Abstract
The papillomavirus E2 regulatory protein has essential roles in viral transcription and the initiation of viral DNA replication as well as for viral genome maintenance. Brd4 has recently been identified as a major E2-interacting protein and, in the case of the bovine papillomavirus type 1, serves to tether E2 and the viral genomes to mitotic chromosomes in dividing cells, thus ensuring viral genome maintenance. We have explored the possibility that Brd4 is involved in other E2 functions. By analyzing the binding of Brd4 to a series of alanine-scanning substitution mutants of the human papillomavirus type 16 E2 N-terminal transactivation domain, we found that amino acids required for Brd4 binding were also required for transcriptional activation but not for viral DNA replication. Functional studies of cells expressing either the C-terminal domain of Brd4 that can bind E2 and compete its binding to Brd4 or short interfering RNA to knock down Brd4 protein levels revealed a role for Brd4 in the transcriptional activation function of E2 but not for its viral DNA replication function. Therefore, these studies establish a broader role for Brd4 in the papillomavirus life cycle than as the chromosome tether for E2 during mitosis.
The papillomaviruses (PVs) are small DNA viruses that are etiologic agents for papillomas and warts in a variety of higher vertebrates including humans. Specific human papillomaviruses (HPVs) have been associated with some human cancers, most notably cervical cancer (58). The papillomaviruses establish long-term, persistent infections of squamous epithelial cells, and the viral life cycle is tightly linked with the differentiation program of the host cell (21). In the infected dividing basal cells of the epithelium, the viral DNA is maintained as a stable plasmid. Vegetative viral DNA replication occurs only in the more differentiated squamous epithelial cells. Bovine papillomavirus (BPV) DNA remains extrachromosomal in transformed rodent cells, a system that has served as a useful model for studying viral genome maintenance (28).
The papillomavirus E2 protein has important roles in regulating viral transcription, in enhancing E1-dependent viral DNA replication, and in genome maintenance (21). E2 is a DNA binding protein that was first identified as a transcriptional activator (46). Subsequent studies established that E2 can also repress some genes, depending upon the location of its cognate binding sites within the promoter region (49). Indeed, E2 functions to repress the promoter directing the E6 and E7 viral oncogenes in the cancer-associated HPV type 16 (HPV16) and HPV18 genomes (38). For viral genome replication, E2 binds the viral helicase E1 and guides it to the origin of replication in the process of initiating origin-dependent viral DNA replication (6, 34). For genome maintenance, E2 has been shown to associate with mitotic chromosomes and, in doing so, to anchor the viral genomes to the host chromosomes during mitosis (4, 22, 31, 35, 44). The structure of E2 resembles that of a prototypic transcription factor, with an amino-terminal transcriptional activation (TA) domain and a carboxy-terminal DNA binding and dimerization domain. The TA domain is necessary for viral DNA replication, interaction with the viral E1 protein, and mediating transcriptional activation. In addition, the TA domain is required for the association of E2 with mitotic chromosomes to ensure the maintenance of the viral DNA in dividing cells (4, 22, 31, 35, 44). Specific mutations in the TA domain have been shown to disrupt the tethering of viral genomes to mitotic chromosomes (1, 5, 57).
We have recently shown that Brd4 (bromodomain-containing protein 4) mediates the association of BPV1 E2 to mitotic chromosomes and that the binding of E2 to Brd4 is conserved among the papillomaviruses (54). Through an interaction of the carboxy-terminal region of Brd4 with the amino-terminal TA domain of E2, this protein complex serves to bridge the viral DNA with cellular mitotic chromosomes (5, 7, 33, 54). Brd4 is a member of the BET family, a group of structurally related proteins characterized by the presence of two bromodomains and one extraterminal domain of unknown function. Bromodomains in general have been shown to interact with acetylated lysines in histones and are involved in chromatin targeting and remodeling (12, 24, 56). Unlike other bromodomain proteins that are released from chromatin during mitosis, BET family members remain bound to chromatin during mitosis (13, 26). Mouse embryos nullizygous for Brd4 die shortly after implantation, suggesting a role for Brd4 in fundamental cellular processes (20). Recently, Brd4 has been shown to influence the general RNA polymerase II-dependent transcription machinery by interacting with the core factors of the positive transcription elongation factor b (P-TEFb) and the Mediator complex (23, 52). In addition, Brd4 binds to acetylated chromatin with preferential binding for acetylated histones H3 and H4 (12). The mechanism that regulates the recruitment of Brd4 to promoters, however, is not yet well understood.
In order to gain insight into the functions of the Brd4/E2 complex, we analyzed a series of E2 single-amino-acid alanine-scanning mutants for their abilities to bind Brd4. These experiments permitted us to map the face of the E2 TA domain involved in binding Brd4, which turned out to be distinct from the region of the E2 TA required for E1 binding and for enhancing E1-dependent viral DNA replication (2, 9, 10, 14, 15, 41). Instead, the amino acids involved in Brd4 binding corresponded well with those necessary for the E2 transcriptional activation function. In combination with functional studies utilizing the C-terminal domain of Brd4 as a dominant negative inhibitor of the E2/Brd4 interaction and with short interfering RNA (siRNA) knockdown experiments of Brd4, we have demonstrated that Brd4 is required to mediate its transcriptional activation function.
MATERIALS AND METHODS
Recombinant plasmids.
The eukaryotic pCMV4-16E2 expression vectors for wild-type (wt) (p3662) and mutant (p3665 to p3688) E2 proteins and pCMV-16E1 (p3692) proteins have been described previously (41). The Escherichia coli pGEX-2T-16E2 expression plasmids (p3798 to p3809) were derived from the wild-type pGEX-2T-16E2 (p3796) plasmid (41). Plasmids containing the full-length human Brd4 (pcDNA4C-Brd4-FL) or the C-terminal domain between amino acids 1047 and 1362 (pcDNA4C-SV40NLS-hBrd4-CTD) and p2x2xE2BS-Luc and p16ori have been described previously (27, 41, 54). The BPV type 1 (BPV1) long control region (LCR) chloramphenicol acetyltransferase (CAT) reporter plasmid (p407-1) and the BPV1 E2 expression plasmid have been previously described (45, 51). The beta interferon (IFN-β) promoter luciferase, the PIN1 promoter luciferase reporter, the interferon regulatory factor 3 (IRF3) expression plasmid, and the E2F expression plasmid have been described previously by Servant et al. and Ronco et al., respectively (39, 40, 42). pSVβ-GAL and pEGFP were purchased from BD Bioscience.
Antibodies.
Rabbit polyclonal Brd4 antibody was raised against an N-terminal Brd4 fragment (amino acids 1 to 470) followed by affinity purification. The HPV16 carboxy terminus of E2 (E2C) antibody has been described previously (41). The anti-XPress-tag antibody was purchased from Invitrogen.
Cell lines and transfections.
The human cervical cancer cell lines HeLa (HPV18 positive) and C33A (HPV negative) were maintained in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal calf serum (HyClone). Cells were tested to be mycoplasma negative (Mycoplasma PCR Elisas; Roche). Plasmid DNAs were introduced into cells by using Fugene 6 transfection reagent (Roche). The NL-3D cell line harboring an E2-inducible β-galactosidase gene was provided by Dan DiMaio and has been previously described (43). Using standard retrovirus production and transfection procedures, pLPCX-HXNCTD or the empty vector pLPCX was used to generate C33A cells stably expressing the Brd4 C-terminal domain (Brd4-CTD) or the empty vector, respectively.
Cellular growth proliferation assay.
