophores are formed exclusively by the symmetric cleavage of β-carotene catalyzed by β-diox.
Figure 5.
Appearance of the deep pseudopupil at 573 nm in the compound eye of living flies after saturating illumination with light of 450 nm. (a) ninaBP315 homozygous flies raised on corn medium. (b) ninaBP315 homozygous flies raised on corn medium supplemented with all-trans retinal. (c) ninaB360d homozygous flies raised on corn medium. (d) ninaB360d homozygous flies raised on corn medium supplemented with all-trans retinal. (e) Visualization of the deep pseudopupil in heterozygous flies. (f) Disappearance of the deep pseudopupil in heterozygous flies after shifting the visual pigments from the metarhodopsin (570-nm) to the rhodopsin (480-nm) state with orange light.
Vitamin A Deficiency Does Not Interfere with Opsin Gene Expression in Drosophila.
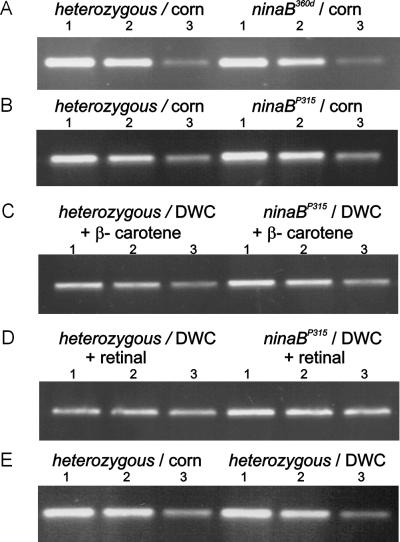
Besides its role as the visual chromophore, vitamin A influences on opsin gene transcription, translation, and the maturation of the visual pigments have been reported (30–32) but are controversial. By using the ninaB mutants, which have a genetically caused vitamin A deficiency, we addressed this question and investigated the impact of this mutation on the regulation of the mRNA levels of the major opsin gene (ninaE) by semiquantitative RT-PCR. In flies raised on standard corn medium (with β-carotene as the source for vitamin A formation), no difference in the mRNA levels isolated from ninaB flies was detectable compared with heterozygous or wt flies (Fig. 6). Then we reared the flies on carotenoid-depleted medium and compared the mRNA levels of ninaE with those of flies reared on the same medium but supplemented with either β-carotene or all-trans-retinal. No differences were observed under these conditions. These results indicate that ninaE mRNA levels are not affected by vitamin A deficiency caused either by a genetic defect in the ninaB mutant or by deprivation of the provitamin in heterozygous and wt flies under the growth conditions applied. Furthermore, the inability to form retinal from β-carotene in the ninaB mutants did not seem to affect the β-diox mRNA levels, indicating that retinoids are not involved in its transcriptional regulation.
Figure 6.
Vitamin A deficiency does not influence the mRNA levels of ninaE (the major opsin) in wild-type flies and ninaB flies. Flies were reared on normal corn medium, DWC (carotenoid-depleted white corn) medium, or DWC medium supplemented with all-trans retinal and β-carotene, respectively. Total RNA was isolated, and RT-PCR was performed as described with three different amounts of total RNA. The RT-PCR products obtained from 100, 33, and 11 ng of total RNA (indicated by the numbers 1, 2, and 3, respectively, above the figures) were separated on an agarose (1.2%) gel containing 1× 90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3 and 0.2 μg⋅ml−1 ethidium bromide. The photographs show the ninaE mRNA levels in heterozygous flies compared with ninaB360d homozygous flies reared on standard corn medium (A), heterozygous flies compared with ninaBP315 homozygous flies reared on standard corn medium (B), heterozygous flies compared with ninaBP315 homozygous flies reared on DWC medium supplemented with β-carotene (C), heterozygous flies compared with ninaBP315 homozygous flies reared on DWC medium supplemented with all-trans retinal (D), and heterozygous flies reared on standard corn medium compared with heterozygous flies reared on DWC medium (E).
