Abstract
We have investigated the antiviral mechanism of a phosphorothioate oligonucleotide, ISIS 5652, which has activity against herpes simplex virus (HSV) in the low micromolar range in plaque reduction assays. We isolated a mutant that is resistant to this compound. Marker rescue and sequencing experiments showed that resistance was due to at least one of three mutations in the UL27 gene which result in amino acid changes in glycoprotein B (gB). Because gB has a role in attachment and entry of HSV, we tested the effects of ISIS 5652 at these stages of infection. The oligonucleotide potently inhibited attachment of virus to cells at 4°C; however, the resistant mutant did not exhibit resistance at this stage. Moreover, a different oligonucleotide with little activity in plaque reduction assays was as potent as ISIS 5652 in inhibiting attachment. Similarly, ISIS 5652 was able to inhibit entry of preattached virions into cells at 37°C, but the mutant did not exhibit resistance in this assay. The mutant did not attach to or enter cells more quickly than did wild-type virus. Strikingly, incubation of wild-type virus with 1 to 2 μM ISIS 5652 at 37°C led to a time-dependent, irreversible loss of infectivity (virucidal activity). No virucidal activity was detected at 4°C or with an unrelated oligonucleotide at 37°C. The resistant mutant and a marker-rescued derivative containing its gB mutations exhibited substantial resistance to this virucidal activity of ISIS 5652. We hypothesize that the GT-rich oligonucleotide induces a conformational change in gB that results in inactivation of infectivity.
Oligonucleotides hold considerable promise for treating viral infections. Although much recent attention has focused on small interfering RNAs, the majority of oligonucleotides that have been studied as antiviral agents to date are modified oligodeoxynucleotides (ODNs) designed to work via an antisense mechanism, such as the licensed anticytomegalovirus drug, fomivirsen (2, 3). However, other ODNs with antiviral activity are not complementary to viral nucleic acid (13, 31, 35). These ODNs are GT rich and have the propensity to form G-quartet structures stabilized by non-Watson-Crick guanine-guanine base pairs (reviewed in references 26 and 33). Certain of these GT-rich ODNs have anti-human immunodeficiency virus (HIV) activity and, evidently, target the HIV envelope protein gp120 (12, 35). Others have been reported to exhibit activity against herpes simplex virus (HSV), but their mechanism(s) of action are not known (13).
HSV most commonly causes genital herpes, cold sores, and corneal keratitis and can cause more severe disease, particularly in the immunocompromised (reviewed in reference 32). The virion consists of an icosahedral nucleocapsid containing the ∼150-kbp double-stranded DNA genome, surrounded by a less-ordered layer of proteins called the tegument and a lipid bilayer envelope that contains a number of virus-encoded glycoproteins (reviewed in reference 25). The replication cycle of HSV begins with the initial attachment of virus to heparan sulfate (HS) moieties on the cell surface (34), which is mediated by glycoprotein C (gC) and gB (reviewed in reference 27). Next, gD binds to one of several cellular receptors. This leads, via as-yet-unknown mechanisms, to fusion of the viral envelope with the plasma membrane and entry of the viral capsid and tegument proteins. Although many of the details of viral entry remain poorly understood, it is widely accepted that fusion requires four of the virion glycoproteins: gB, gD, and the heterodimer gH-gL (reviewed in 28 and 29). Pharmacological agents that target glycoproteins may be useful for a better understanding of viral attachment and entry.
In this study, we investigated the effects of a GT-rich ODN on HSV infection. ISIS 5652 (5652) is a 20-base phosphorothioate ODN with the sequence T2(G4T2)3, which has antiviral activity against HSV during plaque reduction assays. Surprisingly, we have found that 5652 causes an irreversible, temperature-dependent loss of infectivity of viral particles that is mediated by gB.
MATERIALS AND METHODS
Cells and viruses.
Vero (African green monkey kidney) cells were obtained from American Type Culture Collection and grown and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum (2% during infection) and 1% penicillin-streptomycin (Invitrogen) at 37°C and 5% CO2. D6 cells, which are derived from Vero cells and complement mutants defective in gB (11), were kindly provided by Prashant Desai (Johns Hopkins Medical School). D6 cells were maintained as Vero cells were, with the addition of 200 μg of G418 (Gibco) per ml to the growth medium during every other passage. HSV type 1 (HSV-1) wild-type strain KOS and the gB-null mutant KΔ4B, also a kind gift from Prashant Desai (11), were propagated on Vero cells or D6 cells, respectively, as previously described (9). During viral infection at 4°C or 20°C, medium was supplemented with 25 mM HEPES buffer (Cellgro).
Oligonucleotides.
The phosphorothioate ODNs 5652 and fomivirsen, also known as ISIS 2922 (2922), were synthesized by Isis Pharmaceuticals and provided through the courtesy of Frank Bennett and Kevin Anderson. The nucleotide sequence of 5652 is 5′-TTGGGGTTGGGGTTGGGGTT-3′. The nucleotide sequence of 2922 is 5′-GCGTTTGCTCTTCTTCTTGCG-3′ (3).
Plaque reduction assays.
