Abstract
Recently, a production system for infectious particles of hepatitis C virus (HCV) utilizing the genotype 2a JFH1 strain has been developed. This strain has a high capacity for replication in the cells. Cyclosporine (CsA) has a suppressive effect on HCV replication. In this report, we characterize the anti-HCV effect of CsA. We observe that the presence of viral structural proteins does not influence the anti-HCV activity of CsA. Among HCV strains, the replication of genotype 1b replicons was strongly suppressed by treatment with CsA. In contrast, JFH1 replication was less sensitive to CsA and its analog, NIM811. Replication of JFH1 did not require the cellular replication cofactor, cyclophilin B (CyPB). CyPB stimulated the RNA binding activity of NS5B in the genotype 1b replicon but not the genotype 2a JFH1 strain. These findings provide an insight into the mechanisms of diversity governing virus-cell interactions and in the sensitivity of these strains to antiviral agents.
Hepatitis C virus (HCV), a member of the Flaviviridae family, has a positive-strand RNA genome (1, 26). The genome encodes a large precursor polyprotein, which is cleaved by host and viral proteases to generate at least 10 functional viral proteins: core, envelope 1 (E1), E2, p7, nonstructural protein 2 (NS2), NS3, NS4A, NS4B, NS5A, and NS5B (6, 8). NS5B is an RNA-dependent RNA polymerase that is crucial for viral genome replication (1, 26). There is genetic heterogeneity within the HCV genome. Currently, these differences are classified into six genotypes that are further segregated into a series of subtypes (4, 23). In Japan, genotype 1b is predominant; roughly 65% of cases of HCV-related chronic hepatitis involve genotype 1b. By comparison, genotype 2a is present in 17% of these patients (13, 23).
Sustained infection of HCV is the major cause of chronic liver diseases such as chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (16). Rarely, HCV causes fulminant hepatitis (13). The predominant treatment for HCV-infected patients is interferon (IFN) or polyethylene glycol-conjugated IFN alone or in combination with ribavirin (19, 20). However, alternative anti-HCV therapies are needed because virus is not eliminated in about half of the treated patients (19, 20). Lohmann et al. have developed the HCV subgenomic replicon system, in which an HCV subgenomic replicon autonomously replicates in Huh-7 cells (HCV replicon cells) (18). This replicon comprises the HCV 5′ untranslated region (5′UTR) containing an internal ribosomal entry site (IRES), the neomycin phosphotransferase gene, the encephalomyocarditis virus (EMCV) IRES, the coding region for HCV NS3 through NS5B, and the HCV 3′UTR (subgenomic replicon), but it lacks the coding region for the core and envelope proteins, as well as p7 and NS2 (Fig. 1). Subsequently, a genome-length (full-genome) replicon has been developed. This construct contains a full-genome length of HCV, including the coding regions for the core protein through NS2 (Fig. 1) (5, 10). We can evaluate HCV replication using these subgenomic or genome-length replicon systems. Previously, we established HCV subgenomic replicon cells carrying HCV genotype 1b NN strain (15, 29). We demonstrated that an immunosuppressant, cyclosporine (CsA), has anti-HCV activity in these cells (29). In addition, we determined the molecular mechanism of the anti-HCV effect of CsA on this replicon; cyclophilin B (CyPB), one of the cellular targets of CsA, is a cellular replication cofactor of the HCV genome (31). CyPB interacts with NS5B to promote its RNA binding activity (for a detailed description, see reference 31). CsA is suggested to suppress HCV genome replication by inhibiting the functional association of CyPB with NS5B. Another group also reported anti-HCV function of CsA using a subgenomic replicon of other genotype 1b strain, HCV-N (22). In this study, we demonstrate that CsA also has a strong anti-HCV activity in other available genotype 1b replicons carrying the Con1 and O strains (12, 18).
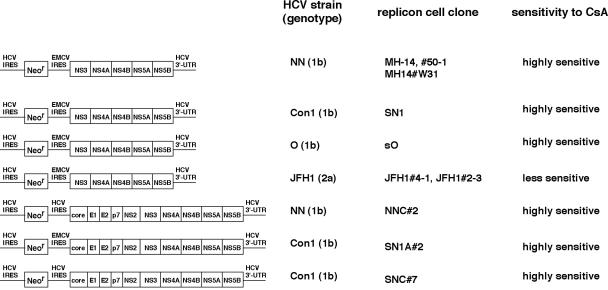
FIG. 1.
Schematic representation of the constructs of HCV subgenomic and genome-length replicon RNA. On the left, the constructs of each replicon RNA are shown. HCV strains, as well as genotypes from which the replicon RNA sequences are derived, are indicated in the second column. The names of replicon cell clones established with each replicon RNA are in the third column. The sensitivity to CsA of each replicon RNA revealed in this study is summarized in the fourth column. The replicon RNAs comprise the HCV 5′UTR, including HCV IRES, the neomycin phosphotransferase gene (Neor), EMCV IRES, or HCV IRES, the coding region for HCV proteins NS3 to NS5B (subgenomic) or core to NS5B (genome length or full genome), and HCV 3′UTR. MH-14 (NN/1b/SG), #50-1 (NN/1b/SG), MH14#W31 (NN/1b/SG), SN1 (Con1/1b/SG), sO (O/1b/SG), JFH1#4-1 (JFH1/2a/SG), and JFH1#2-3 (JFH1/2a/SG) cells carry subgenomic replicons, while NNC#2 (NN/1b/FL), SN1A#2 (Con1/1b/FL), and SNC#7 (Con1/1b/FL) cells have genome-length replicons. NNC#2 (NN/1b/FL) and SNC#7 (Con1/1b/FL) cells contain the replicon RNA without EMCV IRES.
Recently, Wakita and colleagues reported that a replicon of HCV genotype 2a JFH-1 strain, which was isolated from a case of type-C fulminant hepatitis, has a much stronger level of replication activity than genotype 1b replicons in Huh-7 cells (13, 27). A production system of infectious viral particles was recently established with this high-replication-competent strain (17, 27, 34). This viral strain may acquire a growth advantage compared with many other strains, although the underlying mechanism is unknown. In this study, we described a characteristic difference in the replication of JFH1 compared to that of genotype 1b replicons.
