Abstract
The alphaherpesvirus envelope protein Us9 is a type II viral membrane protein that is required for anterograde spread of bovine herpesvirus 5 (BHV-5) infection from the olfactory receptor neurons to the brain. In a rabbit seizure model, Us9-deleted BHV-5 failed to invade the central nervous system (CNS) following intranasal infection. However, when injected directly into the olfactory bulb, retrograde-spread infection from the olfactory bulb (OB) to the piriform cortex and other areas connected to the OB was not affected. In contrast to BHV-5, wild-type BHV-1 failed to invade the CNS following intranasal infection. In this study, we show that mature BHV-1 Us9 is a 30- to 32-kDa protein, whereas mature BHV-5 Us9 is an 18- to 20-kDa protein. In vitro, BHV-1 Us9 is expressed at 3 h postinfection (hpi), whereas BHV-5 Us9 is expressed at 6 hpi. Despite these differences, BHV-1 Us9 not only complemented for BHV-5 Us9 and rescued the anterograde-spread defect of the BHV-5 Us9-deleted virus but conferred increased neurovirulence and neuroinvasiveness in our rabbit seizure model. Rabbits infected with BHV-5 expressing BHV-1 Us9 showed severe neurological signs at 5 days postinfection, which was 1 to 2 days earlier than BHV-5 wild-type or Us9-reverted BHV-5 virus. The data underscore the importance of both Us9 genes for virion anterograde transport and neuroinvasiveness. However, Us9 is not the determinant of the differential neuropathogenesis of BHV-1 and BHV-5.
Bovine herpesvirus 5 (BHV-5) is a neurovirulent alphaherpesvirus that causes fatal encephalitis in calves (4, 17). BHV-1 is associated with abortions, respiratory infections (subtype 1.1), and genital infections (subtype 1.2) in cattle (25) but does not frequently cause encephalitis. Both BHV-1 and BHV-5 establish latency in the trigeminal ganglion (TG), following intranasal and conjunctival inoculation (3, 22). The viruses share 85% DNA homology but differ in their ability to cause neurological disease in calves (4). In a rabbit seizure model, BHV-1.1 and BHV-5 infections are distinguished by their differential neuropathogenesis (11). For example, following intranasal inoculation, nasal swabs yield higher quantities of BHV-1 than of BHV-5 (20). However, only BHV-5 invades the central nervous system (CNS) via the olfactory pathway. When rabbits are inoculated intranasally, BHV-5 invades the brain via the olfactory pathway and produces acute neurological signs that are comparable to those seen in calves (20). The neural spread and neuronal damage are localized in the olfactory bulb and areas connected to the olfactory bulb (anterior olfactory nucleus, piriform-entorhinal cortex, frontal-cingulate cortex, hippocampus-dentate gyrus, amygdala, dorsal raphe, and locus coeruleus (20). Neurons expressing viral proteins are detected within TGs of the infected rabbits, but further invasion of the virus to second-order neurons in the trigeminal pathway of the pons and medulla does not occur (20). In BHV-1-inoculated rabbits, the virus does not invade the CNS, and neurological signs do not develop. However, like BHV-5-infected rabbits, BHV-1-infected rabbits have infected neurons in TGs (11, 20). When inoculated intracerebrally into the olfactory bulb, BHV-1 and BHV-5 replicate within the olfactory bulb neurons with similar efficiency, and the rabbits show severe neurological signs (S. I. Chowdhury, unpublished data). Envelope proteins glycoprotein E (gE) and Us9 are conserved in all members of the neurotropic alphaherpesviruses. gE is a type I transmembrane protein, whereas Us9 is a type II tail-anchored membrane protein. Both in BHV-5 and pseudorabies virus (PRV), it is clear that gE and Us9 mutants infect and replicate within primary presynaptic neurons but do not spread to secondary postsynaptic neurons or are defective in the anterograde spread to secondary postsynaptic neurons (7, 13, 19, 23, 24). Although the mechanism in each case is different, the evidence suggests that gE and Us9 are essential for the anterograde axonal transport of the virus and/or anterograde transsynaptic spread. Furthermore, deletion of either gE or Us9 has a profound effect on the neurovirulence properties of the virus.
Glycoprotein E is required for the efficient axonal localization of capsids, tegument, and certain viral glycoprotein (9). A prevailing hypothesis also predicts that gE is required at cell junctions and synaptic junctions to promote cell-to-cell and anterograde neuronal spread, respectively (14-16, 21).
Us9 is required for the entry and transport of envelope glycoproteins in axons (19, 23, 24). Infection of primary neuronal cultures with PRV Us9 mutants demonstrated that in the absence of Us9, the entry and transport of envelope glycoproteins in axons are affected (19, 23, 24). BHV-5 Us9-deleted virus infects the first-order olfactory receptor neuron (ORN) but is not transported to the bulb. Virus-specific antigens were detected within the cell bodies of ORNs of rabbits infected with Us9-deleted BHV-5, but they were not detected within the axons of the ORN or in the bulb (13). Like Us9-deleted BHV-5, BHV-1 failed to enter the bulb by anterograde transport. However, BHV-1 or BHV-5 can infect the ORN, and virus-specific antigen is present within the ORN cell body and axons (13; data not shown). Predicted amino acid sequence comparisons of BHV-1 and BHV-5 Us9 demonstrated that there are differences between the two Us9 genes; however, there is significant similarity in areas containing putative tyrosine kinase and casein kinase II phosphorylation motifs (13). Similar domains in PRV Us9 are important for anterograde transport of the virus within the neurons (8). In this study, our goal has been to identify and characterize BHV-1 Us9 (Us9-1) and test its ability to complement for BHV-5 Us9 protein with respect to neuropathogenesis. To this end, we constructed a BHV-1 Us9-deleted virus and determined that BHV-1 Us9 is a 32-kDa protein, which is absent in the Us9-deleted BHV-1. We have also constructed a BHV-5 recombinant virus expressing BHV-1 Us9 and determined that BHV-1 Us9 rescues the anterograde transport and neurovirulence defects of BHV-5 Us9-deleted virus.
MATERIALS AND METHODS
Virus strains and cell lines.
BHV-1 Cooper (Colorado-1) strain obtained from the American Type Culture Collection (Rockville, MD) and BHV-5 TX-89 strain (17) were used. The viruses were propagated and titrated in Madin-Darby bovine kidney (MDBK) cells as previously described (10).
Production of anti-BHV-1 Us9-specific polyclonal rabbit and goat serum.
To produce antibody against BHV-1 Us9, the entire BHV-1 Us9 open reading frame (ORF) was expressed in bacteria as a glutathione S-transferase (GST) fusion protein. For this, the entire BHV-1 Us9 ORF coding region was amplified by long PCR using a forward primer that introduced a BamHI site directly upstream of the start codon, allowing for an in-frame fusion with the GST open reading frame and a reverse primer that introduced an EcoRI site 116 bp downstream of the Us9 termination codon. The 598-bp BamHI-EcoRI PCR-generated fragment containing the complete BHV-1 Us9 ORF was ligated into pGEX-4T-2 (Amersham Biotech), generating the pGST-US91 expression vector. Expression of the GST-Us9 fusion protein was induced by the addition of IPTG (isopropyl-Β-d-thiogalactopyranoside) to a culture of BL21 cells transformed with the pGST-US91 plasmid. The fusion protein was then bound to glutathione-Sepharose beads, and the recombinant Us9 protein was cleaved away from the GST tag using thrombin protease following the manufacturer's protocol (Amersham Biotech). The rUs9 protein was isolated as a completely soluble form and used to immunize goats and/or rabbits (Cocalico Biologicals, Reamstown, PA).