Approximately 2 × 106 C33A cells were cotransfected with combinations of 0.6 μg pBABE-puro, 0.4 μg pEGFP, 2 μg pCMV-E2, and 1.5 μg pcDNA4C-Brd4-CTD. Twenty-four hours after transfection, cells were split into groups of 3 × 104 cells per 35-mm dish and put under puromycin selection (0.3 μg/μl). Cells were trypsinized and counted daily in triplicate.
alamarBlue assay.
Approximately 6 × 103 C33A cells stably transfected with the Brd4-CTD were incubated with Dulbecco's modified Eagle's medium containing 10% alamarBlue (Biosource) solution for 0, 2, 4, 6, 8, 20, 22, 26, and 32 h. Fluorescence measurements were performed with a Cytofluor multiwell plate reader (PerSeptive Biosystems) by excitation at 530 nm and emission at 590 nm. The fluorescence emission intensity units were plotted as a function of incubation time.
BrdU incorporation assay.
The bromodeoxyuridine (BrdU) incorporation assay was performed as recommended by the manufacturer (BrdU Cell Proliferation ELISA; Roche). In brief, stably transfected C33A cells were split into groups of 500, 2 × 103, 6 × 103, and 14 × 103 cells per 96-well. Cells were incubated for 2 h with BrdU, followed by fixation for 30 min and incubation for 1 h with anti-BrdU peroxidase-conjugated antibody. After the addition of luminol (substrate), chemiluminescence was measured with a luminometer and expressed as a function of cell number.
RNA interference.
For knockdown of Brd4 expression, the pSUPER vector system was used (OligoEngine). Using the manufacturer's protocol, siRNA expression plasmids were constructed by annealing oligonucleotides containing the siRNA-expressing sequence [for siRNA-Brd4(NT), 5′-GACACTATGGAAACACCAG-3′; for siRNA-Brd4(CT), 5′-GCGGGAGCAGGAGCGAAGA-3′] and cloning them in the BglII/HindIII sites of the vector. The control siRNA-green fluorescent protein (GFP) construct was a gift from B. Lilley and targeted the sequence 5′-GCAAGCTGACCCTGAAGTTC-3′ (50). siRNA-induced silencing was determined by indirect immunofluorescence of Brd4. Cells were cotransfected with 2 μg siRNA and 0.13 μg enhanced GFP (EGFP) expression plasmids. After 36 h, cells were plated onto coverslips and fixed with 3% paraformaldehyde 24 h later. Staining of Brd4 with an anti-Brd4 antibody was performed as described previously (54). Alexa Fluor 594 goat anti-rabbit antibody (Molecular Probes) was used as a secondary antibody. Cells were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) and examined with a Leica DMLB epifluorescence microscope.
GST pull-down.
Glutathione-S-transferase (GST) E2 fusion proteins were expressed in E. coli BL21. Proteins were affinity purified with glutathione-Sepharose 4B beads (Amersham) according to the manufacturer's recommendations. Proteins were eluted from the columns with 10 mM glutathione and dialyzed overnight in a solution containing 150 mM NaCl, 50 mM Tris-HCl, pH 8.0, and 1 mM dithiothreitol (DTT). The purity of the GST fusion proteins was confirmed by sodium dodecyl sulfate (SDS) gel electrophoresis and Coomassie staining.
35S-labeled Brd4-CTD was generated by using the T7-TNT-coupled rabbit reticulocyte lysate system (Promega). Briefly, GST pull-downs were performed as follows. A total of 0.5 μg of each GST-E2 fusion protein and 15 μl of the in vitro-translated Brd4-CTD were incubated in 500 μl binding buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 4 mM MgCl2, 2 mM DTT, 0.5% NP-40, 2% nonfat dry milk) for 60 min at 4°C. Twenty microliters of 50% GST Sepharose slurry equilibrated in binding buffer was added, and incubation continued for 30 min. Beads were sedimented and washed three times with 1 ml of binding buffer without dry milk. The samples were analyzed by SDS-polyacrylamide gel electrophoresis, and the labeled proteins were visualized by autoradiography.
Coimmunoprecipitation and Western blot analysis.
For coimmunoprecipitation, 1 × 107 transiently transfected C33A cells were harvested 48 to 72 h after transfection, and soluble proteins were extracted as described previously (41a). The extract (1.5 ml) was mixed with 30 μl of 50% protein A agarose slurry (Invitrogen) in immunoprecipitation-binding buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 50 mM NaCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT) and precleared overnight at 4°C. The extracts were subsequently incubated with 4 μl of affinity-purified anti-E2C antibody (41) for 4 h at 4°C, followed by the precipitation of the antigen-antibody complexes with 30 μl 50% protein A agarose slurry (Invitrogen). The beads were washed three times with immunoprecipitation-binding buffer, and bound proteins were eluted with 30 μl sample buffer. Aliquots were resolved on an 8% SDS-polyacrylamide gel. Proteins were transferred onto a polyvinylidene difluoride membrane and blotted with anti-Xpress mouse monoclonal antibody (Invitrogen) and anti-HPV16 E2 rabbit polyclonal antibody (41). As secondary antibodies, fluorescence (680 nm and 750 nm)-labeled anti-mouse and anti-rabbit antibodies were used (Molecular Probes). Western blots were visualized and quantitated with an Odyssey infrared imaging system (Leicor).
Transient papillomavirus DNA replication assay.
A transient papillomavirus DNA replication assay was performed according to a protocol described previously by Del Vecchio et al. (11). C33A cells were cotransfected with the papillomavirus origin-containing plasmid (p16ori) and plasmids expressing E1 and E2 (pCMV-16E1 and pCMV-16E2, respectively). Around 48 h after the transfection, low-molecular-weight DNA was prepared using the Hirt method followed by phenol-chloroform extraction and ethanol precipitation (19). Since DNA replicated in eukaryotic cells is not methylated on adenine residues and is resistant to DpnI digestion, the replicated DNA is distinguished from input DNA by DpnI digestion. The digested samples were then analyzed by Southern blotting using a probe encompassing the PV origin of replication labeled with 32P using a random prime labeling kit (Stratagene). The blot was washed twice with a low-stringency buffer (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS) for 1 h at room temperature and once with a high-stringency buffer (0.1× SSC, 0.1% SDS) for 1 h at 63°C. Blots were dried and visualized by autoradiography. Quantification was performed using a Storm PhosphorImager (Molecular Dynamics).
Reporter assays.
Approximately 2 × 106 C33A cells were transfected with 0.7 μg p2x2xE2BS-Luc and 1.4 μg pCMV-E2. To determine or normalize transfection efficiencies, 0.5 μg pSVβ-GAL and 0.4 μg pEGFP were cotransfected. To test the influence of Brd4-CTD on E2 transcriptional activation, cells were cotransfected with 0.0014 μg, 0.014 μg, 0.14 μg, and 1.4 μg of pcDNA4C-Brd4-CTD. For rescue experiments with the full-length Brd4 protein, 0.7 μg pcDNA4C-Brd4-CTD and 0.7 μg or 2.3 μg pcDNA4C-Brd4-FL were used. Forty-eight hours after transfection, cells were lysed, and luciferase activities were measured according to the manufacturer's protocol (Luciferase assay system; Promega). Luciferase activities were normalized to the β-galactosidase activity of pSVβ-GAL cotransfected with the reporter plasmid (Luminescent β-Gal Detection Kit II; BD Bioscience).
For the CAT assays, approximately 1 × 106 CV1 cells were cotransfected with 0.15 μg each of the BPV1 LCR CAT reporter plasmid (p407-1) and the E2 expression plasmid (p2450) plus or minus the Brd4-CTD plasmid. Forty-eight hours after transfection, cells were lysed, and CAT expression was measured using a colorimetric CAT enzyme-linked immunosorbent assay (Roche). Results were normalized to the protein concentration in the lysates.