Here, we report that JFH1 replication is less sensitive to CsA than genotype 1b strains, although the interaction of CyPB with NS5B is observed with this replicon. However, genome replication and RNA binding activity of NS5B are independent of CyPB. We have exploited a chemical compound to demonstrate how strain diversity can be generated by underlying differences in the mechanisms of the virus-cell interaction. These findings provide important insight into the mechanisms that mediate the efficacy of antiviral agents.
MATERIALS AND METHODS
Cell culture.
Huh-7 cells were cultured in Dulbecco's modified Eagle medium (Invitrogen) with 10% fetal bovine serum, nonessential amino acids (Invitrogen), and l-glutamine (Invitrogen). MH-14, #50-1, MH14#W31, SN1, sO (formerly named 1B2R1), JFH1#4-1, and JFH1#2-3 cells (12, 13, 15, 18, 29), carrying subgenomic replicons, and NNC#2, SN1A#2, and SNC#7 cells, carrying full-genome replicons, were cultured in the above medium supplemented with 300- to 500-μg/ml G418 (Invitrogen). In the assay measuring the response to CsA, NIM811, or PSC833 (Fig. 2, 3, and 4), we seeded small numbers of each replicon cells (7 × 103 to 15 × 103 cells/12-well plate) and treated with each drug. Culture medium was changed every 3 days (CsA, NIM811, or PSC833 was supplemented in the fresh medium for the treatment groups). We did not perform any passages in the assay period. At day 7, the cells were 70 to 90% confluent. A schematic representation of the constructs of HCV replicon RNAs, the name of HCV strains from which the replicon RNA sequences are derived, and the name of replicon cell clones used in this study are summarized in Fig. 1. Since many replicon clones were used in this study, we list “strain/genotype/length of the replicon construct” in parentheses after the names of each cell clone in Results and in the figure legends to avoid confusion between names: for example, MH-14 (NN/1b/SG), JFH1#4-1 (JFH1/2a/SG), and SN1A#2 (Con1/1b/FL) cells. The designations SG and FL indicate subgenomic and full-genome replicons, respectively.
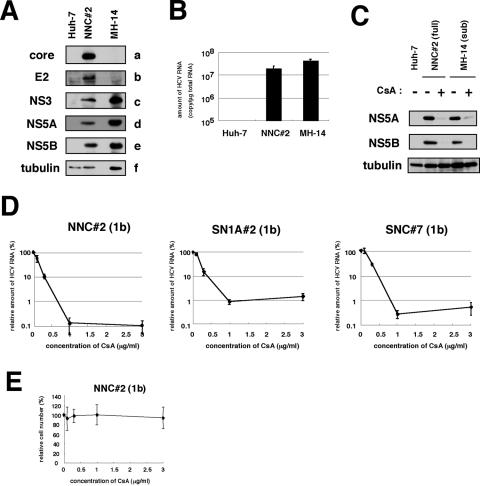
FIG. 2.
CsA suppressed the replication of HCV genome, irrespective of the presence of the structural proteins. (A) Detection of HCV proteins from NNC#2 (NN/1b/FL) genome-length replicon. Core (a), E2 (b), NS3 (c), NS5A (d), NS5B (e), and tubulin (f) in Huh-7, NNC#2 (NN/1b/FL), and MH-14 (NN/1b/SG) cells analyzed by immunoblot analysis are shown. (B) HCV RNA in Huh-7, NNC#2 (NN/1b/FL), and MH-14 (NN/1b/SG) cells quantified by real-time RT-PCR analysis. The data represent the means of three independent experiments. (C) CsA decreased the production of HCV proteins in NNC#2 (NN/1b/FL), as well as in MH-14 (NN/1b/SG) cells. After treatment with 1-μg/ml CsA (+) for 5 days or without treatment (−), total-cell lysates of NNC#2 (NN/1b/FL) and MH-14 (NN/1b/SG) cells, together with Huh-7 cells as a negative control, were recovered to examine the production of HCV NS5A (top), NS5B (middle), and tubulin as an internal control (bottom) by immunoblot analysis. The same result was obtained at day 7 after treatment. (D) The sensitivity to CsA of HCV genome-length replicon was almost the same as that of the subgenomic replicon. HCV RNA was quantified by real-time RT-PCR analysis using total RNA from NNC#2 (NN/1b/FL), SN1A#2 (Con1/1b/FL), and SNC#7 (Con1/1b/FL) cells treated with various concentrations of CsA for 7 days. The relative amount of HCV RNA was plotted against the concentration of CsA (in micrograms per milliliter). (E) Effect of CsA on cell proliferation. NNC#2 (NN/1b/FL) cells were treated with various amount of CsA for 7 days. Cell numbers were counted, and cell numbers relative to those of cells without treatment were plotted against the concentration of CsA.
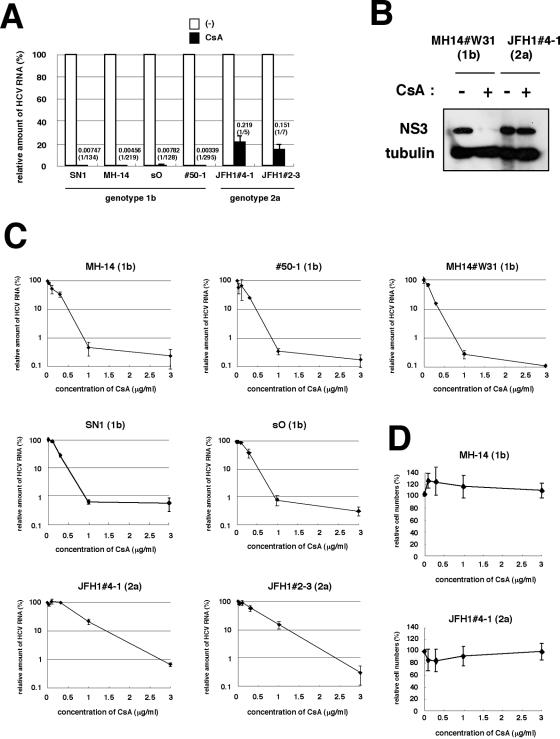
FIG. 3.