Construction of a BHV-1Us9Δ EGFP vector.
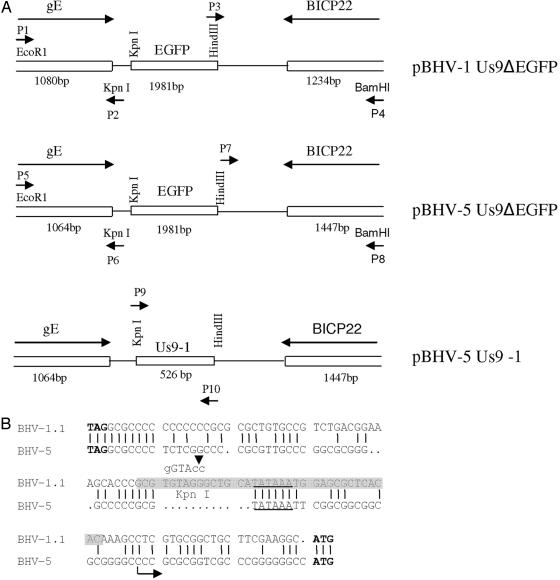
The BHV-1 Us9 gene is flanked by upstream gE and downstream BICP22 sequences. To delete the BHV-1 Us9 ORF coding region, Us9 upstream and downstream flanking sequences were amplified by long PCR using a XL-Long PCR kit (Applied Biosystems, Foster City, Calif.) as described previously (1, 2). These PCR-generated, 1,080-bp Us9-upstream and 1,234-bp Us9 downstream fragments incorporated EcoRI (P1)/KpnI (P2) and HindIII(P3)/BamHI(P4) at their respective 5′ and 3′ ends (Fig. 1A and Table 1). After the nucleotide sequences of both the inserts (Iowa State Core Facility) were verified, the sequences were assembled in correct order in plasmid pBHV-1ΔUS9 5′ 3′ (not shown). In pBHV-1ΔUS9 5′ 3′, the entire Us9 ORF is deleted, and the KpnI and HindIII sites are flanked by Us9 upstream and downstream sequences, respectively. To insert the enhanced green fluorescence protein (EGFP) gene cassette, a 1,981-bp fragment containing the cytomegalovirus immediate early promoter sequence, EGFP ORF sequence, and simian virus 40 poly(A) sequences were amplified using primer pairs P11-P12 (Table 1) and plasmid pEGFP/C1-collapse as described previously (1). The primer pairs P11-P12 incorporated KpnI and HindIII, respectively, which allowed the insertion of the KpnI/HindIII-digested EGFP gene-specific fragment (PCR generated) into the KpnI/HindIII sites of BHV-1 Us9 deletion plasmid pBHV-1ΔUS9 5′ 3′ constructed above. In the resulting plasmid, pBHV-1Us9ΔEGFP, the EGFP gene is flanked on the left by Us9 upstream sequences and on the right by Us9 downstream sequences required for homologous recombination (Fig. 1A).
FIG. 1.
Schematic illustration of the construction of recombinant plasmids. (A) Construction of the BHV-1 Us9 deletion plasmid (pBHV-1Us9ΔEGFP), the BHV-5 Us9 deletion plasmid (pBHV-5Us9ΔEGFP), and the Us9-exchanged BHV-5 plasmid (pBHV5Us9-1). (B) Alignment showing the BHV-1 and BHV-5 Us9 upstream nucleotide sequences containing the respective putative promoter region. The gE stop (TAG) and Us9 start (ATG) codons are in boldface type. The putative TATAAA boxes are underlined. The highlighted region in the BHV-1.1 sequence shows the forward primer sequence (P9) used to amplify the BHV-1 Us9 ORF (Table 1), and the sequence above indicates the KpnI sites incorporated within the forward P9 primer sequence. The arrow (at 27 bp upstream of the Us9 start codon) in the BHV-5 Us9 sequence marks the deletion site in the BHV-5 Us9-deleted virus.
TABLE 1.
List of primers used for PCR reactions
| Primer | Name | Sequence |
|---|---|---|
| BHV-1 Us9 deleted | ||
| Us9 upstream | Forward-P1 | 5′ GAGTTTTTCGACGAGAATTCCTTCTCGGCCAGCATCGAC 3′ |
| Reverse-P2 | 5′ GCCGCACGAGGTACCGTGTGAGCGCTCAATTTATATGCAG 3′ | |
| Us9 downstream | Forward-P3 | 5′ CCGCGAGTAAAGCTTGTCTAATTTTTTCCGCACGC 3′ |
| Reverse-P4 | 5′ CATCGACGTCAGGATCCTCTTCCGCTGCATCGCCA 3′ | |
| BHV-5 Us9 deleted | ||
| Us9 upstream | Forward-P5 | 5′ GGCTGCAGTCCGAATTCTTCGACGAAGCGC 3′ |
| Reverse-P6 | 5′ GCGGGGTACCGCGCCGCCGCCGAATTTATACG 3′ | |
| Us9 downstream | Forward-P7 | 5′ CGCGGGCGAAAGCTTCACGGAGGACTGCTTC 3′ |
| Reverse-P8 | 5′ GCGACGTGAGCGGATCCGACTCCAGCAGCGAG 3′ | |
| BHV-1 Us9 ORF | Forward-P9 | 5′ CGCGGGTACCGCTGCATATAAATGGAGCGCTCACAC 3′ |
| Reverse-P10 | 5′ GGCCTCAAGCTTGGGGGTCTCGGGGCCTCAGGGCCG 3′ | |
| GFPa | Forward-P11 | 5′ CGAACTGAGGTACCTACAGCGTGAGC 3′ |
| Reverse-P12 | 5′ CGCGTTAAGCTTCATTGATGAGTTTGG 3′ |
Amplified by using modified plasmid pEGFP-C1 (1).
Construction of pBHV-5 Us9 deletion vector.
Using similar strategies described above for the BHV-1 Us9 deletion vector, pBHV-5ΔUS9 5′ 3′ was constructed. For long PCR, primer pairs P5-P6 and P7-P8 (Fig. 1A and Table 1) amplified 1,064-bp BHV-5 Us9 upstream and 1,447-bp BHV-5 Us9 downstream sequences, respectively. In addition, they incorporated EcoRI/KpnI (P5-P6) and Hind III/BamHI (P7-P8) sites (Fig. 1A). In the resulting plasmid, the 503-bp BHV-5 Us9 gene sequence, 23 bp upstream of the Us9 start codon (Fig. 1B) and up to 75 bp of Us9 downstream (with respect to the stop codon) (13), was deleted. However, the Us9 upstream promoter sequence containing a putative TATAAA box (Fig. 1B) and the Us9 downstream sequence containing a putative poly(A) site, located 313 bp downstream of the Us9 stop codon (13), were preserved. The EGFP gene cassette in pBHV-1 Us9 ΔEGFP was released as a 1,981-bp fragment and inserted into the KpnI/HindIII sites of plasmid pBHV-5ΔUS9 5′ 3′, resulting in pBHV-5Us9Δ EGFP (Fig. 1A).