RESULTS
Analysis of Brd4 binding to a series of HPV16 E2 mutants.
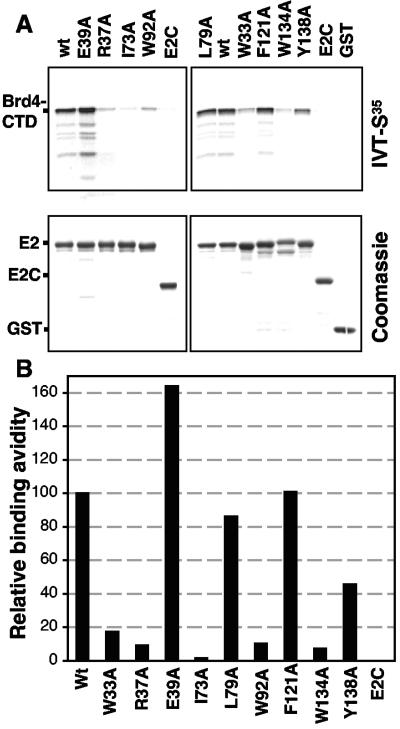
The N-terminal TA domain of E2 mediates the binding to Brd4 (54). To further characterize this interaction, we assayed single-amino-acid alanine substitution mutants within the HPV16 E2 transactivation domain for their abilities to bind Brd4. Specific amino acid mutants within the domain were selected for analysis based on the structure of this region (3, 17) and a previous analysis of amino acids conserved within this domain among different papillomaviruses (41). Our goal was to map the amino acids on the surface of the E2 transactivation domain required for Brd4 binding and to determine whether Brd4 binding correlated with any other E2 functions. In order to assess Brd4 binding, GST pull-down and coimmunoprecipitation assays were performed with individual E2 mutants (Fig. 1). For the GST pull-down assays, GST-tagged wild-type and mutant E2 proteins were expressed and purified from E. coli. Each protein was incubated with an aliquot of in vitro-translated, 35S-labeled Brd4-CTD, and bound material was separated using glutathione-Sepharose. Bound proteins were separated on SDS-polyacrylamide gels, and radioactive proteins were visualized and quantified by using a PhosphorImager (Fig. 1). In parallel, equal amounts of the GST-E2 proteins were run on separate gels as input controls and visualized by Coomassie blue staining (Fig. 1A, bottom). In these experiments, the carboxy terminus of E2 (E2C), spanning amino acid 153 to 365, served as a negative control, since it lacks the TA domain. In the experiment shown, wt E2 bound 28% of the input Brd4-CTD. We found that the E39A, L79A, and F121A mutants bound Brd4 comparably to wt E2. In contrast, I73A did not bind Brd4, and the W33A, R37A, W92A, and W134A mutants bound less than 20% of Brd4 compared to wild-type E2. In these experiments, intermediate binding of Brd4 was observed for the Y138A mutant.
FIG. 1.
Binding of HPV16 E2 transactivation domain mutants to the Brd4 C-terminal domain. (A) GST-tagged E2 (wild type or mutant) was expressed and purified from E. coli. Equal amounts of the proteins were incubated with in vitro-translated (IVT) Brd4-CTD and glutathione-Sepharose beads. After extensive washings, the proteins were separated by SDS-polyacrylamide gel electrophoresis. wt E2 served as the positive control, and the C-terminal 171 amino acids of E2 (E2C) and GST alone (GST) served as negative controls. Upper panel, autoradiogram of 35S-labeled Brd4-CTD bound to the indicated GST-E2 fusion protein; lower panel, Coomassie-stained gel of the input GST fusion proteins used in the binding experiment. (B) The bound 35S-labeled Brd4-CTD was quantified with a PhosphorImager, with wt E2 binding set to 100%.
We next examined the capacity of the E2 mutants to bind Brd4 in vivo by examining coimmunoprecipitations of the E2 mutants and Xpress-tagged Brd4-CTD that were coexpressed in C33A cells. Protein complexes were isolated using an antibody directed to the C terminus of E2, and binding was quantitated by an Odyssey infrared imaging system using an anti-Xpress antibody (Leicor) (data not shown). In agreement with the GST pull-down experiments, the I73A mutant did not bind Brd4, and the W33A, R37A, and W92A mutants were significantly impaired (<10% of wild-type E2) in their ability to bind the Brd4-CTD. The E39A, K68A, L79A, E90A, T93A, F121A, D122A, Y138A, and Y178A mutants efficiently bound the Brd4-CTD at levels greater than 40% of that of wt E2. A summary of the Brd4 binding results from both experiments is shown in Table 1. These binding studies revealed a nearly complete correlation between amino acids important for transcriptional activation and Brd4 binding. In contrast, no correlation was seen between Brd4 binding and E1 binding or DNA replication. For example, the E2 I73A mutant does not bind Brd4 and is inactive in transcriptional activation but has wild-type activities for E1 binding and viral DNA replication. On the other hand, the E2 E39A mutant has a high affinity for Brd4 and can activate transcription but is defective for E1 binding and viral DNA replication.
TABLE 1.
Characterization of different HPV16 E2 alanine substitution mutations for transcriptional activation, DNA replication, and E1 and Brd4 binding functionsa
| Mutation | Transactivation | Brd4 binding | DNA replication | E1 binding |
|---|---|---|---|---|
| wt | +++ | +++ | +++ | +++ |
| W33A | + | + | + | + |
| R37A | − | − | + | +++ |
| E39A | +++ | ++/+++ | − | − |
| K68A | ++ | ++ | ++ | +++ |
| I73A | − | − | +++ | +++ |
| L79A | +++ | +++ | +++ | ++ |
| E90A | +++ | +++ | +++ | +++ |
| W92A | − | − | + | + |
| T93A | +++ | ++ | ++ | +++ |
| F121A | +++ | +++ | + | + |
| D122A | +++ | +++ | ++ | + |
| W134A | − | − | + | + |
| Y138A | ++ | +++ | + | ++ |
| Y178A | +++ | ++/+++ | + | + |
The −, +, ++, and +++ quantitations indicated for the transactivation, DNA replication, and E1 binding columns are from Table 1 in reference 41 (Sakai et al). Brd4 binding relative to wild-type E2. +, 10 to 20%; ++, 20 to 45%; +++, greater than 45%; −, less than 10%.
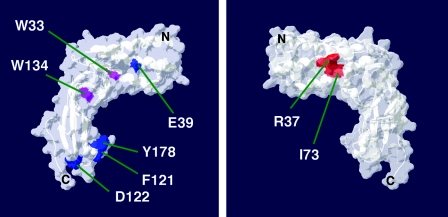
Figure 2 shows a structural model of the HPV16 E2 transactivation domain (3) in which the amino acids R37 and I73, which are important for Brd4 binding and transactivation, have been colored in red. Residues E39, F121, D122, and Y178, which are required for E1 binding and viral DNA replication, are indicated in blue. W33A and W134A (shown in purple) are significantly impaired in each E2 function tested, and therefore, it is possible that mutation of these residues might affect the overall structure of E2. The structural model shows clearly that amino acids required for Brd4 binding and for transcriptional activation cluster on one side of the E2 surface, whereas amino acids necessary for E1 binding and viral DNA replication map to a different side of the E2 protein. These data suggested that Brd4 could be involved in the transcriptional activation function of E2 and that, conversely, it was unlikely to be important for its viral DNA replication function.
FIG. 2.