Replication of a genotype 2a strain, JFH1, was less sensitive to CsA. (A) Sensitivity to CsA of HCV genotype 1b and JFH1 replicons. SN1 (Con1/1b/SG), MH-14 (NN/1b/SG), sO (O/1b/SG), #50-1 (NN/1b/SG), JFH1#4-1 (JFH1/2a/SG), and JFH1#2-3 (JFH1/2a/SG) cells, carrying HCV subgenomic replicon, were treated with 1-μg/ml CsA for 7 days. HCV RNA titers were quantified by real-time RT-PCR analysis, and the relative amounts are shown. The bars represent the means of three independent experiments. White bars, no treatment; black bars, 1-μg/ml CsA. The numbers above the black bars indicate fold difference of the titer with 1-μg/ml CsA treatment compared to no treatment. (B) Levels of NS3 and tubulin as an internal control in MH14#W31 (NN/1b/SG) and JFH1#4-1 (JFH1/2a/SG) cells without (−) or with (+) 1-μg/ml CsA treatment for 5 days were detected by immunoblot analysis. (C) HCV RNA was quantified and plotted as described in the legend to Fig. 2D with genotype 1b replicon cells such as MH-14 (NN/1b/SG), #50-1 (NN/1b/SG), MH14#W31 (NN/1b/SG), SN1 (Con1/1b/SG), and sO (O/1b/SG) cells and JFH1-carrying replicon cells such as JFH1#4-1 (JFH1/2a/SG) and JFH1#2-3 (JFH1/2a/SG) cells. (D) Effect of CsA on cell proliferation. The growth of MH-14 (NN/1b/SG) and JFH1#4-1 (JFH1/2a/SG) cells were examined as described in the legend for Fig. 2E.
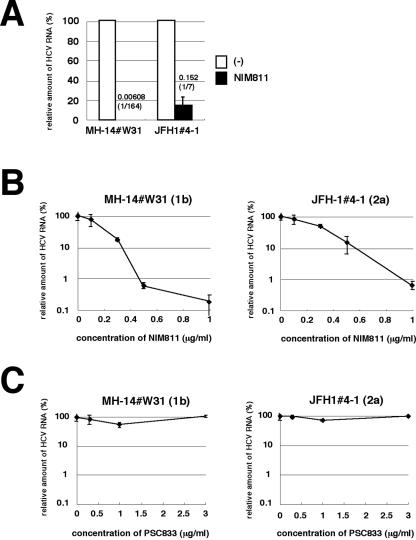
FIG. 4.
JFH1 replication was less sensitive to a CsA derivative, NIM811. (A) MH14#W31 (NN/1b/SG) and JFH1#4-1 (JFH1/2a/SG) cells were treated with 0.5-μg/ml NIM811 for 7 days. HCV RNA titers were quantified as described in the legend to Fig. 3A. White bars, no treatment; black bars, 0.5-μg/ml NIM811. (B and C) HCV RNA in replicon cells treated with various concentrations of NIM811 (B) or PSC833 (C) for 7 days was quantified and plotted against the concentration of NIM811 (B) or PSC833 (C) (in micrograms per milliliter) as described in the legend to Fig. 3C.
Establishment of replicon cells.
MH-14, #50-1, sO, JFH1#4-1, and JFH1#2-3 cells were described previously (12, 13, 15, 29). The replicon RNAs were produced using a MEGAscript T7 kit (Ambion) from pMH14, pSN1, pNNC, pSN1A, and pSNC plasmids for the establishment of the MH14#W31, SN1, NNC#2, SN1A#2, and SNC#7 replicon cells, respectively. For the establishment of MH14#W31, we transfected RNA into the Huh-7 cell strain which was identical to the parental cells of JFH1#4-1 and JFH1#2-3. Each replicon RNA was transfected into Huh-7 cells, following the selection with the medium in the presence of 500- to 1,000-μg/ml G418 for around 4 weeks. The resultant cell colonies were isolated and expanded. The HCV RNA titers in cell clones carrying JFH1 replicons were not significantly different from those in established cell clones carrying genotype 1b replicons.
Plasmid construction.
pSN1, the sequence of which is derived from I377NS3-3′ (18), was prepared essentially as described previously (15). pSN1A was generated by inserting the region from the core to NS2 of pM1LE (15) into the upstream coding region for NS3 in pSN1. To obtain pSNC, the EMCV IRES of pSN1A was replaced by the HCV IRES. pNNC was produced by inserting the coding region from NS3 to NS5B of pM1LE into pSNC.
Real-time reverse transcription-PCR (RT-PCR) analysis.
The 5′UTR of HCV genome RNA was quantified using the ABI PRISM 7700 sequence detector (Applied Biosystems) as described previously (29).
Immunoblot analysis.
Immunoblot analysis was performed as described previously (30). The primary antibodies used in this study were anti-core, anti-E2 (kindly provided by M. Kohara, Tokyo Metropolitan Institute of Medical Science), anti-NS3, anti-NS5A (a generous gift from A. Takamizawa, Osaka University), anti-NS5B (NS5B-6; kindly provided by I. Fukuya, Osaka University), anti-CyPA (Upstate Cell Signaling), anti-CyPB (Affinity BioReagents), and anti-tubulin (Oncogene).
Immunoprecipitation assay and RNA-protein binding precipitation assay.
Immunoprecipitation and RNA-protein binding precipitation were performed as described previously (30, 31).
RNA interference technique.
The condition of small interfering RNA (siRNA) used in this study was described previously (31). Transfection was performed using siLentFect (Bio-Rad), according to the manufacturer's protocol.
Isolation of replication complex.
The HCV replication complex was isolated from cells by treatment with 50-μg/ml digitonin at 27°C for 5 min, following treatment with 0.3-μg/ml proteinase K at 37°C for 5 min as described previously (31).
Purification of recombinant GST-fused CyPB protein.
Glutathione S-transferase (GST) and GST-fused CyPB (GST-CyPB) protein expression was induced in transformed BL21 cells (Amersham) with 1 mM isopropyl-β-thiogalactopyranoside (IPTG). The cell lysate was incubated with glutathione-Sepharose resin (Amersham) and washed extensively. The recombinant protein was eluted by glutathione (pH 8.0) and subsequently dialyzed.
In vitro RNA binding assay.
In vitro-translated 35S-labeled NS5B proteins and poly(U)-Sepharose (Amersham) or protein G-Sepharose (Amersham) resin as a negative control were incubated in the presence of recombinant GST-CyPB protein at 4°C for 1 h. After being washed, precipitates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by imaging analyzer.