Construction of Us9 exchanged BHV-5 vector pBHV-5Us9-1 (containing the BHV-1 Us9 ORF).
A 526-bp fragment containing the BHV-1 Us9 promoter (Fig. 1B) and the Us9 ORF coding sequences were amplified by PCR using primer pairs P9-P10 (Table 1), which incorporated KpnI/HindIII restriction sites at 5′ and 3′ends of the amplified fragment, respectively (Fig. 1A; Table 1). The PCR-generated fragment was cloned into the KpnI/HindIII sites of pBHV-5ΔUS9 5′ 3′, resulting in pBHV-5Us9-1 (Fig. 1A). The plasmid pBHV-5Us9-1 contained the BHV-1 Us9 promoter (Fig. 1B) and the Us9 ORF sequences, which were flanked by the 1,064-bp BHV-5 Us9 promoter and the BHV-5 gE sequences upstream and by 1,447 bp of the BHV-5 BICP22 sequence downstream (Fig. 1A). Therefore, in pBHV-5 Us9-1, BHV-1 Us9 should be under the control of its own promoter (Fig. 1B). The insertion of the BHV-1 Us9 promoter and the Us9 ORF coding sequences in the pBHV-5Us9-1 plasmid were confirmed by sequencing (Iowa State University).
Generation of recombinant viruses. (i) BHV-1Us9Δ.
To generate BHV-1 Us9-deleted recombinant BHV-1Us9Δ, linearized pBHV-1Us9ΔGFP (Fig. 1A) and full-length wild-type BHV-1 DNA were cotransfected in MDBK cells using Lipofectamine (Gibco BRL, Life Technologies, Inc., Grand Island, N.Y.) as described previously (10). Recombinant viruses expressing GFP were plaque purified as described previously (1). The recombinant viruses were characterized further by immunoblotting and/or immunoprecipitation using anti BHV-1Us9-specific goat and/or rabbit polyclonal antibodies.
(ii) BHV-5 Us9ΔEGFP recombinant.
To generate BHV-5 Us9-deleted recombinant BHV-5 Us9ΔEGFP, linearized pBHV-5 Us9ΔEGFP (Fig. 1A) and full-length wild-type BHV-5 DNA were cotransfected; GFP-expressing recombinant viruses were plaque purified and further characterized by the methods described above.
(iii) BHV-5 Us9-1 recombinant.
To generate Us9-exchanged BHV-5 recombinant BHV-5 Us9-1 virus, linearized pBHV-5Us9-1 (Fig. 1A) and full-length BHV-5 Us9ΔEGFP DNA were cotransfected. Recombinant viruses with the BHV-1 Us9 incorporated and GFP deleted were identified as nonfluorescent plaques. These recombinant viruses were plaque purified and analyzed by Western blotting and/or immunoprecipitation for the expression of BHV-1 Us9.
Preparation of radiolabeled mock-and virus-infected cell lysates and immuno-precipitation.
For [35S]methionine-cysteine and [32P]phosphate labeling of virus-infected cell proteins, monolayers of MDBK cells were infected at a multiplicity of infection (MOI) of 5. The growth medium was replaced by serum-free methionine and cysteine-deficient medium at 1 h postinfection (hpi) (for 3-h samples) or at 4 hpi (for 6-h samples). After 2-h starvation with the methionine- and cysteine-deficient medium, the cells were pulse-labeled for 30 min with [35S]methionine-cysteine (50 μCi/ml; Amersham) and washed twice with serum-free medium after being labeled. The cells were either harvested immediately after being washed or incubated at 37°C in complete growth medium further for an additional 10 h and then harvested. After harvesting, the cell pellets were resuspended separately in an extraction buffer to make a 40% suspension (wt/vol) and processed for immunoprecipitation as described earlier (12). For phosphate labeling, the growth medium was replaced by serum-free phosphate-deficient medium at 1 hpi (BHV-1) or 4 hpi (BHV-5). [32P]orthophosphate (100 μCi/ml; Amersham) was added at 3 hpi (BHV-1) or 6 hpi (BHV-5). Cells were harvested after 16 h postinfection and processed for immunoprecipitation as described previously (12). Radioactively labeled infected-cell lysates were immunoprecipitated with Us9-specific goat and/or rabbit antibody developed above.
SDS-PAGE and Western blot analysis.
Cellular extracts were electrophoresed, unless otherwise mentioned in the figure legend, on a 4 to 20% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to a nitrocellulose membrane. Proteins were visualized by using rabbit or goat polyclonal antibodies as described previously (10).
PAA treatment.
To determine the kinetic class of BHV-1 Us9 in its parental BHV-1 virus and in BHV-5 Us9-1 virus, the sensitivity of their expression in the presence of phosphonoacetic acid (PAA) was tested as described previously (13). As a control, the expression of BHV-5 Us9, a known γ2 protein, was tested (13).
Animal experiments.
Four-week-old male New Zealand White rabbits (Myrtles Rabbitry, Thomson Station, TN) weighing 500 to 600 g each were used. All procedures were approved by the Kansas State University Animal Care and Use Committee.
Intranasal infection-immunohistochemical processing.
To compare the neuropathogenic properties of Us9-exchanged BHV-5 Us9-1 and BHV-5 Us9Δ viruses, a rabbit seizure model described previously (11, 20) was used. Five rabbits were infected with BHV-5 Us9Δ or wild-type BHV-5 viruses and 17 rabbits (10 rabbits in the first experiment and 7 rabbits in the second experiment) were infected with BHV-5 Us9-1 virus. In each case, rabbits were infected intranasally with 2 × 107 PFU/nostril of the respective virus into the paranasal sinuses as described previously (11, 20).
For the group infected with BHV-5 Us9-1 and the BHV-5 wild type, rabbits were euthanized when they showed severe neurological signs or at 12 days p.i. (dpi). For the group infected with BHV-5 Us9Δ, rabbits were euthanized at 12 dpi. Rabbits were euthanized by intramuscular injection of ketamine (45 mg per kg of body weight) and xylazine (10 mg/kg), followed by exsanguination. To determine the neural spread of the virus, the animals were euthanized and perfused transcardially, and the brain and TG were processed for immunohistochemistry as described previously (20). For the virus isolation, rabbits were euthanized, and each brain was dissected and processed for virus isolation as described previously (11).
Nasal swabs.
To determine virus replication in the naso-olfactory epithelium, the amount of virus shed in the nasal epithelium was quantitated by plaque assay at 3 and 5 dpi from five animals infected with the BHV-5 Us9Δ or BHV-5 Us9-1-exchanged virus, as described previously (12).
RESULTS
Identification and characterization of BHV-1 Us9.