A model of the HPV16 E2 transactivation domain (PDB accession number 1DTO) was visualized with the Swiss-PdbViewer program (16). Shown are the two opposite surfaces of the TA domain. Amino acids important for Brd4 binding (Fig. 1) and transcriptional activation but not E1 binding are indicated in red. Residues important for E1 binding but neither Brd4 binding nor the transactivation function are shown in blue. Amino acids for which mutants defective for all E2 functions as summarized in Table 1 are shown in purple.
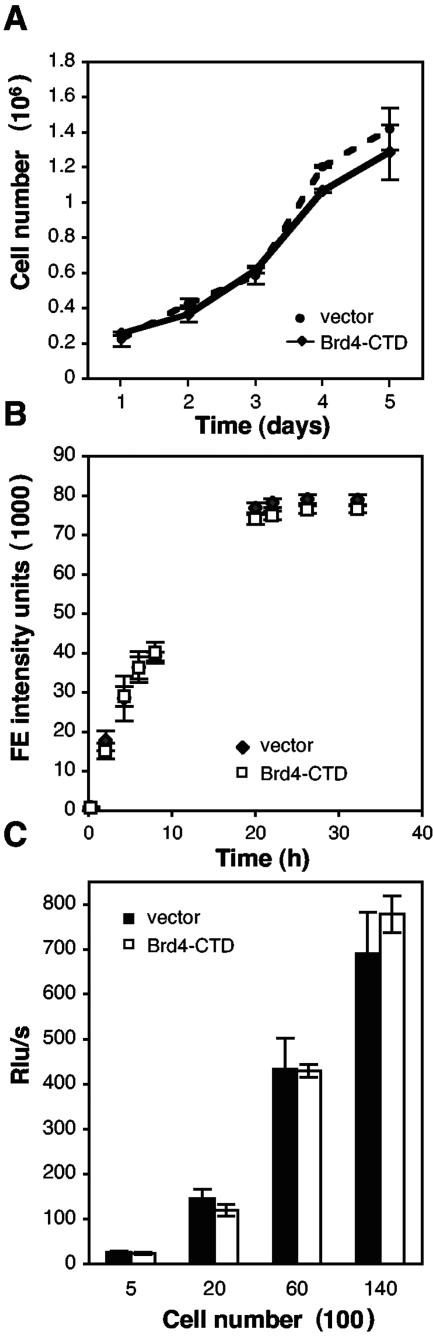
The Brd4-CTD does not influence the growth properties of C33A cells.
To test the potential role of Brd4 binding on the E2 transcriptional activation and viral DNA replication functions, we performed E2-dependent transcriptional reporter assays and transient DNA replication assays in the presence or absence of the Brd4-CTD, a dominant-acting negative inhibitor of Brd4/E2 binding. We first asked whether expression of the Brd4-CTD influenced cellular proliferation or cellular DNA replication of C33A cells by an examination of its effect on cellular growth rates, by an alamarBlue assay, and by BrdU incorporation. No difference in the growth rates of C33A cells transfected with empty vector or transfected with a Brd4-CTD expression vector was observed (Fig. 3A). To determine whether expression of the Brd4-CTD affected cellular metabolic activity, C33A cells transfected with Brd4-CTD were compared to C33A cells transfected with an empty vector using an alamarBlue reduction assay (Fig. 3B). Like MTT [3-(4,5-dimethyldiazol-2-yl)-2,5-diphenyl tetrazolium bromide], alamarBlue is reduced by metabolic intermediates such as NADPH, FADH, and NADH and changes from an oxidized nonfluorescing state to a reduced fluorescing state. We found no evidence that Brd4-CTD expression affected the metabolic state of C33A cells. Since a change in cellular DNA replication could potentially mask changes measured in a viral DNA replication assay, we performed a BrdU incorporation assay to examine the effect of Brd4-CTD on cellular DNA replication (Fig. 3C) and found no difference in C33A cells expressing the Brd4-CTD compared to the vector control cell line. Furthermore, it should be noted that the Brd4-CTD has no effect on E2 levels of expression (54).
FIG. 3.
Brd4-CTD does not affect proliferation, viability, or DNA replication of C33A cells. (A) C33A cells were cotransfected with a plasmid expressing Brd4-CTD or an empty vector and a puromycin resistance plasmid. Cells were split and placed under puromycin selection. Cells from triplicate plates were counted each day with a hemocytometer. (B) The cellular metabolic activity of C33A cells stably transfected with Brd4-CTD or empty vector control was determined by an alamarBlue assay. Equal numbers of cells were incubated with 10% alamarBlue reagent. Fluorescence emission (FE) was measured after 0, 2, 4, 6, 8, 20, 22, 26, and 32 h and plotted against the incubation time. Each point represents the mean value of nine measurements. (C) BrdU incorporation was assayed in 500, 2 × 103, 6 × 103, and 14 × 103 C33A cells stably expressing the Brd4-CTD or vector alone. Cells were labeled for 2 h with BrdU, and incorporation was detected with a peroxidase-conjugated anti-BrdU antibody. Chemiluminescence was measured with a luminometer. The relative light units (Rlu)/second were plotted in correlation to the number of cells plated. Each point represents the mean of six measurements.
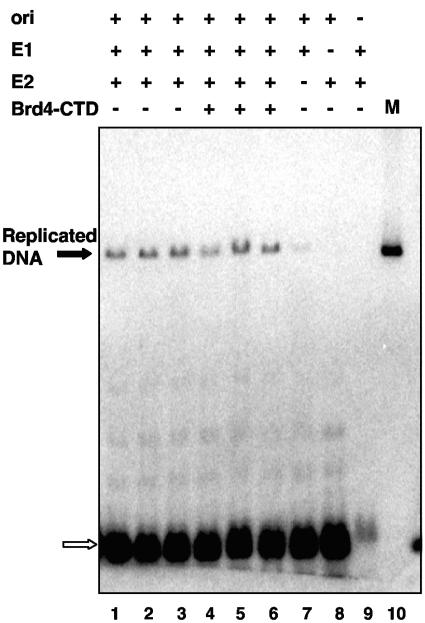
The E2 viral DNA replication function is independent of Brd4.
Some of the E2 mutants, such as the E39A, F121A, Y138A, and Y178A mutants, bound Brd4 well but are incapable of supporting viral DNA replication, suggesting that viral DNA replication is not dependent upon the ability of E2 to bind Brd4. In order to test the role of Brd4 binding on the E2 viral DNA replication function directly, we analyzed the effect of the Brd4-CTD in viral DNA replication assays. The viral DNA replication activity was measured after cotransfection of a papillomavirus origin-containing plasmid (p16ori), E1 and E2 expression plasmids, and either a Brd4-CTD expression plasmid or an empty vector. As negative controls, replication assays without E1 or E2 or without the origin-containing plasmid were performed. As shown in Fig. 4, we observed no significant inhibition of the E2-enhanced p16ori-dependent plasmid replication by the Brd4-CTD. In addition, transfection of a full-length Brd4 expression plasmid had no effect on E2-enhanced p16ori-dependent plasmid replication (data not shown). Furthermore, we found that the Brd4-CTD did not compete E1 binding to E2 in vitro (data not shown). We therefore conclude that E2 binding to Brd4 is not required for its DNA replication function.
FIG. 4.