RESULTS
CsA suppressed the replication of HCV full-genome replicon.
We and another group have reported an anti-HCV activity of CsA using subgenomic replicons (22, 29). HCV structural proteins, especially the core protein, have multiple functions. These proteins interact with many cellular factors and modulate a variety of cellular functions (32). Potentially, these viral proteins could diminish or circumvent the suppression of HCV genome replication by CsA. Core protein and E2 reportedly modulate the activity of IFN signaling (9, 25). To test this possibility, we established a full-genome HCV replicon system with cells transfected with the NN strain (NNC#2 cells [NN/1b/FL]) (Fig. 1). HCV RNA and protein productions were confirmed by real-time RT-PCR and immunoblot analysis (Fig. 2A and B). In addition, we confirmed that this replication was not due to the integration of the replicon construct into the cellular genome (data not shown). Similarly, we generated other full-genome replicon cells carrying sequences from the Con1 strain at the nonstructural coding region of the replicon RNA (SN1A#2 [Con1/1b/FL]) and SNC#7 (Con1/1b/FL) cells (Fig. 1). The replicon of SN1A#2 (Con1/1b/FL) cells possessed the EMCV IRES upstream of the open reading frame for HCV proteins, while that of SNC#7 (Con1/1b/FL) cells contained the HCV IRES (Fig. 1). SNC#7 (Con1/1b/FL) cells exhibited almost the same response as that of SN1A#2 (Con1/1b/FL) cells to CsA treatment (Fig. 2D). Consistent with a previous report (22), the EMCV IRES was not responsible for the anti-HCV activity of CsA. We compared the sensitivity to CsA of full-genome replicons with that of subgenomic replicons. CsA strongly decreased the production of HCV proteins in both the full-genome replicon, NNC#2 (NN/1b/FL) cells and the subgenomic replicon, MH-14 (NN/1b/SG) cells (Fig. 2C). Real-time RT-PCR analysis also revealed a dramatic reduction of the RNA level of full-genome replicons in NNC#2 (NN/1b/FL), SN1A#2 (Con1/1b/FL), and SNC#7 (Con1/1b/FL) cells (Fig. 2D). The 50% inhibitory concentrations (IC50) of CsA in NNC#2 (NN/1b/FL), SN1A#2 (Con1/1b/FL), and SNC#7 (Con1/1b/FL) cells awere estimated to be 0.13, 0.19, and 0.24 μg/ml, respectively. The 90% inhibitory concentrations (IC90) of CsA in these cells were 0.68, 0.94, and 0.81 μg/ml, respectively. The CsA dose-response curves of full-genome replicons and subgenomic replicons were similar (i.e., compare SN1A#2 or SNC#7 [Con1/1b/FL] versus SN1 [Con1/1b/SG], NNC#2 [NN/1b/FL] versus MH-14, #50-1, or MH14#W31 [NN/1b/SG]) (Fig. 3C). These results demonstrate that CsA suppresses the replication of full-genome replicons and subgenomic replicons to almost the same extent. Since CsA concentrations of up to 3 μg/ml did not affect the proliferation of any replicon cells (Fig. 2E and data not shown), the effect of CsA on replication is not due to the cytotoxic effect. In addition, we observed the reduction of production of infectious viral particles in the presence of 3-μg/ml CsA (data not shown) using the viral production system with full-genome JFH1 RNA (27).
The JFH1 replicon was less sensitive to CsA than were genotype 1b replicons.
We compared the sensitivity of HCV replication to CsA in several subgenomic replicon cells. We used MH-14 (NN/1b/SG) and #50-1 (NN/1b/SG) cells carrying subgenomic replicons with HCV NN strain (15, 29), SN1 (Con1/1b/SG) cells carrying the Con1 subgenomic replicon (18), and sO (O/1b/SG) cells bearing the subgenomic O strain (12) as genotype 1b replicon-containing cells. We also employed JFH1#4-1 (JFH1/2a/SG) and JFH1#2-3 (JFH1/2a/SG) cell clones carrying the JFH1 subgenomic replicon (13). Treatment of CsA (1 μg/ml; 7 days) drastically decreased HCV RNA in all the subgenomic replicon cells carrying the HCV genotype 1b strain. HCV RNA levels in SN1 (Con1/1b/SG), MH-14 (NN/1b/SG), sO (O/1b/SG), and #50-1 (NN/1b/SG) cells decreased to 1/134, 1/219, 1/128, and 1/295, respectively (Fig. 3A). Genotype 1b replicon cells appeared highly sensitive to CsA. In contrast, the effect of CsA on HCV RNA levels in replicon cells containing sequences from the JFH1 strain was limited to 1/5 to 1/7 (Fig. 3A). These results of the response to CsA were reproduced in further additional cell clones.
The cellular characteristics of Huh-7 cell strains differ among laboratories. To exclude the possibility that differences between Huh-7 cell strains influence the sensitivity to CsA, we established genotype 1b replicon cells based on the identical Huh-7 cell strain, which were used as parental cells of JFH1#4-1 (JFH1/2a/SG) and JFH1#2-3 (JFH1/2a/SG) cells. The response of the corresponding replicon cells, MH14#W31 (NN/1b/SG), to CsA was almost the same as that of SN1 (Con1/1b/SG), MH-14 (NN/1b/SG), sO (O/1b/SG), and #50-1 (NN/1b/SG) cells (Fig. 3C). Thus, the difference in sensitivity of JFH1 and genotype 1b strains to CsA can be attributed to the characteristic differences of the HCV strains, not to the parental Huh-7 cell strain. In addition, the reduction of NS3 protein in JFH1#4-1 (JFH1/2a/SG) cells following treatment with CsA was less prominent than that in MH14#W31 (NN/1b/SG) cells (Fig. 3B).