Plaque reduction assays, which entail virus titrations in which various concentrations of drugs are added following virus adsorption, were performed as described previously (9) with modifications. Briefly, 200 PFU of virus diluted in medium was added to a Vero cell monolayer and incubated for 90 min at 37°C to allow virus adsorption. Cells were then overlaid with medium containing 1.2% methyl cellulose and various concentrations of ODNs and incubated at 37°C for 2 days. The plates were then fixed and stained, and the plaques were counted.
Selection of 5652-resistant virus.
Selection of a 5652-resistant mutant was accomplished by passaging wild-type KOS in increasing concentrations of 5652 using a protocol similar to that previously used to isolate drug-resistant human cytomegalovirus mutants (4, 21, 30). Briefly, subconfluent Vero cells were pretreated with 1 μM 5652 (approximate 50% effective dose [ED50] in plaque reduction assays) and incubated overnight at 37°C, similar to the protocol used in antiviral assays and mutant selections with 2922 (4, 21). Cells were then washed once with DMEM and infected with wild-type KOS at a multiplicity of infection between 0.01 and 0.1. Virus and cells were then incubated at 37°C for 1 h to allow for viral adsorption. The inoculum was removed, and cells were washed with DMEM, overlaid with fresh medium containing the same concentration of 5652 used during pretreatment, and incubated at 37°C until complete cytopathic effect was observed. Progeny virus was then harvested as previously described (9). This process was repeated using 2, 4, 8, 16, and 32 μM 5652. Following the passage at a concentration of 32 μM, virus was plaque purified three times. The purified mutant virus was named 5652r-1.
Cosmids and plasmids.
Cosmids scKOS 10, 20, 22, 26, and 32 derived from the KOS genome were constructed, mapped, and kindly provided by Jeff Leary (SmithKline Beecham, now GlaxoSmithKline). A library of cosmids of 5652r-1 DNA was constructed following a modification of a previously described protocol (10) by cloning viral DNA into the BamHI site of a SuperCos1 cosmid vector (Stratagene) modified to contain a PacI site. This modified vector (SuperCosP1) was constructed by Anthony Griffiths by annealing complementary oligonucleotides (sequence of one strand, GGCCGAATTCGCGGCGCGGCGCGCCTTAATTAAGGATCCGGCGCGCCTTAATTAAGCGGCCGCGAATTC) and ligating the double-stranded DNA into NotI-digested SuperCos1. Briefly, SuperCosP1 DNA was digested with XbaI, dephosphorylated with calf intestinal alkaline phosphatase, and then digested with BamHI, which resulted in a mixture of 2 fragments, each terminating with a cos site. Viral DNA, prepared as described previously (24) from 5652r-1-infected Vero cells, was partially digested with MboI, dephosphorylated with calf intestinal alkaline phosphatase, and ligated into the prepared SuperCosP1 DNA, according to the manufacturer's instructions. A Gigapack III XL kit (Stratagene) was used to package ligated DNA into phage, and the resulting phage preparation was used to infect Escherichia coli strain VCS257. Ampicillin-resistant clones were characterized by digestion with PacI to release insert, digestion with HindIII for analysis of the location of the cosmid, and Southern blot hybridization using probes from KOS. In this fashion, overlapping cosmids A3, A4, A6, A8, and D6 were identified that span the entire 5652r-1 genome.
To generate pG4.7, cosmid A4 was cleaved with KpnI, and a 4.7-kb KpnI fragment including the entire UL27 (gB) gene was purified and cloned into the vector pGEM7Z (Promega).
Viral genetics.
Mixtures of KOS and 5652r-1 cosmids were cotransfected into Vero cells using Superfect transfection reagent (QIAGEN) according to the manufacturer's instructions. Progeny virus was harvested and tested in plaque reduction assays to determine if the virus was sensitive or resistant to 5652.
To determine if mutations in gB were sufficient to confer the resistant phenotype, two marker-rescued derivatives were constructed. K/B was obtained by cotransfecting D6 cells with KΔ4B infectious DNA, prepared from D6 cells as previously described (18), and the wild-type cosmid scKOS20, which contains gB, using Effectene transfection reagent (QIAGEN). 5/B was constructed in a similar fashion by cotransfection of KΔ4B DNA and the mutant plasmid pG4.7. In each case, virus was harvested from the transfected cells and titrated on both D6 and Vero cells to confirm that the replication deficiency of KΔ4B had been rescued. Plaques that formed on Vero cells were picked and amplified. Each virus was plaque purified a total of three times on Vero cells. Viral, cosmid, and plasmid DNAs were sequenced following amplification of sequences including gB using PCR (100 ng template DNA, 160 μM concentrations of each deoxynucleoside triphosphate [NEB], 2.4 μM concentrations of each primer, 6.25% dimethyl sulfoxide [Sigma], 1× Pfu buffer [Stratagene], 2.5 U Pfu Turbo DNA polymerase [Stratagene]) with the following primers: 5′-CCTTCGACGTGGAGGAAG, which is ∼180 bp from the poly(A) site of UL27, and 5′-CGTGGTCTACGACCGAGACGT, which is ∼80 bp from the UL27 translation start codon. The PCR product was sequenced (Molecular Biology Core Facilities, Dana-Farber Cancer Institute, Boston, MA) and compared to a PCR product that was amplified from HSV-1 strain KOS DNA in the same manner. Sequencing confirmed that 5/B contained the three gB mutations found in 5652r-1 and pG4.7 and no other gB mutations, while K/B contained no changes in gB relative to KOS and scKOS20.