Brd4-CTD does not influence papillomavirus DNA replication. Shown are data from a transient in vivo replication assay of an HPV16 origin-containing plasmid (p16ori). C33A cells were transfected with p16ori along with plasmids expressing E1 and/or E2 and a plasmid expressing the Brd4-CTD or vector control. Each assay was done separately in triplicate. Low-molecular-weight DNA was harvested by the Hirt method 48 h following transfection. DNA was digested with DpnI, and DNA was analyzed by Southern blot hybridization using an HPV16 ori probe. As negative controls, the replication assays were performed without E2 (lane 7), E1 (lane 8), or p16ori (lane 9). As a marker (M), the p16ori plasmid was used without transfection (lane 10). Replicated p16ori DNA is indicated by a solid arrow. Digested p16ori DNA is indicated by the open arrow.
The Brd4-CTD inhibits the E2 transcriptional activation function.
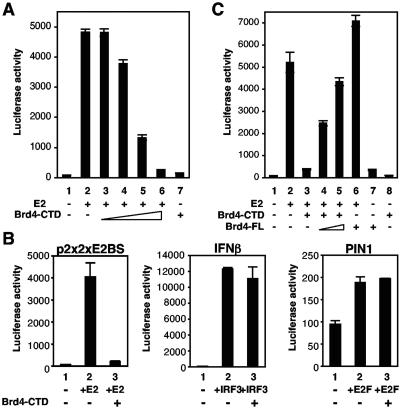
The mutational analysis of the N-terminal domain of E2 described above indicated that Brd4 binding correlated with transcriptional activation functions of the E2 protein. To examine whether Brd4 indeed plays a fundamental role in E2-dependent transcriptional activation, we next tested whether the Brd4-CTD could affect E2-dependent transcriptional activation. C33A cells were transfected with an E2-responsive reporter plasmid (p2x2xE2BS-Luc), which contains four E2 binding sites, the E2 expression plasmid, and increasing amounts of a Brd4-CTD expression plasmid. By itself, E2 enhanced luciferase expression from the reporter plasmid by 60-fold (Fig. 5A, compare lanes 1 and 2). Expression of the Brd4-CTD inhibited the transcriptional activation function of E2 in a dose-dependent manner (lanes 3 to 6). The Brd4-CTD alone had no significant effect on the expression of luciferase from the reporter plasmid in the absence of E2 (lane 7).
FIG. 5.
Role of Brd4 in E2-dependent transcriptional activation. (A) Expression of Brd4-CTD inhibits E2 transcriptional activation. C33A cells were transfected with p2x2xE2BS-Luc, an E2-dependent luciferase reporter plasmid (columns 1 to 7). Coexpression of wt E2 activates the reporter plasmid. Luciferase activity was also measured in the presence of increasing amounts of Brd4-CTD (0.0014 μg for column 3 to 1.4 μg for column 6 in 10-fold increments). The luciferase activities were normalized for transfection efficiency determined by the β-galactosidase activity expressed in the cotransfected cells. The data represent the averages of three experiments. (B) Inhibition of transcriptional activation by Brd4-CTD is specific for E2. C33A cells were transfected with E2 (p2x2xE2BS)-, IRF-3 (IFN-β)-, or E2F (PIN1)-dependent luciferase reporter plasmids. Luciferase activity was stimulated by cotransfection of the corresponding activator plasmid for E2, IRF-3, or E2F, respectively. In addition, each assay was performed by cotransfection of the Brd4-CTD (1.4 μg)-expressing plasmid. The luciferase activities were normalized for transfection efficiency by the β-galactosidase activity expressed in the cotransfected cells. The data represent the averages of three experiments. (C) Full-length Brd4 can rescue transcriptional activation inhibition by Brd4-CTD. C33A cells were transfected with an E2-dependent luciferase reporter (columns 1 to 8) and the E2 expression plasmid (columns 2 to 6). Cells were cotransfected with 0.7 μg of Brd4-CTD (column 3 to 5, 8) and 0.7 μg (column 4) or 2.3 μg (columns 5 to 7) of full-length Brd4, respectively. The luciferase activities were normalized for transfection efficiency as determined by the β-galactosidase activity expressed in the cotransfected cells. The data represent the averages of three experiments.
To address the specificity of the inhibition of E2 transactivation by the Brd4-CTD, two additional reporter plasmids that are not responsive to E2 were tested: an IFN-β promoter luciferase reporter plasmid that responds to IRF3 and a PIN1 promoter luciferase reporter plasmid that responds to the transcription factor E2F. In contrast to the strong inhibition of E2 transcriptional activation by the Brd4-CTD, no inhibition by the Brd4-CTD was observed for either IRF3 activation of the IFN-β promoter or E2F activation of the PIN1 promoter (Fig. 5B).
Brd4 is required for E2 transcriptional activation.
To determine whether the inhibition of E2 transcriptional activation by the Brd4-CTD was due to competition of the full-length Brd4 protein or to some other cellular factor that bound to the same region of the E2 TA domain, we tested whether the full-length Brd4 protein (Brd4-FL) could rescue this inhibition. As shown in Fig. 5C, the full-length protein was able to rescue the inhibition by Brd4-CTD in a dose-dependent manner. Therefore, we conclude that Brd4 has an essential role in the E2 transcriptional activation function.
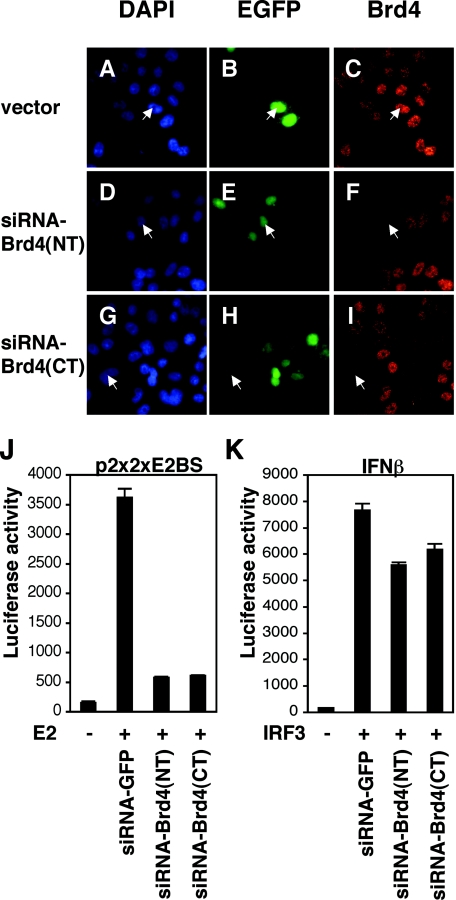
To further examine the requirement for Brd4 in E2 transcriptional activation, we employed siRNAs to knock down endogenous Brd4 expression in C33A cells. Constructs that generated siRNAs directed against the N terminus [siRNA-Brd4(NT)] or the C terminus [siRNA-Brd4(CT)] of Brd4 were constructed using pSUPER vectors. These plasmids were then tested for their abilities to knock down Brd4 protein levels by immunofluorescence. C33A cells were transfected with either the empty vector or one of the siRNA-expressing plasmids along with an EGFP expression plasmid at a ratio of 15:1 (Fig. 6A to I). The cells were stained with an anti-Brd4 antibody and counterstained with DAPI. The siRNA-Brd4(NT) and siRNA-Brd4(CT) expression plasmids each significantly decreased the Brd4-specific signal in the transfected cells (Fig. 6F and I). In contrast, cells transfected with the empty vector did not show any difference in Brd4 levels compared to that from nontransfected cells (Fig. 6C). We next tested whether knockdown of Brd4 using the siRNA-Brd4(NT) and siRNA-Brd4(CT) expression plasmids could inhibit E2 transcriptional activation. The Brd4 siRNA expression plasmids, as well as a GFP siRNA expression plasmid as the negative control, were cotransfected with the E2-dependent luciferase reporter plasmid (p2x2xE2BS). Each of the Brd4 siRNA constructs, either alone or in combination, strongly inhibited E2-dependent transcriptional activation by around 85% compared to the GFP siRNA control (Fig. 6J). Since two independent siRNA constructs for Brd4 strongly inhibited E2 transcriptional activation, we conclude that the result is unlikely a consequence of off-target effects of the siRNA constructs. Furthermore, to test specificity, we examined the effects of these two siRNAs on the IRF3 activation of the IFN-β luciferase reporter and found minimal effects (Fig. 6K). Taken together, these results show that Brd4 is required for papillomavirus E2 transcriptional activation and specifically mediates its transcriptional activation function.