We examined the dose-response curve of HCV RNA against the concentration of CsA (Fig. 3C). The effect of CsA in genotype 1b replicons plateaued at around 1 μg/ml, while in the dose-response curve in JFH1 replicon, the inhibition was not yet saturated (Fig. 3C). As concentrations of CsA up to 3 μg/ml did not affect the proliferation rate of any replicon cells (Fig. 3D and data not shown), the effect of CsA on replication was not due to the cytotoxic effect. The IC50 of CsA in MH-14 (NN/1b/SG), #50-1 (NN/1b/SG), MH14#W31 (NN/1b/SG), SN1 (Con1/1b/SG), sO (O/1b/SG), JFH1#4-1 (JFH1/2a/SG), and JFH1#2-3 (JFH1/2a/SG) cells were estimated to be 0.15, 0.18, 0.16, 0.20, 0.25, 0.67, and 0.43 μg/ml, respectively. The IC90 was 0.86, 0.82, 0.76, 0.88, 0.92, 2.77, and 2.39 μg/ml, respectively. A similar dose-response curve in the JFH1 replicon was obtained by a transient replication assay with the luciferase reporter driven from a JFH1 replicon construct (data not shown) (14).
JFH1 replicon was less sensitive to a CsA derivative, NIM811.
Analysis of several CsA derivatives has revealed that the anti-HCV effect of CsA on the genotype 1b replicon is mediated by the inhibition of CyP (31). We examined the sensitivity of JFH1 replicon to CsA derivatives. CsA is known to have three major cellular targets: CyP, calcineurin (CN)/NF-AT, and P glycoprotein (P-gp) (28, 31). A CsA derivative, NIM811, inhibits CyP and P-gp but not CN/NF-AT, while another derivative, PSC833, inhibits P-gp but neither CyP nor CN/NF-AT (31). The decrease of HCV RNA in MH14#W31 (NN/1b/SG) cells with NIM811 treatment (0.5 μg/ml; 7 days) was more than an order of magnitude greater than that in JFH1#4-1 (JFH1/2a/SG) cells (Fig. 4A). The slope of the dose-response curve of NIM811 treatment of the JFH1 replicon was gentler than that of genotype 1b (Fig. 4B). The IC50 of NIM811 in MH14W#31 (NN/1b/SG) and JFH1#4-1 (JFH1/2a/SG) cells were 0.17 and 0.30 μg/ml, respectively. The IC90 were 0.46 and 0.93 μg/ml, respectively. In contrast, PSC833, which does not inhibit CyP, did not alter HCV RNA level in either genotype 1b or the JFH1 replicon (Fig. 4C). Thus, a CyP inhibitor was less effective at suppressing the replication of the JFH1 replicon than genotype 1b replicons.
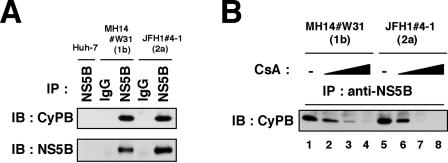
Interactions between CyPB and JFH1 NS5B.
Previously, we have shown that CyPB interacts with NS5B to promote HCV genome replication and that CsA inhibits this binding in a genotype 1b replicon (31). Here, we examined the association between CyPB and NS5B in a JFH1 replicon. Immunoprecipitation analysis revealed an interaction of CyPB with NS5B in JFH1#4-1 (JFH1/2a/SG) cells (Fig. 5A). This interaction was dissociated following the treatment of CsA, as observed with the genotype 1b replicon (Fig. 5B).
FIG. 5.
Interaction of HCV NS5B with CyPB in the JFH1 replicon. (A) Coimmunoprecipitation of endogenous CyPB with NS5B. Lysates from MH14#W31 (NN/1b/SG), JFH1#4-1 (JFH1/2a/SG), and Huh-7 cells as a negative control were used for immunoprecipitation with normal mouse immunoglobulin G (IgG) or anti-NS5B antibody (NS5B), followed by immunoblot analysis with either anti-CyPB (top) or anti-NS5B antibodies (bottom). IP, antibodies used for immunoprecipitation. (B) The interaction of CyPB with NS5B in JFH1 replicon was disrupted by CsA treatment. Coimmunoprecipitation between CyPB and NS5B was analyzed with MH14#W31 (NN/1b/SG) or JFH1#4-1 (JFH1/2a/SG) cells treated without CsA (lanes 1 and 5) or with CsA (0.3 μg/ml in lanes 2 and 6, 1 μg/ml in lanes 3 and 7, and 3 μg/ml in lanes 4 and 8).
Role of CyPB in replication of the JFH1 replicon.
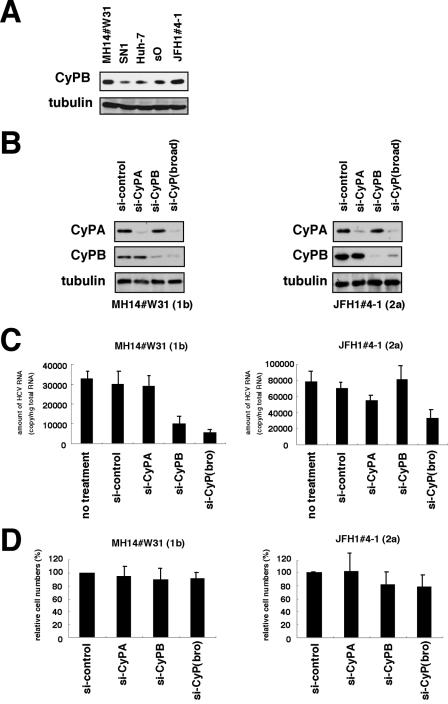
Although we observed some differences of expression levels of endogenous CyPB among the replicon cells in the immunoblot analysis (Fig. 6A), there was no particular correlation between endogenous CyPB expression levels and replication sensitivity to CsA among cells. CyPB reportedly regulates HCV genome replication of the genotype 1b replicon (31). We then explored the requirement of CyPB for the replication of JFH1 replicon with RNA interference. Transfecting siRNAs designed to recognize several CyP subtypes [si-CyP(broad)] (Fig. 6B) reduced HCV RNA to <1/5 in MH14#W31 (NN/1b/SG) cells (Fig. 6C). Specific knockdown of CyPB but not CyPA (Fig. 6B) decreased HCV RNA in MH14#W31 (NN/1b/SG) cells, consistent with a previous report (Fig. 6C) (31). In contrast, HCV RNA in JFH1#4-1 (JFH1/2a/SG) cells was not altered following the suppression of either endogenous CyPA or CyPB (Fig. 6B and C). We observed a weak decrease of HCV RNA levels (around one-half) with si-CyP(broad) (Fig. 6C). These data suggests the possibility that the replication of the JFH1 replicon is independent of CyPB, in contrast to the genotype 1b replicon. In the previous study, it was reported that the doubling time, saturation density, and response to cell confluence of the replicon cells carrying JFH1 were different from those in cells carrying a genotype 1b replicon, suggesting the possibility that the coupling relationship between the replication and cell growth was different between genotype 1b and the JFH1 replicon (21). The introduction of either si-CyPB or si-CyP(broad), however, had little effect on cell growth in MH14#W31 (NN/1b/SG) or JFH1#4-1 (JFH1/2a/SG) cells (Fig. 6D). And we did not observe cells being confluent in the experiment period. The above results suggest that the different response to si-CyPB in the two lines is independent of the conditions of cell growth.
