Abstract
During a productive infection, the prototype strain of the parvovirus minute virus of mice (MVMp) induces dramatic morphological alterations in permissive A9 fibroblasts, culminating in cell lysis at the end of infection. These cytopathic effects (CPE) result from rearrangements and destruction of the cytoskeletal micro- and intermediate filaments, while other structures such as the nuclear lamina and particularly the microtubule network remain protected throughout the infection (J. P. F. Nüesch et al., Virology 331:159-174, 2005). In order to unravel the mechanism(s) by which parvoviruses trigger CPE, we searched for NS1 interaction partners by differential affinity chromatography, using distinct NS1 mutants debilitated specifically for this function. Thereby, we isolated an NS1 partner polypeptide, whose interaction with NS1 correlated with the competence of the viral product for CPE induction, and further identified it by tandem mass spectrometry and Western blotting analyses to consist of the catalytic subunit of casein kinase II, CKIIα. This interaction of NS1 with CKIIα suggested interference by the viral protein with intracellular signaling. Using permanent cell lines expressing dominant-negative CKIIα mutants, we were able to show that this kinase activity was indeed specifically involved in parvoviral CPE and progeny particle release. Furthermore, the NS1/CKIIα complex proved to be able to specifically phosphorylate viral capsids, indicating a mediator function of NS1 for CKII activity and specificity, at least in vitro. Altogether our data suggest that parvovirus-induced CPE is mediated by NS1 interference with intracellular CKII signaling.
The autonomous parvovirus minute virus of mice (MVMp) consists of a small icosahedral nonenveloped particle with a single-stranded 5.1-kb linear DNA as a genome. This DNA codes for two structural (VP) proteins and at least four nonstructural (NS) proteins. Among the nonstructural regulatory proteins, only the large 83-kDa polypeptide NS1 is essential for productive infection of all cells, while the three 24-kDa NS2 polypeptides are dispensable in some permissive cell lines. A productive infection of mouse A9 fibroblasts with MVMp culminates in cell lysis and progeny virus release (for a review, see reference 8). The NS1 protein was found to be endowed with cytotoxic function(s) and to be sufficient to alter the morphology of the host cells (6) and eventually cause their death (3). Interestingly, this activity was potentiated as a result of transformation of the target cell with oncogenes (24). The mechanisms of NS1-induced cytotoxicity and the processes of cell lysis and release of infectious progeny particles, however, are poorly understood.
MVMp infection of A9 cells leads to characteristic alterations of the host cell morphology, which may facilitate virus replication and the eventual release of progeny particles. Early during infection, subnuclear structures termed APAR bodies are formed, which serve as replication centers for viral DNA amplification (2, 11). At later time points, MVM also induces significant changes within the cytoplasm, leading to a collapse of the infected A9 cells, which round up and detach from the culture dish prior to cytolysis (3). These morphological alterations, termed cytopathic effects (CPE), could be assigned to the activity of NS1 (6) and were shown to mainly affect micro- and intermediate filaments of the cytoskeleton network, while the nuclear lamina and microtubules remained intact throughout infection (32).
The 83-kDa regulatory protein NS1 is a key player in virus propagation. It is endowed with many properties and enzymatic activities, including ATP binding and hydrolysis, oligomerization, site-specific DNA binding to a cognate recognition motif, site- and strand-specific nicking, helicase function, and the capacity for promoter trans regulation. This allows this multifunctional protein to control a variety of processes necessary for progeny particle production, including viral DNA amplification and gene expression regulation (reviewed in reference 25). Some of the latter functions may also take the cellular genome as a target and may contribute in this way to the cytotoxic activities of NS1 (19). Moreover, the ability of NS1 to physically interact with a variety of host cell proteins, including members of the DNA replication machinery (5) and transcriptional regulators (15, 20), may represent another “scavenging” mechanism through which NS1 negatively interferes with essential cellular processes. In particular, it has been postulated that NS1 interferes with intracellular signaling, thereby accounting for the changes observed in the phosphorylation pattern during infection, not only of NS1 itself (7), but also of host cell factors such as tubulin (32) and tubulin-associated-protein (1). Little is known about the molecular mechanisms of NS1-induced cell perturbation. Interestingly, recent investigations analyzing NS1 regulation by phosphorylation for “late” toxic functions showed that the cytopathic activity of NS1 could be dissociated, at least in part, from its roles in virus production (6, 12). These observations suggest that NS1 is endowed with a genuine cytotoxic function(s) which is likely to actively take part in progeny virus release and spreading, rather than being a mere side effect of the virus multiplication process.