FIG. 6.
Expression of siRNAs against Brd4 inhibits E2-dependent transcriptional activation. (A to I) C33A cells transfected with an empty vector (A to C) or two different siRNA expression constructs, siRNA-Brd4(NT) and siRNA-Brd4(CT) (D to I), were stained with an anti-Brd4 antibody (C, F, and I). Transfected cells were visualized by cotransfection of an EGFP expression plasmid (B, E, and H). Cells were also counterstained with DAPI to identify nuclei (A, D, and G). (J) C33A cells were cotransfected with an E2-responsive reporter plasmid (p2x2xE2BS-Luc), an E2 expression plasmid, and plasmids expressing siRNAs against GFP (column 2) as a control or Brd4 (columns 3 and 4). (K) C33A cells were transfected with an IRF-3 (IFN-β)-dependent luciferase reporter plasmid, and luciferase activity was stimulated by cotransfection of an IRF-3 expression plasmid. Luciferase activities were measured in the presence of the control siRNA against GFP (column 2) or siRNAs against Brd4 (columns 3 and 4). The luciferase activities of both experiments were normalized for transfection efficiency by the β-galactosidase activity expressed in the cotransfected cells. The data represent the averages of three experiments.
Brd4 is required for E2 activation of an authentic papillomavirus promoter.
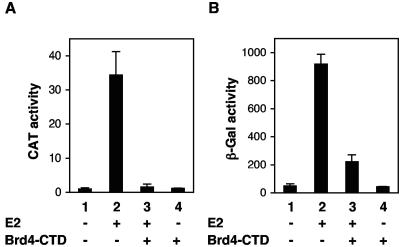
The experiments shown in Fig. 5 and 6 employed a heterologous promoter containing four E2 binding sites. To address whether Brd4 is also required for the E2-dependent activation of an authentic E2-responsive papillomavirus promoter, we examined the effect of the Brd4-CTD on the E2 activation of the BPV1 LCR P7940 early promoter, which can be strongly activated by E2 (18, 45, 48). The BPV1 LCR CAT reporter plasmid p407-1 was transfected along with a BPV1 E2 expression plasmid plus or minus the Brd4-CTD in CV1 cells. By itself, E2 activated the promoter reporter plasmid by approximately 35-fold (Fig. 7A, compare lanes 1 and 2). Coexpression of the Brd4-CTD nearly completely inhibited the transcriptional activity of E2 on this natural target promoter (Fig. 7A, lane 3). The Brd4-CTD had no significant effect on the LCR promoter basal activity (Fig. 7A, lanes 1 and 4).
FIG. 7.
Brd4 is required for the E2-dependent transcriptional activation of the viral LCR. (A) E2 transactivation of an episomal LCR reporter plasmid depends on Brd4. CV1 cells were transfected with a CAT reporter plasmid with the reporter gene under the control of the BPV1 LCR. Cotransfection of BPV1 E2 induced the expression of the CAT gene. CAT gene expression was also measured after cotransfection of E2 and Brd4-CTD or Brd4-CTD alone. The CAT expression levels were determined with a CAT enzyme-linked immunosorbent assay. The data represent the averages of three experiments. (B) E2 transactivation of a cellular integrated LCR requires Brd4. NL-3D cells (43) with an integrated β-galactosidase gene downstream of the BPV1 LCR were cotransfected with BPV1 E2 and Brd4-CTD alone or in combination. The β-galactosidase (β-Gal) activity was measured with a luminescent β-galactosidase assay. The data represent the averages of three experiments.
To examine whether Brd4 is also important for the activation of an integrated viral promoter, we examined the effect of the Brd4-CTD on the E2 activation of an integrated lacZ gene under the control of the BPV1 LCR (43). In this cell line, expression of the BPV1 E2 induced the β-galactosidase activity19-fold (Fig. 7B, compare lanes 1 and 2). The E2 induction was significantly reduced by coexpression of the Brd4-CTD (lane 3). The Brd4-CTD by itself had little effect on the basal expression of β-galactosidase (lane 4). Knockdown of Brd4 protein expression by siRNA also significantly reduced the level of β-galactosidase activity induced by E2 in this cell line (data not shown). These experiments show that the E2 activation of authentic papillomavirus promoters is dependent upon Brd4 and independent of the episomal state compared to the integrated state of the viral DNA.
DISCUSSION
The PV E2 proteins have well-characterized regulatory functions that affect viral transcription, viral DNA replication, and long-term plasmid maintenance. Our laboratory has identified Brd4 as the cellular mitotic chromosome-associated factor that mediates the chromosome binding of E2 (54). In addition, recent studies have shown that stable PV-based plasmid maintenance by E2 in Saccharomyces cerevisiae requires Brd4 and that Brd4 binding to E2 is necessary for the mitotic chromosome localization of E2 (5, 7). Furthermore, we have recently shown that blocking the interaction of E2 with Brd4 enhances viral genome loss and enhances the phenotypic reversion of bovine papillomavirus-transformed cells (55).
In this study, we have found that Brd4 is also required for the transcriptional activation function of E2. This was first suggested by our protein interaction studies using a functionally well characterized series of alanine-scanning point mutants of the N-terminal TA domain of HPV16 E2 (41). Mutants defective for their ability to transactivate an E2-responsive promoter were impaired in binding to Brd4 (Table 1). Visualization of amino acids necessary for Brd4 binding on the structural model of the HPV16 E2 transactivation domain revealed that these amino acids are localized on a different face of E2 than that involved in the binding of E1 (Fig. 2). As expected, E1/E2 binding was not competed by the Brd4-CTD, and the Brd4-CTD did not significantly affect viral DNA replication (data not shown and Fig. 4).
Alanine substitution of amino acids R37 and I73 of HPV16 E2 resulted in proteins that were defective for transcriptional activation and for Brd4 binding (Fig. 1). Interestingly, for HPV31 E2, conservative substitution mutants of the analogous residues R37K and I73L were also defective for E2-dependent transcriptional activation, yet the I73L mutation did not severely impact the viral genome maintenance function as described previously (47). It is not clear why the isoleucine-to-leucine mutation of amino acid 73 for HPV31 E2 should be defective for its transactivation function and not the plasmid maintenance function. However, the HPV31 E2 mutants have not yet been assayed directly for Brd4 binding. It is therefore possible that the conserved isoleucine-to-leucine mutation in HPV31 E2 might not completely disrupt the Brd4/E2 interaction, and as such, the mutation might result in a partial phenotype. To that end, a close examination of Fig. 3 in a previous report by Stubenrauch et al. (47) indicates that the plasmid maintenance function of the I73L mutant might be slightly lower than that of wt E2 protein. Nevertheless, at this point, we cannot rule out that, in contrast to BPV1, the HPVs might require another factor together with or in addition to Brd4 for their E2-mediated viral genome maintenance functions.