FIG. 6.
CyPB in HCV replication of genotype 1b and JFH1. (A) Expression level of endogenous CyPB protein (top) and tubulin as an internal control (bottom) in MH14#W31 (NN/1b/SG), SN1 (Con1/1b/SG), sO (O/1b/SG), JFH1#4-1 (JFH1/2a/SG), and Huh-7 cells. (B) Knockdown of endogenous CyP proteins. MH14#W31 (NN/1b/SG) and JFH1#4-1 (JFH1/2a/SG) cells were transfected with siRNA specific for CyPA (si-CyPA), CyPB (si-CyP), a broad range of CyP subtypes [si-CyP(broad)], or a randomized siRNA (si-control). At 72 h posttransfection, CyPA (top), CyPB (middle) and tubulin as an internal control (bottom) were detected in total cell lysates of MH14#W31 (NN/1b/SG) (left) and JFH1#4-1 (JFH1/2a/SG) (right) cells by immunoblot analysis. (C) Depletion of CyPB did not affect HCV replication of JFH1 replicon. At 5 days posttransfection, HCV RNA titers in MH14#W31 (NN/1b/SG) (left) and JFH1#4-1 (JFH1/2a/SG) (right) cells were quantified by real-time RT-PCR analysis. no treatment, treatment with only the transfection reagent in the absence of siRNA. (D) Effect of siRNA on cell proliferation. Cell numbers of MH14W#31 (NN/1b/SG) and JFH1#4-1 (JFH1/2a/SG) cells treated with siRNA for 5 days were counted. Relative cell numbers were indicated.
The role of CyPB in the RNA binding activity of JFH1 NS5B.
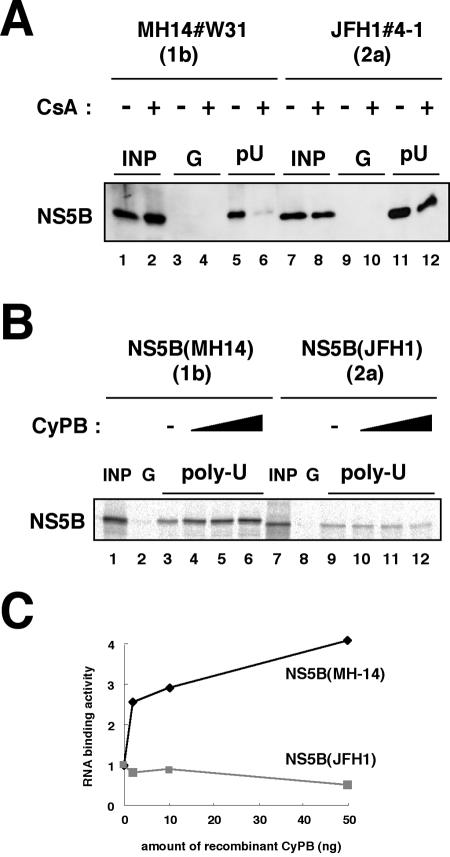
CyPB regulates HCV genome replication of a genotype 1b replicon by promoting the RNA binding activity of NS5B (31). We examined the effect of CyPB on the RNA binding activity of NS5B in JFH1. NS5B in the replication complex was isolated from cells by treatment with digitonin-proteinase K, as described previously (31). This fraction was incubated with poly(U) RNA-Sepharose or protein G-Sepharose as a negative control for the detection of RNA binding NS5B in the replication complex. RNA-bound NS5B in this fraction from MH14#W31 (NN/1b/SG) cells was decreased drastically following treatment with CsA (Fig. 7A, lanes 5 and 6). However, the reduction of RNA binding of NS5B in the replication complex of JFH1#4-1 (JFH1/2a/SG) cells was not as prominent (Fig. 7A, lanes 11 and 12). We confirmed this result by an in vitro RNA binding assay, in which in vitro-synthesized NS5B was incubated with poly(U) RNA-Sepharose, together with recombinant GST-CyPB. The addition of recombinant GST-CyPB increased the binding of genotype 1b NS5B to poly(U) RNA (Fig. 7B and C). However, this augmentation of RNA binding was not observed with NS5B from the JFH1 strain (Fig. 7B and C). From the above results, it is suggested that the RNA binding of JFH1 NS5B is free from regulation by CyPB.
FIG. 7.
RNA binding capacity of JFH1 NS5B was independent of CyPB. (A) An RNA-protein binding precipitation assay was performed using MH14#W31 (NN/1b/SG) cells (lanes 1 to 6) and JFH1#4-1 (JFH1/2a/SG) cells (lanes 7 to 12) as described in Materials and Methods. MH14#W31 (NN/1b/SG) and JFH1#4-1 (JFH1/2a/SG) cells preincubated without (lanes 1, 3, 5, 7, 9, and 11) or with (lanes 2, 4, 6, 8, 10, and 12) CsA were treated with digitonin, followed by digestion with proteinase K to isolate the replication complex. This fraction was then incubated with poly(U) RNA-Sepharose (lanes 5, 6, 11, and 12) or protein G-Sepharose as a negative control (lanes 3, 4, 9, and 10). Precipitates were detected by immunoblot analysis with anti-NS5B antibody. INP, one-sixth of the amount of cell lysate used in the precipitation assay; G and pU, samples with protein G-Sepharose and poly(U)-Sepharose, respectively. (B) An in vitro RNA binding assay was performed as described in Materials and Methods. In vitro-synthesized NS5B of MH-14 (lanes 1 to 6) or JFH1 (lanes 7 to 12) with the rabbit reticulocyte lysate in the presence of [35S]methionine was incubated with protein G-Sepharose (lanes 2 and 8) or poly(U)-Sepharose in the absence (lanes 3 and 9) or presence of various amounts of purified recombinant GST-CyPB (2 ng in panels 4 and 10, 10 ng in panels 5 and 11, and 50 ng in panels 6 and 12). The resultant precipitates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by the detection of radiolabeled protein. (C) The density of the bands of NS5B in the RNA binding fraction was quantified and plotted against the amount of the recombinant GST-CyPB (in nanograms). Solid line, NS5B of MH-14; faint line, NS5B of JFH1.