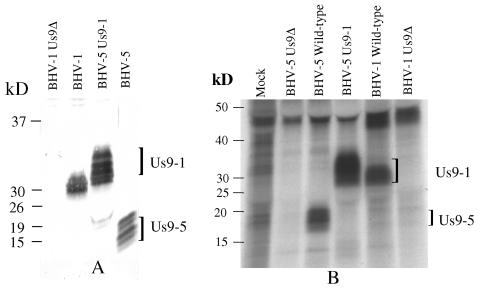
To identify BHV-1 Us9, the Us9 ORF coding region was expressed as a GST fusion protein. Bacterially expressed BHV-1 Us9 was purified as described in Materials and Methods and used for generating goat and rabbit antisera. As shown in Fig. 2A, the BHV-1 Us9-specific antibody precipitated proteins with approximate molecular masses of 28 to 32-kDa in BHV-1 and 15 to 20 kDa in BHV-5. The BHV-1 Us9-specific bands were absent in bovine cells productively infected with the BHV-1 Us9-deleted mutant (Fig. 2A). These results demonstrated that the antibody specifically recognized Us9 encoded by BHV-1 and BHV-5. BHV-1 and BHV-5 Us9 ORFs consisted of 144 and 134 amino acids (aa), respectively (13). The corresponding predicted molecular masses were 14.7 and 13.7 kDa, respectively (13); however, the respective BHV-1 and BHV-5 Us9-specific bands recognized by the Us9-specific antibody were of a higher molecular mass, suggesting that the respective Us9 proteins are processed posttranslationally to a higher-molecular-mass protein.
FIG. 2.
Identification of BHV-1 Us9 and characterization of BHV-1 Us9 expression by BHV-5Us9-1 (Us9-exchanged BHV-5). (A) Immunoprecipitation-immunoblotting analysis of BHV-1-, BHV-5 Us9-1-, and BHV-5-infected cell lysates were immunoprecipitated with goat anti-BHV-1 Us9 antibody. After SDS-PAGE of the immunoprecipitated samples, they were immunoblotted with rabbit anti BHV-1 Us9 antibody. (B) Autoradiograph showing phosphorylation of Us9 expressed by BHV-1-, BHV-5-, and BHV-5 Us9-1-infected MDBK cells. MDBK cells were infected with the viruses designated above for each lane and were labeled with 32P for 10 h starting at 3 hpi (BHV-1, Us9-deleted BHV-1, and BHV-5 Us9-1) or at 6 hpi (BHV-5 and BHV-5 Us9 deleted). The lysates were immunoprecipitated with goat anti-Us9 antibody as before.
Us9 posttranslational modifications.
Since the HSV-1 and PRV Us9 homologues are phosphorylated (5, 6), we tested whether this was true for BHV-1 and BHV-5 Us9. BHV-1- and BHV-5-infected cells were labeled overnight with [32P]orthophosphate beginning at 3 hpi and 6 hpi, respectively. The immunoprecipitation results shown in Fig. 2B demonstrate that the 30- to 32-kDa and 18- to 20-kDa BHV-1 and BHV-5 Us9-specific bands, respectively, were phosphorylated (Fig. 2B). However, the 28- to 30-kDa and the 13- to 15-kDa BHV-1 and BHV-5 Us9 bands detected immediately after pulse-labeling (Fig. 3A) were not phosphorylated (Fig. 2B). As a control for this experiment, no Us9-specific phosphorylated bands were detected in the Us9-deleted BHV-1-infected or Us9-deleted BHV-5-infected cell lysates.
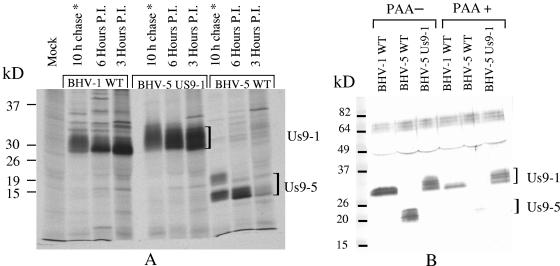
FIG. 3.
(A) Autoradiograph showing a time course of Us9 expression in BHV-1, BHV-5 Us9-1, and BHV-5. Virus-infected cell proteins were labeled with [35S]methionine-cysteine at 3 or 6 hpi. Cell monolayers were washed twice with serum-free Dulbecco's modified Eagle's medium, and detergent extracts were prepared. Radiolabeled lysates were immunoprecipitated with goat anti-Us9 antibody. After labeling and washing of cell monolayers as above, 10-h chase samples (*) were further incubated for 10 h at 37°C in complete growth medium without labeled cysteine and methionine. (B) Immunoblots showing BHV-1 and BHV-5 Us9 expression in untreated (PAA−) and PAA-treated (PAA+) BHV-1-, BHV-5-, and BHV-5 Us9-1-infected cell lystaes. MDBK cells were infected with the viruses indicated above. Infected cell lysates were immunoprecipitated with goat anti-Us9 antibody. Western blots containing immunoprecipitates were immunoblotted with rabbit anti-Us9 antibody.
To determine the kinetics of BHV-1 and BHV-5 Us9 processing, pulse-chase and immunoprecipitation experiments were performed as described previously (12). The results of pulse-chase analysis showed that with the exception of the earlier time course of BHV-1 Us9 expression (Fig. 3A), the kinetics of BHV-1 and BHV-5 Us9 processing was similar (data not shown).
Time course of BHV-1 Us9 expression.
During the course of this study, we noticed that BHV-1 Us9 is expressed at earlier times postinfection relative to the BHV-5-encoded Us9. To confirm this finding, BHV-1- and BHV-5-infected cells were pulse-labeled for 30 min with [35S]methionine-cysteine at 3 and 6 h postinfection, respectively. Radioactive Us9 was immunoprecipitated with the Us9-specific antiserum and analyzed by SDS-PAGE. The levels of radioactive BHV-1 Us9 were higher at 3 hpi than at 6 hpi. In the case of BHV-5, Us9 expression was barely detectable at 3 hpi and required 6 h to reach the comparable level of Us9 protein expressed by BHV-1 at 3 hpi (Fig. 3A). At 3 h postinfection, the predominant BHV-1 Us9 specific band was 28 to 30 kDa, whereas at 6 h postinfection, the predominant BHV-5 Us9-specific band was 13 to 15 kDa (Fig. 3A).
To determine the processing and stability of the protein, the pulse-labeled protein was chased overnight by incubation for an additional 10 h at 37°C. Immunoprecipitation results of 10-h chase samples showed that both the proteins were stable and processed to a diffused and slightly larger protein after an overnight chase (Fig. 3A). After a 10-h chase, BHV-1 Us9 was processed to a diffuse 30- to 32-kDa band, and the BHV-5 Us9 was processed to an 18- to 20-kDa band (Fig. 3A). These results indicate that relative to BHV-5 Us9, the BHV-1 Us9 protein was expressed at earlier times postinfection.
Previously, we reported that BHV-5 Us9 is a γ2 protein (13). To determine the kinetic class of BHV-1 Us9 (Us9-1), Us9-1 expression in the presence and absence of PAA was determined. As documented in Fig. 3B, BHV-5 Us9 expression was blocked in the presence of PAA (sensitive), while BHV-1 Us9 expression was reduced slightly but not blocked by PAA. Taken together, these results indicate that BHV-1 Us9 is an early (β)/late (γ1) protein, while BHV-5 Us9 is a γ2 protein.
Construction and characterization of BHV-5-expressing BHV-1 Us9 (BHV-5 Us9-1).