Two recent studies used a mutational analysis of the E2 TA domain to identify E2 mutants defective for localization to mitotic chromosomes. Baxter and colleagues used mutant BPV1 E2 proteins to study the mitotic chromosome binding activity and binding to Brd4 (5). In their study, they used a combination of multiple-site point mutations as well as a series of single-amino-acid substitution mutants (R37A, E39A, R68A, and I73A). Consistent with our results, they found that E39A and R68A bound mitotic chromosomes at wild-type levels, whereas I73A was nearly completely excluded from the mitotic chromosome (5). Abroi et al. also examined a series of BPV1 E2 TA domain mutants for a variety of functions (1). Their study predated our publication of the E2/Brd4 interaction, so it did not include an analysis of Brd4 binding. The results reported previously by Abroi et al. differed from those of Baxter et al. The discrepancy between the findings of those two groups could perhaps be explained by the different experimental conditions used in each study. As reported previously by Zheng et al., lowering the temperature or using agents that promote protein folding may increase the ability of some mutant E2 proteins to associate with mitotic chromosomes (57). In our studies, we were able to avoid the difficulties noted in those previous reports by using the Brd4-CTD as a dominant negative inhibitor of the Brd4/E2 interaction and by siRNA knockdown experiments.
Based on the correlation we observed between Brd4 binding and the transcriptional activation capacity of the E2 TA mutants, we tested whether the Brd4-CTD could inhibit E2 transactivation of an E2-responsive promoter. We found a dose-dependent inhibition of the E2 transactivation activity by the Brd4-CTD, with nearly complete inhibition at the highest concentration (Fig. 5A, lanes 3 to 6). This inhibition was specific for E2 transactivation and was rescued by coexpression of the full-length Brd4 protein (Fig. 5B and C). The importance of Brd4 in the E2 transcriptional activation function was further validated by the Brd4 knockdown experiments employing siRNAs to Brd4 (Fig. 6). The transactivation function of E2 plays an important role in the BPV1 life cycle through the activation of the LCR early promoter (18, 45, 48). The requirement for Brd4 was demonstrated in transient transfection experiments as well as in a cell line harboring an integrated copy of a BPV1 LCR β-galactosidase reporter gene. Thus, the E2-dependent activation of an authentic viral target promoter is dependent upon Brd4 and is independent of the chromosomal status of the LCR (Fig. 7).
E2 is an essential regulatory factor for the papillomaviruses. Of the various E2 functions, its transcriptional activities are perhaps the least well understood at a mechanistic level. E2 can either activate or repress a promoter containing E2 binding sites, depending upon the number and position of the binding sites within the promoter region (21). The E2 protein has been shown to bind a number of general cellular transcription factors such as TFIIB and TATA binding protein; transcriptional coactivators AMF-1 (activation domain-modulating factor 1), p/CAF, and p300/CBP; and the nucleosome assembly protein NAP-1 (8, 29, 30, 36, 37, 53). It has also been shown that a direct interaction between the transcription factor Sp1 and E2 brings distantly bound E2 to the promoter region through the formation of stable DNA loops (32). Interestingly, even though E2 interacts with a large number of cellular proteins over its E2 TA domain, until now, no complete correlation between the transcriptional activation domain and the binding of another protein on the E2 TA domain has been demonstrated, as there is for Brd4. The association sites of E2 TA with cellular transcription cofactor AMF-1 (amino acids 134 to 216) and TFIIB (amino acids 74 to 134), for example, are distinct from the transcriptional activation domain, indicating that Brd4 is a mediator of the E2-dependent transcriptional activation (8, 36, 53). Brd4 has recently been shown to be a component of the P-TEFb complex and to interact with subunits of the Mediator complex (20, 23, 25, 52). It therefore seems reasonable to surmise that Brd4 may link the transcription factor E2 with the P-TEFb and Mediator complexes, thus connecting E2 to the general transcription machinery. It is possible that Brd4 regulates the recruitment of the transcription machinery to specific genes through interactions with certain transcription factors such as E2 but not with others.
In this study, we were able to demonstrate that Brd4 mediates the transcriptional activation function of E2. How does this result go along with previous work showing that the Brd4/E2 interaction is required for the maintenance of the BPV1 viral genome? By immunocytochemistry, chromatin immunoprecipitation, and fluorescence in situ hybridization, we have shown that the viral genome maintenance function of E2 is mediated by binding of Brd4 on mitotic chromosomes (54, 55). Brd4 is also necessary for stable papillomavirus plasmid maintenance in S. cerevisiae (7). It is not clear at this point whether Brd4 functions other than that of the physical tethering of E2 to mitotic chromosomes play a role in papillomavirus genome maintenance. Nevertheless, for BPV1, the Brd4/E2 interaction has at least two roles in the viral life cycle: during interphase, it is required to mediate the E2 transcriptional activation function, and during mitosis, it serves to tether the viral genomes to mitotic chromosomes. Given the importance of this interaction for the papillomavirus life cycle, it further highlights the interaction between E2 and Brd4 as a potential target for the development of antiviral therapeutics.
Acknowledgments
We are very grateful to J. Kim, G. Martinez-Noel, A. Nishimura, J. Altreuter, A. Chan, and J. DeMasi in the Howley laboratory for assistance and discussions and to V. A. Erdmann, M. Schweiger, and M. Hirsch-Kauffmann for helpful suggestions. We are grateful to Grace Gill and Karl Münger for a critical reading of the paper and helpful discussions.
This work has been supported by grants (P01CA050661 and R01CA116720) from the National Cancer Institute to P.M.H. M.-R.S. is a fellow of the Studienstiftung des Deutschen Volkes associated with the Free University of Berlin. J.Y. has been supported by a fellowship from Charles A. King Trust, Bank of America, cotrustees.