Attachment assays.
The effects of compounds on attachment were assessed using a modification of a previously described assay (19). Briefly, 200 PFU of virus and various concentrations of ODNs or heparin (Sigma) were added to a 4°C prechilled Vero cell monolayer and incubated at 4°C for 3 h, unless otherwise indicated. Cells were then washed with phosphate-buffered saline (PBS) three times to remove compounds and unattached virus, overlaid with medium containing 1.2% methylcellulose, and incubated at 37°C for 2 days. Plates were then fixed and stained, and the plaques were counted. To confirm that incubation at 4°C allowed only viral attachment and not entry, cells to which virus had been preattached at 4°C were treated with acidic glycine (0.1 M glycine, 0.14 M NaCl, pH 3) to inactivate virus. This reproducibly resulted in 100% inhibition of plaque formation (data not shown).
Entry assay.
To assess the effect of compounds on entry, a modification of a previously described assay (19) was used. Briefly, 200 PFU of virus was added to a 4°C prechilled Vero cell monolayer and incubated at 4°C for 3 h to allow for viral attachment. Cells were then washed with PBS three times to remove unattached virus. To assay the effect of 5652 on entry, various concentrations of 5652, diluted in medium, were added to the cells, and the temperature was shifted to 37°C for 2 h prior to inactivation of extracellular virus. To measure the time course of virus entry, cells were incubated in the absence of inhibitor at 37°C for various times prior to inactivation of extracellular virus. To inactivate extracellular virus (and remove any 5652 in the entry assay), 1 ml of acidic glycine was added to each cell monolayer and incubated at 20°C for 60 s. Cells were then subsequently washed with PBS three times to return the pH to neutral, overlaid with medium containing 1.2% methylcellulose, and incubated at 37°C for 2 days. In parallel, the same amount of virus was allowed to attach to cells for 3 h at 4°C as above, and the monolayer was overlaid with methylcellulose containing medium; the number of plaques produced was set at 100%. Plates were then fixed and stained, and the plaques were counted.
Virucidal assay.
Virus was diluted in PBS to a concentration of 5 × 106 PFU/ml. Various concentrations of ODN were then added, and the virus-ODN sample was then incubated at either 4, 20, or 37°C for various lengths of time. After incubation, the sample was diluted with medium to reduce the concentration of ODN to those not active in attachment or plaque reduction assays. Virus was then titrated on Vero cells. To determine if the loss of infectivity was reversible, samples were initially brought through the virucidal assay. After diluting out the ODN, the sample was reincubated at 37°C. After the indicated amount of time, these samples were titrated on Vero cells.
RESULTS
5652 inhibits HSV plaque formation.
We began investigating the anti-HSV effects of 5652 by performing plaque reduction assays using the wild-type HSV-1 strain KOS. For 5652, the reduction of plaques was dose dependent, and the dose that reduced plaque formation by 50% (ED50) was 1 to 2 μM (Fig. 1). Very similar results were obtained in plaque reduction assays using the wild-type HSV-2 strain HG52 (data not shown). In contrast, a different ODN, 2922, was much less active with an ED50 of >15 μM (supplemental Fig. 1 available at http://coen.med.harvard.edu).
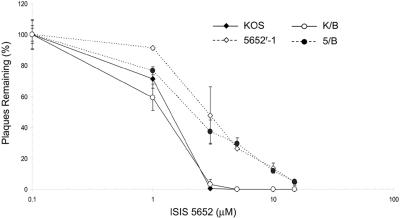
FIG. 1.
Susceptibilities of viruses to 5652. Plaque reduction assays were performed with wild-type KOS (filled diamonds), 5652r-1 (open diamonds), K/B, the marker-rescued derivative containing wild-type gB (open circles), and 5/B, the marker-rescued derivative containing the three gB mutations of 5652r-1 (closed circles). Results presented are the means of a single experiment done in duplicate and are representative of three independent experiments. Error bars represent standard errors of the means, unless these standard errors were too small to be visible on the plots.
Isolation of a 5652-resistant mutant.
Drug-resistant mutants can be invaluable in understanding the mechanism of action of antiviral drugs (8). We serially passaged KOS in increasing concentrations of 5652, as described in Materials and Methods, and obtained a 5652-resistant virus, 5652r-1. In plaque reduction assays, 5652r-1 reproducibly exhibited modest resistance to 5652, with an ED50 approximately twofold higher than that of KOS (Fig. 1). Applying a paired t test to the data in Fig. 1 and those from two other independent experiments, the ED50 for the mutant was significantly different from that of KOS (P = 0.00073).
Mutation(s) in UL27 confer resistance to 5652.