The Us9-exchanged BHV-5 (BHV-5 Us9-1) was characterized by immunoblotting and immunoprecipitation with BHV-1 Us9-specific sera raised in rabbits and/or goats. As shown in Fig. 2A and 3A, BHV-1 Us9 expressed by the BHV-5 Us9-1 virus had a higher molecular mass (30 to 34 kDa) which was slightly larger than the BHV-1 Us9 (30 to 32 kDa). Interestingly, the amount of Us9 expressed by BHV-5 Us9-1 virus was two- to fourfold larger than the amounts of Us9 expressed by the wild-type BHV-1 and BHV-5 viruses (Fig. 2A and 3A).
To determine the time course of BHV-1 Us9 expression in the BHV-5 Us9-1 virus, BHV-5 Us9-1 virus-infected cells were pulse-labeled for 30 min at 3 hpi and 6 hpi or chased for 10 h as above. Based on the relative levels of expression, the amount of Us9-1 protein expression in the context of the BHV-5 backbone was at least two- to fourfold larger than Us9 expression in its parental BHV-1 background (Fig. 3A). As noted in Materials and Methods and Fig. 1B, in BHV-5 Us9-1 virus, the BHV-1 Us9 promoter sequence and Us9-1 ORF coding sequences were placed immediately downstream of the BHV-5 Us9 upstream sequences. Therefore, Us9-1 expressed by BHV-5 should be under the control of the BHV-1 Us9 promoter. As expected, the time course of Us9-1 expression in the BHV-5 background was similar to that of BHV-1. However, the amount of Us9-1 expression was consistently two- to fourfold larger than that of Us9-1 and Us9-5 expressed by their authentic BHV-1 and BHV-5 viruses, respectively (Fig. 2A and 3A). Since the BHV-5 Us9-1 promoter sequence including the putative TATAAA box was not removed from the BHV-5 Us9-1 virus, having two TATAAA boxes might have caused the increased amount of Us9-1 expression in BHV-5 Us9-1 virus.
Results of pulse-chase analysis (data not shown) indicated that the kinetics of Us9-1 processing-maturation in the BHV-5 Us9-1 virus was similar to that of the parental BHV-1 (data not shown). Results of the PAA experiment showed that as with BHV-1, Us9-1 expressed by BHV-5 was not blocked by PAA (Fig. 3B). Results of the phosphorylation experiment showed that the processed Us9-1 bands in both BHV-5 (approximate molecular mass, 30 to 34 kDa) and BHV-1 (approximately molecular mass, 30 to 32 kDa) were phosphorylated; however, the phosphorylated form of Us9-1 expressed by the BHV-5 Us9-1 virus displayed higher intensity and migrated with a molecular mass ranging from 30 to 34 kDa instead of 30 to 32 kDa (Fig. 3B). This could be due to the increased amount of Us9-1 protein expressed in the BHV-5 Us9-1 virus-infected cells, increased phosphorylation of the protein in BHV-5 Us9-1, or both.
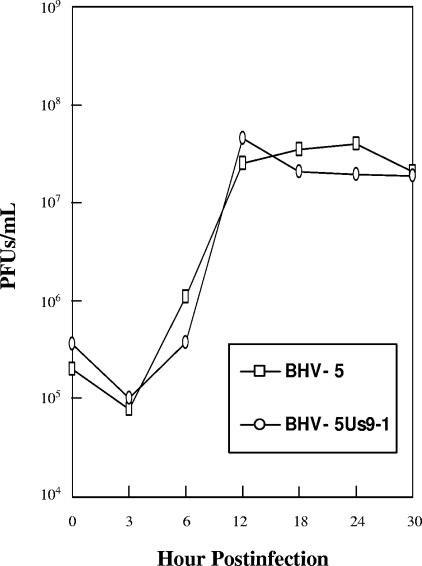
To determine the relative growth kinetics of BHV-5 and BHV-5 Us9-1 virus, an in vitro growth kinetics assay was performed. The results showed that BHV-5 wild type and BHV-5 Us9-1 grew with similar kinetics (Fig. 4).
FIG. 4.
One-step growth curve of BHV-5 and recombinant BHV-5 Us9-1 viruses in MDBK cells. Confluent MDBK cells were infected at an MOI of 5 PFU per cell with viruses. After 1 h of adsorption at 4°C, residual input viruses were removed. The cultures were washed three times with PBS, and 5 ml of medium was added into each flask before further incubation (37°C). At indicated time intervals, replicate cultures were frozen. Virus yields were determined by titration on MDBK cells. Each data point represents the average of duplicate samples obtained from separate infections.
Pathogenicity of BHV-5 Us9-1 virus after intranasal infection.
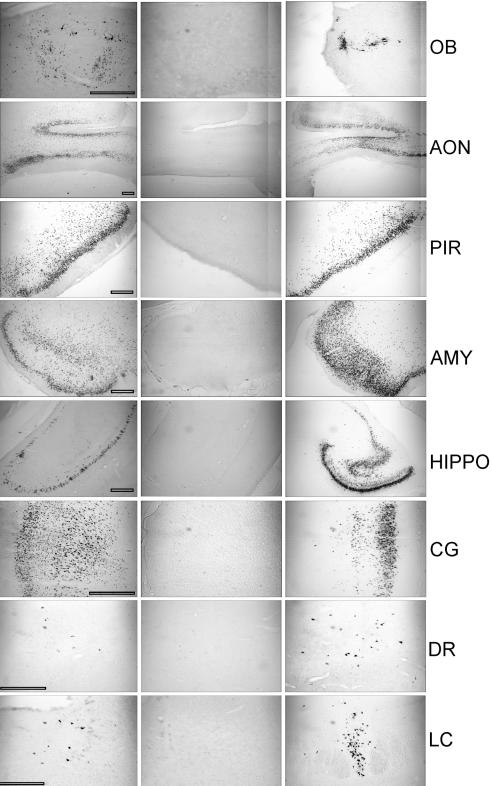
In the first experiment, 7 out of 10 rabbits infected with BHV-5 Us9-1 showed severe neurological signs between 5 to 11 days postinfection and were euthanized. Four animals were processed for immunohistochemistry, and three were processed for virus isolation. None of the five rabbits infected with BHV-5 Us9-deleted virus showed neurological signs; they were euthanized at 12 dpi. In the second experiment, five of seven rabbits infected with BHV-5 Us9-1 virus showed severe neurological signs between 5 to 10 days postinfection and were euthanized. Three were processed for virus isolation, and two were processed for immunohistochemistry. Significant amounts of virus were detected in the olfactory bulb (357 PFU/g), anterior cortex (4,336 PFU/g), and posterior cortex (2,342 PFU/g) of rabbits infected with BHV-5 US9-1 virus ( Table 2), which was comparable to levels detected in rabbits infected with wild-type BHV-5 (20). The spread of BHV-5 Us9-1 in the CNS was detected by immunohistochemistry and was compared to wild-type BHV-5 or Us9-deleted BHV-5-infected rabbits. The results are shown in Fig. 5A to C and summarized in Table 3. As noted above, BHV-5 Us9-1-infected rabbits showed severe neurological signs as early as 5 dpi. In the case of wild-type BHV-5, the earliest time point for the onset of severe neurological signs was like that seen in a previous study, at 7 dpi (20). Based on immunohistochemistry and virus isolation results, BHV-5 Us9-1 virus spread to all the areas of the brain in a fashion as similar to that of wild-type BHV-5 (Tables 2 and 3). Furthermore, the number of neurons infected by BHV-5 Us9-1 was similar or more than those detected in rabbits infected with the wild type (Table 3 and Fig. 6). These results indicate that the BHV-5 Us9-1 virus demonstrated increased neurovirulence and similar but faster anterograde neural spread properties relative to those of wild-type BHV-5.