Conclusions
In summary, the molecular analyses of the ninaB Drosophila mutants revealed that their vitamin A deficient phenotype indeed is caused by a mutation in the β-diox gene and identified it as the gene encoding the key enzyme for vitamin A formation. In Drosophila, the ninaB mutation led to a vitamin A deficiency accompanied by a dramatically reduced content of visual pigments, but no other defects became phenotypically manifest. In the databases, several cDNAs encoding putative β-diox are found, including chicken (20), mouse (AJ278064), and human homologues (AF284900). The results obtained by the analyses of Drosophila ninaB flies indicate that these vertebrate homologues likely exert the same in vivo function. However, it remains to be investigated whether the β-diox function in vertebrates is encoded by a single gene or by a small gene family. In vertebrates, a mutation comparable with ninaB probably would cause fatal effects considering the multiple vitamin A functions in development and cell differentiation as well as in vision. The molecular identification of the key enzyme for vitamin A formation will now open up further avenues of research. This research will include tissue specificity of vitamin A formation, the regulation of vitamin A homeostasis, and, especially in vertebrates, the impact of vitamin A formation on cell differentiation and developmental processes mediated by retinoic acid.
The significant similarity among plant and animal carotenoid-cleaving enzymes indicates that these enzymes belong to a diverse and widespread class of dioxygenases. In plants, a variety of carotenoid-cleavage products has been described, including all-trans-retinal in algal rhodopsins. The sequence homologies between algal and animal opsin genes (33) as well as genes involved in carotenoid cleavage in plants and animals support the hypothesis (34) that the various rhodopsin-based phototransduction systems most likely arose from a common ancestor.
Acknowledgments
We thank Prof. William L. Pak (Purdue University, West Lafayette, IN) for the gift of the ninaBP315 and ninaB360d mutant fly stocks and Beate Ziser for her technical assistance. M.F.W. was supported by a grant from Boehringer Ingelheim Fonds.
Abbreviations
- β-diox
β,β-carotene-15,15′-dioxygenase
- RT
reverse transcription
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ276682).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031576398.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031576398
References
- 1.Zuker C S. Proc Natl Acad Sci USA. 1996;93:571–576. doi: 10.1073/pnas.93.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stryer L. Proc Natl Acad Sci USA. 1996;93:557–559. doi: 10.1073/pnas.93.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pak W L. Nature (London) 1970;227:518–520. doi: 10.1038/227518b0. [DOI] [PubMed] [Google Scholar]
- 4.Hotta Y, Benzer S. Nature (London) 1969;222:354–356. doi: 10.1038/222354a0. [DOI] [PubMed] [Google Scholar]
- 5.O'Tousa J E, Baehr W, Martin R L, Hirsh J, Pak W L, Applebury M L. Cell. 1985;40:839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- 6.Shieh B H, Stamnes M A, Seavello S, Harris G L, Zuker C S. Nature (London) 1989;338:67–70. doi: 10.1038/338067a0. [DOI] [PubMed] [Google Scholar]
- 7.Bloomquist B T, Shortridge R D, Schneuwly S, Perdew M, Montell C, Steller H, Rubin G, Pak W L. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- 8.Masai I, Okazaki A, Hosoya T, Hotta Y. Proc Natl Acad Sci USA. 1993;90:11157–11161. doi: 10.1073/pnas.90.23.11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith D P, Shieh B H, Zuker C S. Proc Natl Acad Sci USA. 1990;87:1003–1007. doi: 10.1073/pnas.87.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wald G. Nature (London) 1968;219:800–807. doi: 10.1038/219800a0. [DOI] [PubMed] [Google Scholar]