DISCUSSION
Until now, we and another group have utilized subgenomic replicons carrying genotype 1b NN and HCV-N strains to demonstrate that CsA suppresses HCV genome replication (22, 29). This study reveals that CsA is effective on full-genome replicons to almost the same extent. In addition, other available genotype 1b replicons carrying the Con1 and O strains also have a high sensitivity to CsA, consistent with our proposal that HCV genotype 1b is highly sensitive to CsA. However, a fulminant-type genotype 2a replicon, JFH1, was less responsive to CsA, although a high dose of CsA suppressed the replication of this strain.
CyPB interacts with genotype 1b NS5B to stimulate its RNA binding activity. In contrast, CyPB binds JFH1 NS5B but does not regulate the function of JFH1 NS5B. This is consistent with a previous speculation that genotype 1b and JFH1 replicons utilize the same cellular factors in distinct manners (21). The NS5B sequence of NN strain has 95.0, 95.9, and 70.4% homology to that of Con1, O, and JFH1, respectively (Fig. 8). The region spanning amino acids (aa) 521 to 591 of NS5B, which is involved in the interaction with CyPB (31), is highly conserved among genotype 1b strains NN, Con1, and O while that of JFH1 has 21 substituted residues in this region. The proline at 540 aa, which is important for CyPB binding (31), is conserved but the adjacent residues such as isoleucine at 539 aa and alanine at 541 aa are replaced by leucine and glutamic acid, respectively, in JFH1. Through molecular interactions, CyPB seems to make the conformation of NS5B of genotype 1b strains but not JFH1 suitable for RNA binding (31). The diverse regulation system of NS5B by CyPB among strains may be due to differences in either the sequence or the entire conformation of NS5B. Further study is important for elucidating the regulation mechanism of RNA binding activity of NS5B by CyPB.
FIG. 8.
Amino acid sequence alignment of NS5B encoded by HCV strains NN, Con1, O, and JFH1. The numbers above the sequence indicate the amino acid numbers. Conserved residues are shown by dashes. The region spanning 521 to 591 aa, which is involved in the interaction with CyPB, is boxed.
Thus, replication in JFH1 replicon is independent of CyPB. Interestingly, human immunodeficiency virus type 1 (HIV-1) strains also have a diversity of CyP dependence on viral proliferation (3, 33). CyPA plays an important role in the life cycle of HIV-1. The interaction of the HIV-1 capsid protein with CyPA that resides within the target cells of infection is critical for HIV-1 replication (7, 24). In peripheral blood mononuclear cells or Jurkat T cells, CsA suppresses the proliferation of HIV-1 group main (M) strain (3). However, certain strains of group outlier (O), such as MVP5180 and MVP9435, are resistant to CsA (3, 33), suggesting the different dependency of the replication on CyPA. Authors have suggested that MVP5180 and MVP9435 clones adapt to replicate independently of CyPA and that this adaptation provides a significant replication advantage for the virus in vivo (3). In vesicular stomatitis virus (VSV) strains, a role for CyPA in virus replication also has been reported (2). CyPA is required for the infection of the VSV-NJ strain but not the VSV-IND strain. These authors proposed that during evolutionary divergence from the ancestral lineages that initially were dependent on CyPA for replication, VSV-IND may have adapted to reduce its dependency on CyPA (2). In the case of HCV, a fulminant type genotype 2a replicon (JFH1) replicates independently of CyPB. It has previously been reported that JFH1 has a much higher competency of replication in the cells than other strains (13). The adaptation to independence from CyPB may contribute to the high capacity of replication of JFH1.
Although the JFH1 replicon is less sensitive to CsA, high concentrations of CsA still suppress replication of the JFH1 replicon. Moreover, the introduction of the siRNA designed to recognize several CyP subtypes [si-CyP(broad)] moderately diminishes HCV RNA in the JFH1 replicon. We suspect that a CyP family member other than CyPB is involved in HCV genome replication. Further analysis is needed on the role of other CyP subtypes.
As there a replicon system for a fulminant-type genotype 1b replicon or chronic-type genotype 2a replicon does not yet exist, we cannot conclude whether chronic-type genotype 2a replicons or fulminant-type replicons are less sensitive to CsA or not. However, there is a clinical report describing cotreatment of patients with chronic hepatitis C with IFN and CsA that resulted in a higher sustained virological rate than with treatment of IFN alone (11). In this report, increase in the sustained virological rate was prominent with patients carrying genotype 1 HCV (51.7% versus 21.9%), while it was relatively weak in patients carrying genotype 2 HCV (66.7% versus 58.3%) (11). Thus, genotype may affect the sensitivity of HCV replication to CsA. However, we cannot exclude the possibility that the diminished sensitivity to CsA is a characteristic only of the fulminant-type genotype 2a strain.
Our results suggest that sensitivity to CsA and replication dependency to CyPB is different among HCV strains. This finding is an important insight into the diversity of the mechanism of HCV genome replication and its sensitivity to antiviral agents.
Acknowledgments
We thank H. Takahashi and M. Hosaka for preparing replicon cells and generating plasmids. We are grateful to A. Takamizawa at Osaka University, I. Fukuya at Osaka University, and M. Kohara at Tokyo Metropolitan Institute of Medical Science for antibodies and R. Bartenschlager at Heiderberg University for the I377/NS3-3′ sequence. We also appreciate Novartis (Basel, Switzerland) for providing the CsA derivatives NIM811 and PSC833.
This work was supported by grants-in-aid for cancer research and for the second-term comprehensive 10-year strategy for cancer control from the Ministry of Health, Labor, and Welfare; grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology; and grants-in-aid from the Research for the Future from the Japanese Society for the Promotion of Science, the Program for Promotion of Fundamental Studies in Health Science of the Organization for Pharmaceutical Safety and Research of Japan, and Research on Health Sciences focusing on Drug Innovation from the Japan Health Sciences Foundation.