TABLE 2.
Summary of clinical signs and virus isolation from the brain segments
| Virus | Animal no. | Neurological signsb | Virus isolation
|
|||
|---|---|---|---|---|---|---|
| PFU (range) | Scorec (no. of rabbits)
|
|||||
| Nose 3 dpi | Olfactory bulb | Anterior cortex | Posterior cortex | |||
| BHV-5 Us9-1a | 17 | Severe (12) | 500 (150-1,150) | ++ (6) | +++ (6) | +++ (6) |
| BHV-5 wild type | 5 | Severe (3) | 950 | ++ (2) | +++ (2) | +++ (2) |
| BHV-5 Us9Δ | 5 | None | 600 | − | − | − |
Data collected from two independent experiments.
Numbers in parentheses indicate number of animals showing clinical signs or brain segments positive for virus isolation.
−, no virus detected; ++, 100 to 990 PFU/g; +++, 1,000 to 10,000 PFU/g.
FIG. 5.
Localization of viral antigen in brain sections. Animals were inoculated intranasally with BHV-5 Us9-1, BHV-5 Us9Δ, and BHV-5 wild-type viruses as described in Materials and Methods. The animals were euthanized when they showed neurological signs for BHV-5 Us9-1 and BHV-5 wild-type viruses and on day 12 for BHV-5 Us9Δ virus. Representative sections of olfactory bulb (OB), anterior olfactory nucleus (AON), piriform cortex (PIR), amygdala (AMY), hippocampus (HIPPO), cingulate cortex (CG), dorsal raphe (DR), and locus coeruleus (LC) are shown. Bars, 500 μm.
TABLE 3.
Summary of viral spread in the brain after intranasal inoculation
| Virus | Presence and location of viral antigena
|
|||||||
|---|---|---|---|---|---|---|---|---|
| OB | AON | PIR | HIPPO | AMYG | CG | LC | DR | |
| BHV-5 Us9-1 | ++++ | ++++ | ++++b | +++ | ++++ | ++++b | + | + |
| BHV-5 WT | ++ | ++++ | ++++ | +++ | ++++ | +++ | + | + |
| BHV-5 Us9Δ | − | − | − | − | − | − | − | − |
Indicated as no labeling (−) or 1 to 25 (+), 30 to 150 (++), 160 to 500 (+++), 550 to 3,000 (++++) labeled neurons per field at a magnification 5×. OB, olfactory bulb; AON, anterior olfactory nucleus; PIR, piriform cortex; HIPPO, hippocampus; AMYG, amygdale; CG, cingulated cortex; LC, locus coeruleus; DR, dorsal raphe. Scores for the BHV-5 Us9-1 are average for six rabbits; for each brain segment, five sections (total, 60 sections) were analyzed. Scores for BHV-5 WT and Us9-deleted virus are average for three rabbits, and for each segment, five sections were counted (15 slides for each section).
Upper end of the scale.
FIG. 6.
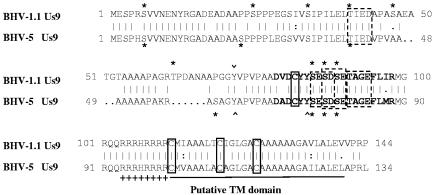
Comparison of the predicted amino acid sequence of the BHV-5 Us9 with the predicted amino acid sequence of BHV-1.1 (13; GenBank accession no. 298199). The sequences were aligned with the GCG Gap program. Conserved cysteine residues are boxed. The transmembrane anchor sequences are shown by a solid boldface line. The acidic domain is in boldface, and the cluster of positively charged amino acids preceding the putative transmembrane domain is indicated by a plus sign. Four caseine kinase II phosphorylation sites are indicated (smaller dashed rectangular boxes). Putative tyrosine kinase-mediated phosphorylation sites (Y) are indicated (∧ or ∨). In addition, serine and threonine residues with potential for phosphorylation are indicated by asterisks (NetPhos 2.0 server; http:/www.cbs.dtu.dk/services/NetPhos/). Note that BHV-1 Us9 has one additional serine and threonine residue each but lacks a tyrosine phosphorylation site.
DISCUSSION
We recently demonstrated that Us9-deleted BHV-5 fails to invade the CNS following intranasal infection of rabbits in a rabbit seizure model and that BHV-5 Us9 is essential for neuroinvasion and/or anterograde transport from the olfactory receptor neurons to the CNS (13). Using the same rabbit model, we demonstrated that BHV-1 does not invade the CNS after intranasal infection of rabbits (12). Comparison of the BHV-1 and BHV-5 predicted Us9 amino acid sequences showed 83% similarity and 77% identity (13). Alignment of BHV-1 and BHV-5 predicted Us9 amino acid sequences showed that acidic domains of the two Us9 proteins containing several potential phosphorylation sites (Fig. 6) (13) are highly conserved in the two proteins. Interestingly, a region in the predicted BHV-1 Us9 amino acid sequence between the residue at aa 44 to 67 differs significantly from the corresponding region of BHV-5 Us9 (13). In BHV-5 Us9, this region lacked 10 amino acid residues, resulting in a shorter Us9 protein for BHV-5 (Fig. 6) (13). As noted above, BHV-1 does not invade the CNS after intranasal infection. Therefore, an important question was whether the observed sequence differences between the two Us9 proteins were responsible for the differential neuropathogenesis of BHV-1 and BHV-5 observed with our rabbit seizure model. The results presented in this study demonstrate that processed BHV-1 Us9 is a 30- to 32-kDa protein, whereas processed BHV-5 Us9 is an 18- to 20-kDa protein. In vitro, BHV-1 Us9 is expressed at 3 hpi, whereas BHV-5 Us9 is expressed at 6 hpi. Despite these substantial differences, BHV-1 Us9 not only complemented BHV-5 Us9 and rescued the anterograde spread defect of the BHV-5 Us9-deleted virus but conferred increased neurovirulence and neuroinvasiveness in our rabbit seizure model. Rabbits infected with BHV-5 expressing BHV-1 Us9 showed severe neurological signs at 5 dpi, or 1 to 2 days earlier than for the BHV-5 wild-type or Us9-reverted BHV-5 viruses.