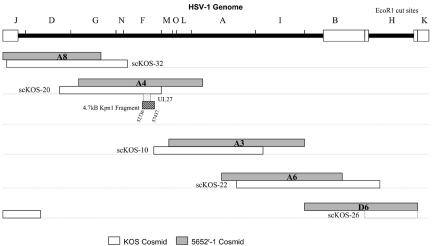
To map the resistance mutation(s) in 5652r-1, we constructed a cosmid library spanning the 5652r-1 genome (Fig. 2). Cotransfection of these cosmids into Vero cells resulted in virus that was resistant to 5652 (supplemental Fig. 2 available at http://coen.med.harvard.edu). We were kindly provided with a corresponding set of cosmids from KOS (Fig. 3). Unfortunately, this set, when cotransfected, did not yield infectious virus. However, when cotransfected with 5652r-1 cosmid D6, infectious virus that was sensitive to 5652 was produced (supplemental Fig. 2 available at http://coen.med.harvard.edu). We then performed cotransfections in which each KOS cosmid was replaced by the corresponding cosmid from 5652r-1. When 5652r-1 cosmid A4 replaced KOS cosmid scKOS-20, the virus that was produced was resistant to 5652 in plaque reduction assays (supplemental Fig. 2 available at http://coen.med.harvard.edu), suggesting that the resistance mutation(s) were contained within the A4 sequences.
FIG. 2.
Locations of wild-type KOS and 5652r-1 cosmids relative to the HSV-1 genome. The HSV-1 genome is depicted in the top line in the prototype arrangement with repeated sequences shown as open boxes and unique sequences as thick, bold lines. EcoRI restriction fragments are indicated above the line. Below this are shown the locations of matched pairs of cosmids, with KOS-derived cosmids (indicated with the scKOS prefix) shown as open bars and the 5652r-1 cosmids (A8, A4, A3, A6, and D6) shown as shaded bars. The location of the UL27 gene is shown as a stippled box below the A4 and scKOS-20 cosmids. Below that, the 4.7-kb KpnI fragment cloned into pG4.7 is shown as a hatched box. The nucleotide location of this fragment relative to the strain 17 sequence (20) is indicated.
FIG. 3.
Effects on attachment. (A) 5652r-1 (dotted line and open diamonds) or KOS (solid line and closed diamonds sometimes hidden behind other points) was attached to cells at 4°C in the presence of various concentrations of 5652. Also, KOS was attached to cells in the presence of various concentrations of 2922 (open squares). Unattached virus was removed, media containing methylcellulose was added, the cells were incubated at 37°C, and plaques were allowed to form. The results presented are the means of a single experiment done in duplicate and are representative of three independent experiments. (B) 5652r-1 (open diamonds) or KOS (closed diamonds) was tested for inhibition of attachment by various concentrations of heparin as described for panel A. (C) 5652r-1 (open diamonds) or KOS (closed diamonds) was allowed to attach to cells at 4°C in the absence of drug for various times prior to removal of unattached virus as described for panel A. The percentage of virus attached was determined by comparison to the number of plaques obtained in a parallel assay in which the same amount of virus was added to cells under the conditions of a plaque reduction assay. The results for the attachment assay with heparin (B) and the time course assay (C) are presented as the means of a single experiment done in duplicate and are representative of two independent experiments. For all assays, error bars represent standard errors of the means unless these standard errors were too small to be visible on the plots.
Because investigators at Isis Pharmaceuticals had obtained data suggesting that 5652 acts early in the HSV infectious cycle (K. Anderson, personal communication) and because GT-rich ODNs evidently target envelope glycoproteins of HIV (12, 35), we sequenced glycoprotein-encoding genes in cosmid A4. The gH (UL22) gene contained no mutations. However, the gB (UL27) gene contained three mutations which would result in amino acid changes in the extracellular domain of gB: V571I, S668N, and E687G.
To determine whether the gB mutations conferred resistance of 5652r-1, we took advantage of a gB deletion mutant virus, KΔ4B, kindly provided by Prashant Desai (11). This mutant does not form plaques on Vero cells but can replicate in complementing D6 cells. Cotransfection of infectious DNA from KΔ4B with a plasmid containing the gB gene from 5652r-1 resulted in progeny virus that could form plaques on Vero cells. One such marker-rescued derivative was isolated, further plaque purified, and sequenced to confirm that it contained the 5652r-1 gB mutations. This virus, 5/B, exhibited resistance to 5652, similar to that of 5652r-1 (Fig. 1). In contrast, a marker-rescued derivative, K/B, constructed by cotransfecting wild-type gB sequences with infectious DNA from KΔ4B exhibited sensitivity to 5652, similar to that of KOS (Fig. 1). Thus, one or more gB mutations in mutant 5652r-1 confer resistance to 5652.
5652 resistance is not manifest at the stage of attachment of virus.
Because gB has a role in both the attachment and entry stages of viral infection (reviewed in references 27 and 28), we investigated whether 5652 was involved in inhibiting one or both of these two processes. To investigate attachment, we tested the inhibition of plaque formation when 5652 was present only during viral attachment at 4°C. 5652 was able to inhibit viral attachment approximately 10 times more potently than it inhibited plaque formation during plaque reduction assays (Fig. 3A) (ED50 of 0.15 μM for KOS during the attachment assay). However, 5652r-1 did not exhibit resistance at this stage (Fig. 3A). Moreover, 2922, which exhibited little activity or antiviral activity in plaque reduction assays, was as potent as 5652 in the attachment assay (Fig. 3A).