REFERENCES
- 1.Abroi, A., I. Ilves, S. Kivi, and M. Ustav. 2004. Analysis of chromatin attachment and partitioning functions of bovine papillomavirus type 1 E2 protein. J. Virol. 78:2100-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abroi, A., R. Kurg, and M. Ustav. 1996. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J. Virol. 70:6169-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antson, A. A., J. E. Burns, O. V. Moroz, D. J. Scott, C. M. Sanders, I. B. Bronstein, G. G. Dodson, K. S. Wilson, and N. J. Maitland. 2000. Structure of the intact transactivation domain of the human papillomavirus E2 protein. Nature 403:805-809. [DOI] [PubMed] [Google Scholar]
- 4.Bastien, N., and A. A. McBride. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124-134. [DOI] [PubMed] [Google Scholar]
- 5.Baxter, M. K., M. G. McPhillips, K. Ozato, and A. A. McBride. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J. Virol. 79:4806-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg, M., and A. Stenlund. 1997. Functional interactions between papillomavirus E1 and E2 proteins. J. Virol. 71:3853-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brannon, A. R., J. A. Maresca, J. D. Boeke, M. A. Basrai, and A. A. McBride. 2005. Reconstitution of papillomavirus E2-mediated plasmid maintenance in Saccharomyces cerevisiae by the Brd4 bromodomain protein. Proc. Natl. Acad. Sci. USA 102:2998-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breiding, D. E., F. Sverdrup, M. J. Grossel, N. Moscufo, W. Boonchai, and E. J. Androphy. 1997. Functional interaction of a novel cellular protein with the papillomavirus E2 transactivation domain. Mol. Cell. Biol. 17:7208-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brokaw, J. L., M. Blanco, and A. A. McBride. 1996. Amino acids critical for the functions of the bovine papillomavirus type 1 E2 transactivator. J. Virol. 70:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper, C. S., S. N. Upmeyer, and P. L. Winokur. 1998. Identification of single amino acids in the human papillomavirus 11 E2 protein critical for the transactivation or replication functions. Virology 241:312-322. [DOI] [PubMed] [Google Scholar]
- 11.Del Vecchio, A. M., H. Romanczuk, P. M. Howley, and C. C. Baker. 1992. Transient replication of human papillomavirus DNAs. J. Virol. 66:5949-5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. USA 100:8758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 20:6537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson, M. K., and M. R. Botchan. 1996. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J. Virol. 70:4193-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossel, M. J., F. Sverdrup, D. E. Breiding, and E. J. Androphy. 1996. Transcriptional activation function is not required for stimulation of DNA replication by bovine papillomavirus type 1 E2. J. Virol. 70:7264-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 17.Harris, S. F., and M. R. Botchan. 1999. Crystal structure of the human papillomavirus type 18 E2 activation domain. Science 284:1673-1677. [DOI] [PubMed] [Google Scholar]
- 18.Haugen, T. H., T. P. Cripe, G. D. Ginder, M. Karin, and L. P. Turek. 1987. Trans-activation of an upstream early gene promoter of bovine papilloma virus-1 by a product of the viral E2 gene. EMBO J. 6:145-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 20.Houzelstein, D., S. L. Bullock, D. E. Lynch, E. F. Grigorieva, V. A. Wilson, and R. S. Beddington. 2002. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 22:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howley, P. M., and D. R. Lowy. 2001. Papillomaviruses and their replication, p. 2197-2229. In D. M. Knipe, P.M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Strauss (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 22.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523-534. [DOI] [PubMed] [Google Scholar]
- 24.Jeanmougin, F., J. M. Wurtz, B. Le Douarin, P. Chambon, and R. Losson. 1997. The bromodomain revisited. Trends Biochem. Sci. 22:151-153. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, Y. W., P. Veschambre, H. Erdjument-Bromage, P. Tempst, J. W. Conaway, R. C. Conaway, and R. D. Kornberg. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 95:8538-8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanno, T., Y. Kanno, R. M. Siegel, M. K. Jang, M. J. Lenardo, and K. Ozato. 2004. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell 13:33-43. [DOI] [PubMed] [Google Scholar]
- 27.Kovelman, R., G. K. Bilter, E. Glezer, A. Y. Tsou, and M. S. Barbosa. 1996. Enhanced transcriptional activation by E2 proteins from the oncogenic human papillomaviruses. J. Virol. 70:7549-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Law, M.-F., D. R. Lowy, I. Dvoretzky, and P. M. Howley. 1981. Mouse cells transformed by bovine papillomavirus contain only extrachromosomal viral DNA sequences. Proc. Natl. Acad. Sci. USA 78:2727-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, D., S. G. Hwang, J. Kim, and J. Choe. 2002. Functional interaction between p/CAF and human papillomavirus E2 protein. J. Biol. Chem. 277:6483-6489. [DOI] [PubMed] [Google Scholar]
- 30.Lee, D., B. Lee, J. Kim, D. W. Kim, and J. Choe. 2000. cAMP response element-binding protein-binding protein binds to human papillomavirus E2 protein and activates E2-dependent transcription. J. Biol. Chem. 275:7045-7051. [DOI] [PubMed] [Google Scholar]
- 31.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, R., J. D. Knight, S. P. Jackson, R. Tjian, and M. R. Botchan. 1991. Direct interaction between Sp1 and the BPV enhancer E2 protein mediates synergistic activation of transcription. Cell 65:493-505. [DOI] [PubMed] [Google Scholar]
- 33.McPhillips, M. G., K. Ozato, and A. A. McBride. 2005. Interaction of bovine papillomavirus E2 protein with Brd4 stabilizes its association with chromatin. J. Virol. 79:8920-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohr, I. J., R. Clark, S. Sun, E. J. Androphy, P. MacPherson, and M. R. Botchan. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699. [DOI] [PubMed] [Google Scholar]
- 35.Piirsoo, M., E. Ustav, T. Mandel, A. Stenlund, and M. Ustav. 1996. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 15:1-11. [PMC free article] [PubMed] [Google Scholar]
- 36.Rank, N. M., and P. F. Lambert. 1995. Bovine papillomavirus type 1 E2 transcriptional regulators directly bind two cellular transcription factors, TFIID and TFIIB. J. Virol. 69:6323-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehtanz, M., H. M. Schmidt, U. Warthorst, and G. Steger. 2004. Direct interaction between nucleosome assembly protein 1 and the papillomavirus E2 proteins involved in activation of transcription. Mol. Cell. Biol. 24:2153-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romanczuk, H., F. Thierry, and P. M. Howley. 1990. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J. Virol. 64:2849-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryo, A., Y. C. Liou, G. Wulf, M. Nakamura, S. W. Lee, and K. P. Lu. 2002. PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol. Cell. Biol. 22:5281-5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakai, H., T. Yasugi, J. D. Benson, J. J. Dowhanick, and P. M. Howley. 1996. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J. Virol. 70:1602-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Schreiber, E., P. Matthias, M. M. Müller, and W. Schaffner. 1989. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 17:6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Servant, M. J., B. ten Oever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276:355-363. [DOI] [PubMed] [Google Scholar]
- 43.Settleman, J., and D. DiMaio. 1988. Efficient transactivation and morphologic transformation by bovine papillomavirus genes expressed from a bovine papillomavirus/simian virus 40 recombinant virus. Proc. Natl. Acad. Sci. USA 85:9007-9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spalholz, B. A., P. F. Lambert, C. L. Yee, and P. M. Howley. 1987. Bovine papillomavirus transcriptional regulation: localization of the E2-responsive elements of the long control region. J. Virol. 61:2128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spalholz, B. A., Y. C. Yang, and P. M. Howley. 1985. Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell 42:183-191. [DOI] [PubMed] [Google Scholar]
- 47.Stubenrauch, F., A. M. Colbert, and L. A. Laimins. 1998. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J. Virol. 72:8115-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szymanski, P., and A. Stenlund. 1991. Regulation of early gene expression from the bovine papillomavirus genome in transiently transfected C127 cells. J. Virol. 65:5710-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thierry, F., and M. Yaniv. 1987. The BPV1-E2 trans-acting protein can be either an activator or a repressor of the HPV18 regulatory region. EMBO J. 6:3391-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tiscornia, G., O. Singer, M. Ikawa, and I. M. Verma. 2003. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl. Acad. Sci. USA 100:1844-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, Y. C., H. Okayama, and P. M. Howley. 1985. Bovine papillomavirus contains multiple transforming genes. Proc. Natl. Acad. Sci. USA 82:1030-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19:535-545. [DOI] [PubMed] [Google Scholar]
- 53.Yao, J. M., D. E. Breiding, and E. J. Androphy. 1998. Functional interaction of the bovine papillomavirus E2 transactivation domain with TFIIB. J. Virol. 72:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349-360. [DOI] [PubMed] [Google Scholar]
- 55.You, J., M. R. Schweiger, and P. M. Howley. 2005. Inhibition of E2 binding to Brd4 enhances viral genome loss and phenotypic reversion of bovine papillomavirus-transformed cells. J. Virol. 79:14956-14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeng, L., and M. M. Zhou. 2002. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513:124-128. [DOI] [PubMed] [Google Scholar]
- 57.Zheng, P. S., J. Brokaw, and A. A. McBride. 2005. Conditional mutations in the mitotic chromosome binding function of the bovine papillomavirus type 1 E2 protein. J. Virol. 79:1500-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.zur Hausen, H. 2001. Oncogenic DNA viruses. Oncogene 20:7820-7823. [DOI] [PubMed] [Google Scholar]