The predicted BHV-1 and BHV-5 Us9 ORFs contain 144 aa and 134 aa, respectively, and the corresponding molecular masses are 14.7 kDa and 13.7 kDa, respectively. However, Us9-specific antibody recognized 28- to 32-kDa and 15- to 20-kDa bands for BHV-1 and BHV-5, respectively. Therefore, BHV-1 Us9, predicted to be 1 kDa larger, acquired a relatively higher molecular mass (30 to 32 kDa) relative to BHV-5 (15 to 20 kDa) after processing. The predicted amino acid sequence alignment of BHV-1 and BHV-5 Us9 sequences revealed that there are several phosphorylation motifs conserved in both the sequences; however, in the BHV-1 Us9 sequence there are at least one to two additional potential phosphorylation sites (Fig. 6). Consistent with this analysis, we determined that Us9 proteins encoded by BHV-1 or BHV-5 were phosphoproteins. It is most likely that the additional 10 aa and the extra phosphorylation sites in the BHV-1 Us9 sequence have contributed to the increase in BHV-1 Us9 molecular mass, relative to its predicted molecular mass.
Us9 gene homologues are found in most of the alphaherpesviruses, including PRV and HSV-1 (5-7, 13). PRV and HSV-1 Us9 ORFs are predicted to encode proteins with an approximate molecular mass of 10.8 kDa and 10.0 kDa, respectively (5-6) However, Us9-specific polyclonal serum precipitated several proteins with molecular masses of from 17 to 20 kDa in PRV and between 12 to 25 kDa in HSV-1 (5-6). In both PRV and HSV-1, the Us9 is phosphorylated (5-6). In addition to phosphorylation, HSV-1 Us9 protein is ubiquitinated (5). In summary, it appears that the Us9 proteins in alphaherpesviruses are phosphorylated, suggesting that phosphorylation of Us9 has a functional role.
The results of time course experiments demonstrated that BHV-1 Us9 is expressed at 3 hpi, while BHV-5 Us9 is expressed at 6 hpi. Consistent with these results, BHV-5 Us9 expression is blocked by PAA and is dependent on ongoing DNA synthesis, whereas BHV-1 Us9 expression is slightly reduced but not blocked by PAA. Therefore, BHV-1 Us9 is a β (early)/γ1 (late) protein, while BHV-5 Us9 is a true late or γ2 protein and more resembles PRV Us9. Nevertheless, the two Us9 proteins are similar in their processing kinetics to larger phosphorylated forms.
To determine if BHV-1 Us9 could replace BHV-5 Us9 with respect to neurovirulence and neuroinvasiveness of BHV-5, we constructed a BHV-5 Us9-1 virus expressing BHV-1 Us9 under the control of the BHV-1 Us9 promoter. Our results demonstrated that the characteristics of Us9-1 expressed by BHV-5 Us9-1 virus including the molecular mass, expression, processing kinetics, and early time course (3 hpi) resembled those of Us9-1 in its parental virus, BHV-1. Interestingly, the amount of Us9-1 protein expressed by the BHV-5 Us9-1 virus has been two- to fourfold higher than that of BHV-1 and BHV-5 Us9. As noted earlier, this could be due to the increased amount of Us9-1 protein expressed in the BHV-5 Us9-1 virus-infected cells, increased phosphorylation of the protein in BHV-5 Us9-1, or both. Since both BHV-5 wild-type and BHV-5 Us9-1 viruses grew with similar kinetics and a similar amount of protein was applied in the immunoprecipitation-immunoblotting experiments, increased levels of Us9-1 expression in the BHV-5 Us9-1 cannot be due to differences in the replication kinetics or loading artifacts. As shown in Fig. 1B, most of the BHV-5 Us9 promoter sequence in the BHV-5 Us9-1 virus, including the TATAAA box, was intact, which preceded the BHV-1 Us9 promoter and the BHV-1 Us9 ORF coding sequences. We believe that the increased levels of Us9-1 expression in the BHV-5 Us9-1-infected cells could be a combined effect of the dual promoter sequences.
To determine if Us9-1 (BHV-1 Us9) could replace Us9-5 (BHV-5 Us9) with respect to anterograde transport, neurovirulence, and neuroinvasiveness, the neuropathogenic properties of the BHV-5 Us9-1 virus in rabbits were tested. In rabbits, BHV-5 Us9-1 virus was extremely neurovirulent, and the onset of severe neurological signs was consistently between 5 to 8 days (2 days early), whereas in BHV-5-infected rabbits, the onset of severe neurological signs was usually from 7 to 11 days. The early onset of neurological signs in BHV-5 Us9-1-infected rabbits might be due to faster anterograde transport. We believe that the early and increased levels of Us9-1 expression in this recombinant virus might have contributed to faster anterograde transport, resulting in the earlier (5 dpi versus 7 dpi in the BHV-5 wild type) onset of severe neurological signs. Within the CNS, BHV-5 Us9-1 virus spread to all areas of the olfactory pathway infected by wild-type BHV-5. Noticeably, the number of neurons infected by BHV-5 Us9-1 virus was higher than with wild-type BHV-5, which may also contribute to enhanced neuropathogenicity of the BHV-5 Us9-1 virus. It is possible that increased expression and phosphorylation of the protein contributed to the increased neurovirulence of the recombinant BHV-5 Us9-1 virus.
Several reports have proven that Us9 is vital for anterograde axonal transport (7, 8, 13, 19, 23, 24). In PRV, the acidic cluster was important for Us9-mediated anterograde spread in vivo, and several serine residues within the acidic cluster were major sites for Us9 phosphorylation (8). Notably, the rate of anterograde transneuronal viral spread was dependent on phosphorylation of two of the serine residues within the acidic domain (8). As shown in Fig. 6 and reported previously (13), tyrosine kinase, three serine residues, and casein kinase II phosphorylation motifs within the acidic domains are conserved in both BHV-1 and BHV-5 Us9 sequences. Consistent with these findings, BHV-1 Us9 complemented the anterograde neuronal transport and neuron invasiveness of BHV-5 in the BHV-5 Us9-1 virus. However, the BHV-5 Us9-1 virus demonstrated increased neurovirulence-neuroinvasiveness in our rabbit seizure model (11, 20). As discussed above, this could be a result of early and increased expression of Us9-1 in BHV-5 Us9-1 virus. Alternatively, the additional 10-aa sequences (between residues 44 and 67) in the BHV-1 Us9 (Fig. 6) might be important for the increased neurovirulence property of BHV-5 Us9-1. Specifically, in BHV-1 Us9, residue T62 within this region is predicted to be a potential threonine phosphorylation site which is absent in the corresponding BHV-5 Us9 sequence (Fig. 6). It remains to be tested whether these additional amino acid sequences are important for Us9-mediated neurovirulence.
Taken together, we suggest that Us9 has an universal role in alphaherpesvirus neuropathogenesis and/or anterograde neuronal transport and that the differences in the predicted Us9 amino acid sequences were not responsible for the observed differential neuropathogenesis of BHV-1 and BHV-5. Consistent with this suggestion, we observed that like BHV-5 (13), BHV-1 infects and replicates within the olfactory receptor neurons and that virus-specific antigens are detectable within the axons of neurons infected by BHV-1 (data not shown).
It is clear that Us9 is important for axonal localization of envelope glycoproteins but is not essential for axonal transport of tegumented capsids (19, 23, 24). This function is conserved in BHV-1 Us9, yet BHV-1 fails to invade the CNS (20). Therefore, there has to be a mechanism that, in addition to axonal localization of viral glycoproteins or tegumented capsids, requires the transport of the virus across the synapse which BHV-1 lacks. Anterograde neuronal transport of the virus also requires glycoprotein E (9, 18). Based on recently proposed models, it is possible that passage of the nonenveloped tegumented capsid requires gE at or near the synapse (14-16). Apparently, BHV-5 is able to cross the synapse leading to the bulb, while BHV-1 cannot. Therefore, it is possible that BHV-5 gE, which is also required for the anterograde axonal transport and CNS invasion of BHV-5, is the differential factor between the two viruses (12). Consistent with this assumption, we determined earlier that BHV-1 gE failed to complement for the BHV-5 gE with respect to neurovirulence.