To investigate the attachment phase further, we assayed the susceptibility of viral attachment to heparin. During initial attachment, gB binds to HS moieties located on cell surface proteoglycans. Heparin, a polyanionic molecule chemically similar to HS, has been shown to inhibit attachment of HSV (22, 34). As expected, then, heparin inhibited the attachment of KOS; however, we did not observe a difference in sensitivity to heparin between KOS and 5652r-1 (Fig. 3B). Thus, the gB mutations in 5652r-1 do not appear to affect the sensitivity of HSV to ODNs or to heparin during the stage of attachment of virus to cells.
We considered the possibility that the mutant might be resistant by being able to attach to cells more rapidly than does KOS. To test this possibility, we assayed the time course of attachment for both viruses. Wild-type and mutant virus attached to cells with very similar kinetics (Fig. 3C). Thus, the gB mutations do not appear to affect attachment of the virus, at least during the initial infection.
5652 resistance is not manifest at the stage of entry.
To test the effect of 5652 during viral entry, we preattached virus to cells at 4°C in the absence of drug, added various concentrations of 5652, and then incubated the mixtures at 37°C for 2 h to permit entry. After inactivating virus that had not entered with acidic glycine, plaques were allowed to form for 2 days and then counted. 5652 was able to inhibit the entry of KOS at 37°C potently (ED50, ∼0.5 μM), but again, 5652r-1 did not exhibit resistance at this stage (Fig. 4A).
FIG. 4.
Effects on entry. Wild-type KOS (closed diamonds) or 5652r-1 (open diamonds) was allowed to attach to cells at 4°C, unattached virus was removed, and the cells were incubated at 37°C in various concentrations of 5652 for 2 h (A) or in the absence of drug for various lengths of time (B). At the end of each time period, virus that had not entered was inactivated, medium containing methylcellulose was added, and plaques were allowed to form. For both assays, the results are presented as the means of a single experiment done in duplicate and are representative of two independent experiments. Error bars represent standard errors of the means unless these standard errors were too small to be visible on the plots.
To test whether the mutant might be resistant by being able to enter cells more rapidly than does KOS, we preattached wild-type or mutant virus to cells and then incubated the mixtures for various times prior to inactivating extracellular virus with acidic glycine. The two viruses entered cells with very similar kinetics (Fig. 4B). Thus, the gB mutations do not appear to affect entry of the virus during initial infection.
5652 has virucidal activity.
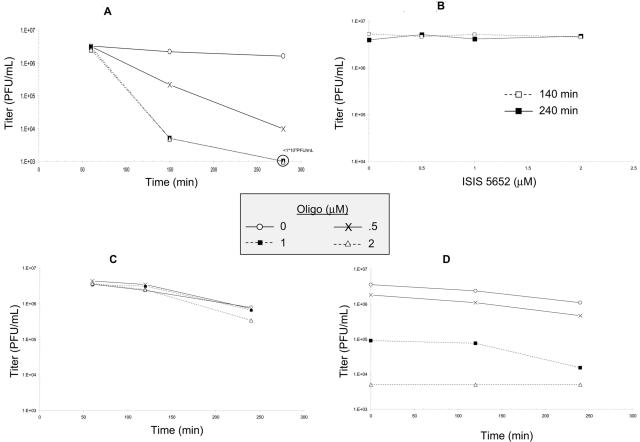
To further investigate the mechanism of action of 5652, we explored the possibility that 5652 was inactivating infectious HSV particles. KOS was incubated with various concentrations of ODNs at 4, 20, or 37°C for various amounts of time. After incubation, the samples were diluted to reduce ODN concentrations well below those that affect virus attachment, and the virus was titrated on Vero cells. Remarkably, we saw a ≥1,000-fold drop in titer when KOS and 5652 were incubated with 1 or 2 μM at 37°C (Fig. 5A), while there was no detectable loss of infectivity at 4°C (Fig. 5B) or 20°C (data not shown). Incubation of the control ODN, 2922, and KOS at 37°C led to little or no loss in infectivity (Fig. 5C).
FIG. 5.
Virucidal activity of 5652. KOS was incubated at 37°C for various lengths of time with either no ODN (open circles), 0.5 μM (×), 1 μM (closed squares), or 2 μM (open diamonds) 5652 (A and D) or 2922 (C). For panels A and C, virus (initial titer of 4.2 × 106 PFU/ml) was diluted to reduce the concentration of 5652 below those that inactivate virus and inhibit virus attachment and titrated on Vero cells. For panel D, following dilution, the virus was then reincubated for various amounts of time (on the x axis) before titration. (B) KOS was incubated at 4°C in various concentrations of 5652 for either 140 min (open squares) or 240 min (closed squares), diluted, and titrated on Vero cells.