REFERENCES
- 1.Bartenschlager, R., and V. Lohmann. 2001. Novel cell culture systems for the hepatitis C virus. Antiviral Res. 52:1-17. [DOI] [PubMed] [Google Scholar]
- 2.Bose, S., M. Mathur, P. Bates, N. Joshi, and A. K. Banerjee. 2003. Requirement for cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype. J. Gen. Virol. 84:1687-1699. [DOI] [PubMed] [Google Scholar]
- 3.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIVCPZGAB but not group O HIV-1 or other primate immunodeficiency viruses. J. Virol. 70:4220-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukh, J., R. H. Purcell, and R. H. Miller. 1994. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc. Natl. Acad. Sci. USA 91:8239-8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frese, M., V. Schwarzle, K. Barth, N. Krieger, V. Lohmann, S. Mihm, O. Haller, and R. Bartenschlager. 2002. Interferon-gamma inhibits replication of subgenomic and genomic hepatitis C virus RNAs. Hepatology 35:694-703. [DOI] [PubMed] [Google Scholar]
- 6.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatziioannou, T., D. Perez-Caballero, S. Cowan, and P. D. Bieniasz. 2005. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 79:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hijikata, M., H. Mizushima, T. Akagi, S. Mori, N. Kakiuchi, N. Kato, T. Tanaka, K. Kimura, and K. Shimotohno. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosui, A., K. Ohkawa, H. Ishida, A. Sato, F. Nakanishi, K. Ueda, T. Takehara, A. Kasahara, Y. Sasaki, M. Hori, and N. Hayashi. 2003. Hepatitis C virus core protein differently regulates the JAK-STAT signaling pathway under interleukin-6 and interferon-gamma stimuli. J. Biol. Chem. 278:28562-28571. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue, K., K. Sekiyama, M. Yamada, T. Watanabe, H. Yasuda, and M. Yoshiba. 2003. Combined interferon α2b and cyclosporin A in the treatment of chronic hepatitis C: controlled trial. J. Gastroenterol. 38:567-572. [DOI] [PubMed] [Google Scholar]
- 12.Kato, N., K. Sugiyama, K. Namba, H. Dansako, T. Nakamura, M. Takami, K. Naka, A. Nozaki, and K. Shimotohno. 2003. Establishment of a hepatitis C virus subgenomic replicon derived from human hepatocytes infected in vitro. Biochem. Biophys. Res. Commun. 306:756-766. [DOI] [PubMed] [Google Scholar]
- 13.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 14.Kato, T., T. Date, M. Miyamoto, M. Sugiyama, Y. Tanaka, E. Orito, T. Ohno, K. Sugihara, I. Hasegawa, K. Fujiwara, K. Ito, A. Ozasa, M. Mizokami, and T. Wakita. 2005. Detection of anti-hepatitis C virus effects of interferon and ribavirin by a sensitive replicon system. J. Clin. Microbiol. 43:5679-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kishine, H., K. Sugiyama, M. Hijikata, N. Kato, H. Takahashi, T. Noshi, Y. Nio, M. Hosaka, Y. Miyanari, and K. Shimotohno. 2002. Subgenomic replicon derived from a cell line infected with the hepatitis C virus. Biochem. Biophys. Res. Commun. 293:993-999. [DOI] [PubMed] [Google Scholar]
- 16.Liang, T. J., and T. Heller. 2004. Pathogenesis of hepatitis C-associated hepatocellular carcinoma. Gastroenterology 127:S62-S71. [DOI] [PubMed] [Google Scholar]
- 17.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 18.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 19.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 20.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto, M., T. Kato, T. Date, M. Mizokami, and T. Wakita. 2006. Comparison between subgenomic replicons of hepatitis C virus genotypes 2a (JFH-1) and 1b (Con1 NK5.1). Intervirology 49:37-43. [DOI] [PubMed] [Google Scholar]
- 22.Nakagawa, M., N. Sakamoto, N. Enomoto, Y. Tanabe, N. Kanazawa, T. Koyama, M. Kurosaki, S. Maekawa, T. Yamashiro, C. H. Chen, Y. Itsui, S. Kakinuma, and M. Watanabe. 2004. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem. Biophys. Res. Commun. 313:42-47. [DOI] [PubMed] [Google Scholar]
- 23.Ohno, O., M. Mizokami, R. R. Wu, M. G. Saleh, K. Ohba, E. Orito, M. Mukaide, R. Williams, and J. Y. Lau. 1997. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J. Clin. Microbiol. 35:201-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sokolskaja, E., D. M. Sayah, and J. Luban. 2004. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J. Virol. 78:12800-12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 26.Tellinghuisen, T. L., and C. M. Rice. 2002. Interaction between hepatitis C virus proteins and host cell factors. Curr. Opin. Microbiol. 5:419-427. [DOI] [PubMed] [Google Scholar]
- 27.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldmeier, P. C., K. Zimmermann, T. Qian, M. Tintelnot-Blomley, and J. J. Lemasters. 2003. Cyclophilin D as a drug target. Curr. Med. Chem. 10:1485-1506. [DOI] [PubMed] [Google Scholar]
- 29.Watashi, K., M. Hijikata, M. Hosaka, M. Yamaji, and K. Shimotohno. 2003. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 38:1282-1288. [DOI] [PubMed] [Google Scholar]
- 30.Watashi, K., M. Hijikata, A. Tagawa, T. Doi, H. Marusawa, and K. Shimotohno. 2003. Modulation of retinoid signaling by a cytoplasmic viral protein via sequestration of Sp110b, a potent transcriptional corepressor of retinoic acid receptor, from the nucleus. Mol. Cell. Biol. 23:7498-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watashi, K., N. Ishii, M. Hijikata, D. Inoue, T. Murata, Y. Miyanari, and K. Shimotohno. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111-122. [DOI] [PubMed] [Google Scholar]
- 32.Watashi, K., and K. Shimotohno. 2003. The roles of hepatitis C virus proteins in modulation of cellular functions: a novel action mechanism of the HCV core protein on gene regulation by nuclear hormone receptors. Cancer Sci. 94:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiegers, K., and H. G. Krausslich. 2002. Differential dependence of the infectivity of HIV-1 group O isolates on the cellular protein cyclophilin A. Virology 294:289-295. [DOI] [PubMed] [Google Scholar]
- 34.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]