Coincidentally, a BHV-1-expressing BHV-5 gE was not neuroinvasive; however, in that virus the Us9 promoter sequence was inadvertently deleted (12). Since in the current study, BHV-1 Us9 restored both the anterograde neuronal transport and neurovirulence of the BHV-5 US9-null virus, it is highly likely that the reported failure of BHV-1-expressing BHV-5 gE to invade the CNS could be due to the lack of Us9. Conversely, the question whether BHV-5 gE constitutes a neurovirulence determinant could be directly addressed by replacing the BHV-1 gE by the BHV-5 gE. These experiments are currently in progress.
Acknowledgments
We thank Samir ElZarkouny for technical assistance in cloning the BHV-1 Us9 sequence in pGEX-4T-2.
This work was supported by USDA grants 00-02103 and 2004-35204-14657 to S.I.C.
Footnotes
Contribution 06-82-J, Kansas Agricultural Experiment Station.
REFERENCES
- 1.Al-Mubarak, A., and S. I. Chowdhury. 2004. In the absence of glycoprotein I (gI), gE determines bovine herpesvirus type 5 neuroinvasiveness and neurovirulence. J. Neurovirol. 10:233-243. [DOI] [PubMed] [Google Scholar]
- 2.Al-Mubarak, A., Y. Zhou, and S. I. Chowdhury. 2004. A glycine-rich bovine herpesvirus 5 (BHV-5) gE-specific epitope within the ectodomain is important for BHV-5 neurovirulence. J. Virol. 78:4806-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashbaugh, S. E., K. E. Thompson, E. B. Belknap, P. C. Schultheiss, S. I. Chowdhury, and J. K. Collins. 1997. Specific detection of shedding and latency of bovine herpesvirus 1 and 5 using a nested polymerase chain reaction. J. Vet. Diagn. Investig. 9:387-394. [DOI] [PubMed] [Google Scholar]
- 4.Belknap, E. B., J. K. Collins, V. K. Ayers, and P. C. Schulteiss. 1994. Experimental infection of neonatal calves with neurovirulent bovine herpes virus type 1.3. Vet. Pathol. 31:358-365. [DOI] [PubMed] [Google Scholar]
- 5.Brandimarti, R., and B. Roizman. 1997. Us9, a stable lysine-less herpes simplex virus 1 protein, is ubiquitinated before packaging into virions and associates with proteasomes. Proc. Natl. Acad. Sci. USA 94:13973-13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brideau, A. D., B. W. Banfield, and L. W. Enquist. 1998. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J. Virol. 72:4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brideau, A. D., J. P. Card, and L. W. Enquist. 2000. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J. Virol. 74:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brideau, A. D., G. Eldridge, and L. W. Enquist. 2000. Directional transneuronal infection by pseudorabiesvirus is dependent on an acidic internalization motif in the Us9 cytoplasmic tail. J. Virol. 74:4549-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ch'ng, T. H., and L. W. Enquist. 2005. Efficient axonal localization of alphaherpesvirus structural proteins in cultured sympathetic neurons requires viral glycoprotein E. J. Virol. 79:8835-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury, S. I., C. S. D. Ross, B. J. Lee, V. Hall, and H.-J. Chu. 1999. Construction and characterization of a glycoprotein E gene-deleted bovine herpesvirus type 1 recombinant. Am. J. Vet. Res. 60:227-232. [PubMed] [Google Scholar]
- 11.Chowdhury, S. I., B. J. Lee, D. Mosier, J.-H. Sur, F. A. Osorio, G. Kennedy, and M. L. Weiss. 1997. Neuropathology of bovine herpesvirus type 5 (BHV-5) meningo-encephalitis in a rabbit seizure model. J. Comp. Pathol. 117:295-310. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhury, S. I., B. J. Lee, A. Ozkul, and M. L. Weiss. 2000. Bovine herpesvirus 5 glycoprotein E is important for the neuroinvasiveness and neurovirulence in the olfactory pathway of the rabbit. J. Virol. 74:2094-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury, S. I., M. Onderci, P. S. Bhattacharjee, A. Al-Mubarak, M. L. Weiss, and Y. Zhou. 2002. Bobine herpesvirus 5 (BHV-5) Us9 is essential for BHV-5 neuropathogenesis. J. Virol. 76:3839-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingwell, K. S., and D. C. Johnson. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 72:8933-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dingwell, K. S., C. R. Brunetti, R. L. Hendricks, Q. Tang, M. Tang, A. J. Rainbow, and D. C. Johnson. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dingwell, K. S., L. C. Doering, and D. C. Johnson. 1995. Glycoprotein E and I facilitate neuron-to-neuron spread of herpes simplex virus. J. Virol. 69:7087-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Offay, J. M., R. E. Mock, and R. W. Fulton. 1993. Isolation and characterization of encephalitic bovine herpesvirus type 1 isolates from cattle in North America. Am. J. Vet. Res. 54:534-539. [PubMed] [Google Scholar]
- 18.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1999. Spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51:237-347. [DOI] [PubMed] [Google Scholar]
- 19.Enquist, L. W. 2002. Exploiting circuit-specific spread of pseudorabies virus in the central nervous system: insights to pathogenesis and circuit tracers. J. Infect. Dis. 186:209-214. [DOI] [PubMed] [Google Scholar]
- 20.Lee, B. J., M. L. Weiss, D. Mosier, and S. I. Chowdhury. 1999. Spread of bovine herpesvirus type5 (BHV-5) in the rabbit brain after intranasal inoculation. J. Neurovirol. 5:473-483. [DOI] [PubMed] [Google Scholar]
- 21.Mettenleiter, T. C. 2003. Pathogenesis of neurotropic herpesviruses: role of viral glycoprotein in neuroinvasion and transneuronal spread. Virus Res. 92:197-206. [DOI] [PubMed] [Google Scholar]
- 22.Rock, D. L., W. A. Hagemoser, F. A. Osorio, and D. E. Reed. 1986. Detection of bovine herpesvirus type 1 RNA in trigeminal ganglia of latently infected rabbits by in situ hybridization. J. Gen. Virol. 67:2515-2520. [DOI] [PubMed] [Google Scholar]
- 23.Tomishima, M. J., and L. W. Enquist. 2001. A conserved α-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomishima, M. J., G. A. Smith, and L. W. Enquist. 2001. Sorting and transport of alpha herpesviruses in axons. Traffic 2:429-436. [DOI] [PubMed] [Google Scholar]
- 25.Wyler, R., M. Engels, and M. Schwyzer. 1989. Infectious bovine rhinotracheitis/vulvovaginitis (BHV-1), p. 1-72. In G. Wittman (ed.), Herpesvirus diseases of cattle, horses and pigs. Kluwer Academic Publishers, Hingham, Mass.