To determine if the inactivation of KOS with 5652 was reversible, samples were treated with 5652 for 4 h at 37°C, diluted to reduce the concentrations of 5652 below those that inactivate virus and inhibit virus attachment, and then reincubated at 37°C for various amounts of time. None of the samples regained infectivity (Fig. 5D). Thus, 5652 exhibits time-dependent, temperature-dependent, irreversible inactivation of infectious HSV; i.e., virucidal activity.
Resistance to virucidal activity due to mutation(s) in gB.
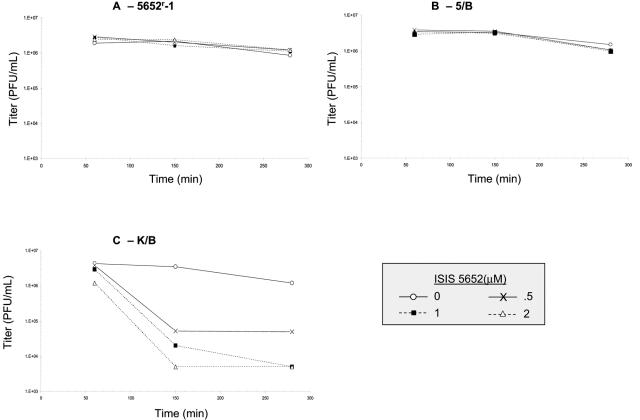
We then tested whether the 5652r-1 mutant was resistant to the virucidal activity of 5652. Even when incubated at 37°C for nearly 5 h in 2 μM 5652, no loss of infectivity was observed (Fig. 6A). To test whether this resistance to the virucidal activity was due to the gB mutation(s), we tested the marker-rescued derivatives 5/B and K/B. Like 5652r-1, 5/B, which contains the three gB mutations, was not detectably inactivated by 5652 (Fig. 6B). However, K/B, the derivative containing wild-type gB, was drastically inactivated by the ODN (Fig. 6C). Thus, the gB mutations confer resistance to the virucidal activity of 5652.
FIG. 6.
UL27 mutations confer resistance to virucidal activity of 5652. 5652r-1 (A), 5/B, which contains the 5652r-1UL27 gene (B), or K/B, which contains the KOS UL27 gene (C), was incubated as described for Fig. 5 with either no ODN (open circles), 0.5 μM (×), 1 μM (closed squares), or 2 μM (open diamonds) 5652. Following dilution, the viruses (initial titers, ∼4 × 106 PFU/ml) were titrated on Vero cells.
DISCUSSION
We investigated the mechanism of action of a phosphorothioate ODN, 5652. This investigation was greatly abetted by the isolation of a mutant resistant to this ODN. Although 5652r-1 exhibited only ∼2-fold resistance in plaque reduction assays, it was highly resistant to the virucidal action of the ODN, exhibiting no inactivation under conditions where the titer of KOS was reduced ≥1,000-fold. Because resistance to the activities of 5652 in plaque reduction and virucidal assays was due to at least one of three gB mutations in the mutant, we conclude that wild-type gB is necessary for the full extent of these activities. Below, we discuss the effects of 5652 and the gB mutations on virus attachment and entry, how 5652 could mediate virucidal activity, and how the mutations might confer resistance to this activity.
Effects of 5652 on attachment and entry.
Although 5652 is more potent in inhibiting viral attachment at 4°C than in plaque reduction assays, it is not at all clear that the former activity contributes to the latter. The plaque reduction assays entail adsorption of the virus to the monolayer in the absence of drug for more than 1 h at 37°C, by which time most virus could attach and enter. A control ODN, 2922, inhibited attachment just as potently as did 5652 but had little or no activity in plaque reduction assays. Similarly, heparin, which is another polyanionic molecule that inhibits virus attachment, also exhibited little or no activity in our plaque reduction assays (data not shown), consistent with its being substantially less potent and efficacious in inhibiting viral spread than attachment (1, 22). Thus, it appears that large, polyanionic compounds such as oligonucleotides and heparin do not interrupt attachment of virus during cell-to-cell spread. The mutant virus, 5652r-1, did not exhibit resistance at the stage of attachment, nor did it exhibit an increased rate of attachment that could explain its resistance. The resistance mutations do not cause substitutions in a basic stretch of gB that has been proposed to mediate heparin sensitivity and attachment to HS moieties (17). We cannot exclude the possibility that inhibition of attachment contributes to the activity of 5652 in plaque reduction assays, but we do not think this is likely.
5652 inhibits viral entry somewhat more potently than it does plaque reduction, but the mutant did not exhibit resistance at this stage, nor did it exhibit an increased rate of entry. The V571I mutation of 5652r-1 is near but distinct from gB mutations shown to affect rate of entry (5, 15). Nevertheless, it may be that the antientry activity of 5652 accounts for much of its activity in plaque reduction assays. This may also explain why the mutant exhibits only very modest resistance in those assays. One might wonder why the mutant did not exhibit resistance in the entry assay, as this assay entails incubation of virus at 37°C with drug. However, virucidal activity requires higher concentrations than those that inhibit entry. Moreover, most viruses have entered cells by the time virucidal activity is evident.
gB-mediated virucidal activity.