- 11.Vogt K. Z Naturforsch B: 2 Naturforsch. 1984;39c:196–197. [Google Scholar]
- 12.Dowling J E, Wald G. Proc Natl Acad Sci USA. 1960;46:587–608. doi: 10.1073/pnas.46.5.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chytil F. In: Retinoids: From Basic Science to Clinical Applications. Livrea M A, Vidali G, editors. Basel: Birkhäuser; 1994. pp. pp.11–19. [Google Scholar]
- 14.Harris W A, Ready D F, Lipson E D, Hudspeth A J, Stark W S. Nature (London) 1977;266:648–650. doi: 10.1038/266648a0. [DOI] [PubMed] [Google Scholar]
- 15.Goodman D S, Huang H S. Science. 1965;149:879–880. doi: 10.1126/science.149.3686.879. [DOI] [PubMed] [Google Scholar]
- 16.Ganguly J, Sastry P S. World Rev Nutr Diet. 1985;45:199–220. [PubMed] [Google Scholar]
- 17.Schwartz S H, Tan B C, Gage D A, Zeevaart J A D, McCarty D R. Science. 1997;276:1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- 18.Qin X, Zeevaart J A D. Proc Natl Acad Sci USA. 1999;96:15354–15361. doi: 10.1073/pnas.96.26.15354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Lintig J, Vogt K. J Biol Chem. 2000;275:11915–11920. doi: 10.1074/jbc.275.16.11915. [DOI] [PubMed] [Google Scholar]
- 20.Wyss A, Wirtz G, Woggon W, Brugger R, Wyss M, Friedlein A, Bachmann H, Hunziker W A. Biochem Biophys Res Commun. 2000;271:334–336. doi: 10.1006/bbrc.2000.2619. [DOI] [PubMed] [Google Scholar]
- 21.Tan B C, Schwartz S H, Zeevaart J A D, McCarty D R. Proc Natl Acad Sci USA. 1997;94:12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephenson R S, O'Tousa J, Scarvarda N J, Randall L L, Pak W L. Symp Soc Exp Biol. 1983;36:477–501. [PubMed] [Google Scholar]
- 23.Giovannucci D R, Stephenson R S. Vision Res. 1999;39:219–229. doi: 10.1016/s0042-6989(98)00184-9. [DOI] [PubMed] [Google Scholar]
- 24.Seki T, Isono K, Ozaki K, Tsukahara Y, Shibata-Katsuta Y, Ito M, Irie T, Katagiri M. Eur J Biochem. 1998;257:522–527. doi: 10.1046/j.1432-1327.1998.2570522.x. [DOI] [PubMed] [Google Scholar]
- 25.von Lintig J, Welsch R, Bonk M, Giuliano G, Batschauer A, Kleinig H. Plant J. 1997;12:625–634. doi: 10.1046/j.1365-313x.1997.00625.x. [DOI] [PubMed] [Google Scholar]
- 26.Stark W S, Johnson M A. J Comp Physiol. 1980;140:275–286. [Google Scholar]
- 27.Franceschini N, Kirschfeld K. Kybernetik. 1971;9:159–182. doi: 10.1007/BF02215177. [DOI] [PubMed] [Google Scholar]
- 28.Stavenga D G. In: Handbook of Sensory Physiology. Autrum H, editor. 7/6A. Heidelberg: Springer; 1979. pp. pp.358–439. [Google Scholar]
- 29.Ostroy S E, Wilson M, Pak W L. Biochem Biophys Res Commun. 1974;59:960–966. doi: 10.1016/s0006-291x(74)80073-2. [DOI] [PubMed] [Google Scholar]
- 30.Ozaki K, Nagatani H, Ozaki M, Tokunaga F. Neuron. 1993;10:1113–1119. doi: 10.1016/0896-6273(93)90059-z. [DOI] [PubMed] [Google Scholar]
- 31.Sun D, Chen D M, Harrelson A, Stark W S. Exp Eye Res. 1993;57:177–187. doi: 10.1006/exer.1993.1113. [DOI] [PubMed] [Google Scholar]
- 32.Shim K, Picking W L, Kutty R K, Thomas C F, Wiggert B N, Stark W S. Exp Eye Res. 1997;65:717–727. doi: 10.1006/exer.1997.0383. [DOI] [PubMed] [Google Scholar]
- 33.Ebnet E, Fischer M, Deininger W, Hegemann P. Plant Cell. 1999;11:1–14. doi: 10.1105/tpc.11.8.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deininger W, Fuhrmann M, Hegemann P. Trends Genet. 2000;16:158–159. doi: 10.1016/s0168-9525(99)01959-9. [DOI] [PubMed] [Google Scholar]