Several possible mechanisms by which 5652 inactivates viral infectivity, such that mutations in gB prevent this virucidal activity, can be envisioned. One possibility is a direct interaction between 5652 and gB causing a conformational change in gB that results in loss of infectivity. The conformational change could result in such tight binding of 5652 to gB as to be irreversible, or the conformational change could irreversibly result in gB losing function. This latter possibility is especially intriguing, given that glycoproteins of other viruses that mediate viral entry undergo such conformational changes when they “spring open” and then close back to mediate membrane fusion. Once the changes have occurred, the virion is no longer infectious (reviewed in references 14 and 36). In this case, the resistance mutations could either lower the affinity of gB for 5652 or prevent the conformational change. However, this latter option would imply differences between how conformational changes occur upon 5652 binding and how such changes occur during the initial entry of virus into cells, as wild-type virus and 5652r-1 enter cells with similar kinetics. Another possible scenario is that 5652 is interacting with a virion component other than gB, possibly another glycoprotein, which in turn interacts with gB. In this case, the gB mutations might alter the interaction with the other virion component or a conformational change.
All three of the gB mutations in 5652r-1 cause substitutions in the extracellular domain of gB. In particular, they affect a region (residues 463 to 791) that was retained in a series of gB nonsense and deletion mutants that inhibited the ability of a wild-type gB plasmid to complement a gB null mutant virus (6). This result has been interpreted as implying that this region is required for oligomerization of gB. Two of the mutations (S668N and E687G) are also within a smaller region (residues 596 to 711) identified in coimmunoprecipitation experiments as mediating oligomerization (16). Thus, in the scenarios above, it is possible that 5652 acts to disrupt gB oligomers directly or indirectly. It is also possible that the resistance mutations could stabilize oligomers against disruption. The various scenarios suggest testable predictions.
There has been some interest in developing virucidal agents against HSV, particularly in combination with virucidal agents against HIV, for preventing genital transmission of these two viruses, especially given the evidence that HSV infections can increase the likelihood of HIV infection (reviewed in reference 7). In this context, it is interesting that ODNs similar to 5652 have anti-HIV activity (23, 31, 35). Although 5652 would be limited by its size, expense, and relatively slow time course of activity, additional studies of this compound may facilitate development of virucidal therapies against HSV as well as further insights into viral glycoproteins and entry.
Acknowledgments
We are grateful to Kevin Anderson and Frank Bennett for supplying ODNs and for encouragement and helpful advice, Prashant Desai for generous provision of KΔ4B and D6 cells, Jeff Leary for kindly providing the scKOS cosmids and communicating unpublished results, and Anthony Griffiths for graciously providing the SuperCosP1 vector and for helpful advice and discussions.
This work was supported by NIH grant A126077 and a grant from Isis Pharmaceuticals.
REFERENCES
- 1.Aguilar, J. S., M. Rice, and E. K. Wagner. 1999. The polysulfonated compound suramin blocks adsorption and lateral diffusion of herpes simplex virus type-1 in Vero cells. Virology 258:141-151. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, K. P., M. C. Fox, V. Brown-Driver, M. J. Martin, and R. F. Azad. 1996. Inhibition of human cytomegalovirus immediate-early gene expression by an antisense oligonucleotide complementary to immediate-early RNA. Antimicrob. Agents Chemother. 40:2004-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azad, R. F., V. B. Driver, K. Tanaka, R. M. Crooke, and K. P. Anderson. 1993. Antiviral activity of a phosphorothioate oligonucleotide complementary to RNA of the human cytomegalovirus major immediate-early region. Antimicrob. Agents Chemother. 37:1945-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biron, K. K., J. A. Fyfe, S. C. Stanat, L. K. Leslie, J. B. Sorrell, C. U. Lambe, and D. M. Coen. 1986. A human cytomegalovirus mutant resistant to the nucleoside analog, 9-[(2-hydroxy-1-(hydroxymethyl)ethoxy)methyl]guanine (BW B759U) induces reduced levels of BW B759U triphosphate. Proc. Natl. Acad. Sci. USA 83:8769-8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185-190. [DOI] [PubMed] [Google Scholar]
- 6.Cai, W., S. Person, C. DebRoy, and B. Gu. 1988. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1: an analysis of linker insertion mutants. J. Mol. Biol. 201:575-588. [DOI] [PubMed] [Google Scholar]
- 7.Celum, C., R. Levine, M. Weaver, and A. Wald. 2004. Genital herpes and human immunodeficiency virus: double trouble. Bull. WHO 82:447-453. [PMC free article] [PubMed] [Google Scholar]
- 8.Coen, D. M. 1991. The implications of resistance to antiviral agents for herpesvirus drug targets and drug therapy. Antivir. Res. 15:287-300. [DOI] [PubMed] [Google Scholar]
- 9.Coen, D. M., H. E. Fleming, Jr., L. K. Leslie, and M. J. Retondo. 1985. Sensitivity of arabinosyladenine-resistant mutants of herpes simplex virus to other antiviral drugs and mapping of drug hypersensitivity mutations to the DNA polymerase locus. J. Virol. 53:477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham, C., and A. J. Davison. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:116-124. [DOI] [PubMed] [Google Scholar]
- 11.Desai, P., F. L. Homa, S. Person, and J. C. Glorioso. 1994. A genetic selection method for the transfer of HSV-1 glycoprotein mutations from plasmid to the viral genome: preliminary characterization of transdominance and entry kinetics of mutant viruses. Virology 204:312-322. [DOI] [PubMed] [Google Scholar]
- 12.Esté, J. A., C. Cabrera, D. Schols, P. Cherepanov, A. Gutierrez, M. Witvrouw, C. Pannecouque, Z. Debyser, R. F. Rando, B. Clotet, J. Desmyter, and E. De Clercq. 1998. Human immunodeficiency virus glycoprotein gp120 as the primary target for the antiviral activity of AR177 (Zintevir). Mol. Pharmacol. 53:340-345. [DOI] [PubMed] [Google Scholar]
- 13.Fennewald, S. M., S. Mustain, J. Ojwang, and R. F. Rando. 1995. Inhibition of herpes simplex virus in culture by oligonucleotides composed entirely of deoxyguanosine and thymidine. Antivir. Res. 26:37-54. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, S. C. 2001. Principles of virus structure, p. 53-85. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 15.Highlander, S. L., D. J. Dorney, P. J. Gage, T. C. Holland, W. Cai, S. Person, M. Levine, and J. C. Glorioso. 1989. Identification of mar mutations in herpes simplex virus type 1 glycoprotein B which alter antigenic structure and function in virus penetration. J. Virol. 63:730-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Highlander, S. L., W. F. Goins, S. Person, T. C. Holland, M. Levine, and J. C. Glorioso. 1991. Oligomer formation of the gB glycoprotein of herpes simplex virus type 1. J. Virol. 65:4275-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laquerre, S., R. Argnani, D. B. Anderson, S. Zucchini, R. Manservigi, and J. C. Glorioso. 1998. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J. Virol. 72:6119-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLean, A. R. 1998. Preparation of HSV-DNA and production of infectious virus, p. 19-25. In S. M. Brown and A. R. MacLean (ed.), Herpes simplex virus protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 19.MacLean, C. A. 1998. HSV entry and spread, p. 9-18. In S. M. Brown and A. R. MacLean (ed.), Herpes simplex virus protocols. Humana Press, Totowa, N.J.
- 20.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 21.Mulamba, G. B., A. Hu, R. F. Azad, K. P. Anderson, and D. M. Coen. 1998. Human cytomegalovirus mutant with sequence-dependent resistance to the phosphorothioate oligonucleotide fomiversen (ISIS 2922). Antimicrob. Agents Chemother. 42:971-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nahmias, A. J., and S. Kibrick. 1964. Inhibitory effect of heparin on herpes simplex virus. J. Bacteriol. 87:1060-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ojwang, J. O., R. W. Buckheit, Y. Pommier, A. Mazumder, K. De Vreese, J. A. Esté, D. Reymen, L. A. Pallansch, C. Lackman-Smith, T. L. Wallace, E. De Clercq, M. S. McGrath, and R. F. Rando. 1995. T30177, an oligonucleotide stabilized by an intramolecular guanosine octet, is a potent inhibitor of laboratory strains and clinical isolates of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 39:2426-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parris, D. S., R. A. F. Dixon, and P. A. Schaffer. 1980. Physical mapping of herpes simplex virus type 1 ts mutants by marker rescue: correlation of the physical and genetic maps. Virology 100:275-287. [DOI] [PubMed] [Google Scholar]
- 25.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 26.Shafer, R. H., and I. Smirnov. 2001. Biological aspects of DNA/RNA quadruplexes. Biopolymers 56:209-227. [DOI] [PubMed] [Google Scholar]
- 27.Shukla, D., and P. G. Spear. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Investig. 108:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spear, P. G. 1993. Entry of alphaherpesviruses into cells. Semin. Virol. 4:167-180. [Google Scholar]
- 29.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan, V., and D. M. Coen. 1991. Isolation of foscarnet-resistant human cytomegalovirus: patterns of resistance and sensitivity to other antiviral drugs. J. Infect. Dis. 164:781-784. [DOI] [PubMed] [Google Scholar]
- 31.Tondelli, L., F. P. Colonna, A. Garbesi, S. Zanella, M. E. Marongiu, S. Corrias, A. G. Loi, and P. La Colla. 1996. Native oligodeoxynucleotides specifically active against human immunodeficiency virus type 1 in vitro: a G-quartet-driven effect? Antimicrob. Agents Chemother. 40:2034-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitley, R. J. 2001. Herpes simplex viruses, p. 2461-2509. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 33.Williamson, J. R. 1994. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 23:703-730. [DOI] [PubMed] [Google Scholar]
- 34.WuDunn, D., and P. G. Spear. 1989. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J. Virol. 63:52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyatt, J. R., T. A. Vickers, J. L. Roberson, R. W. J. Buckheit, T. Klimkait, E. DeBaets, P. W. Davis, B. Rayner, J. L. Imbach, and D. J. Ecker. 1994. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human immunodeficiency virus envelope-mediated cell fusion. Proc. Natl. Acad. Sci. USA 91:1356-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young, J. A. T. 2001. Virus entry and uncoating, p. 87-103. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.