To further assess this possibility and to determine molecular mechanisms underlying NS1-induced cytotoxicity, we took advantage of previous reports analyzing biochemically characterized NS1 mutants for their cytotoxic potential, particularly their ability to induce morphological alterations in the host cell (6, 12, 17). To be competent for CPE induction in permissive A9 cells, MVMp NS1 needs an intact ATP-binding site and it has to oligomerize. In addition, mutagenesis at distinct potential or known protein kinase C (PKC) phosphorylation sites debilitates NS1 for CPE induction. Interestingly, an entirely nuclear NS1 variant was found to lose its toxic activity, while keeping the ability to drive viral DNA amplification (17, 27, 30). Therefore, the viral protein may be regulated for CPE induction through both posttranslational modifications and intracellular distribution. The finding that only limited enzymatic activities of NS1 were required for CPE induction and the observation that CPE-negative mutants clustered in distinct areas within the NS1 coding sequence (6) suggested that some of the multiple, yet discrete, NS1 regions involved in cytotoxicity may represent domains through which the viral product interacts with cellular polypeptides. To isolate partner proteins involved in toxicity and to obtain sufficient amounts of these proteins for identification, we developed a purification procedure ending in differential affinity chromatography steps using wild-type and mutant phosphorylated NS1 proteins which were associated in constitutive dimers due to an N-terminally fused glutathione S-transferase (GST) polypeptide (27). Thereby, cellular proteins were sought, which had no or little affinity for a given NS1 nontoxic mutant, while interacting with the wild-type or otherwise mutated NS1 polypeptide. This technique led to the identification of casein kinase II alpha (CKIIα), which specifically interacts with the NS1 helicase domain so that the disruption of the complex correlates with the loss of NS1 cytotoxicity. Moreover, the ability of NS1-CKIIα to target novel phosphorylation of MVM capsids suggests that NS1 could mediate the observed cytoskeleton filament alterations by interfering with host cell CKII signaling.
MATERIALS AND METHODS
Antibodies and reagents.
Antibodies specific for tropomyosin were purchased from Sigma (T3651 and T2780), and those for vimentin and CKIIα were purchased from Santa Cruz Biotechnology (S-20 and C-18). Rabbit antiserum recognizing the viral nonstructural protein NS1 (anti-NS1C) (9) was previously obtained using specific peptides corresponding to the 16 C-terminal-end amino acids of NS1, while the monoclonal anticapsid antibodies EIIF3 (kindly provided by Tattersall and Cotmore) and B7 (14) were produced with preparations of empty capsids. Horseradish peroxidase (HRP)-conjugated anti-rabbit and anti-mouse antibodies were purchased from Promega, while HRP-conjugated anti-goat immunoglobulin Gs (IgGs) were obtained from Santa Cruz Biotechnology. Fluorescent dye-labeled secondary antibodies were from Dianova. Glutathione-Sepharose beads and columns were purchased from Pharmacia Amersham, and phosphocellulose P11 and DE52 anion-exchange chromatography beads were purchased from Whatman.
Cells and viruses.
A9 fibroblasts, derivatives thereof, and CV-1 and BSC-40 cells were maintained as monolayers in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS). HeLa-S3 cells were cultured using Spiner cultures in S-MEM containing 10% FCS. MVMp was propagated in adherent A9 cells, and virus stocks were prepared by repeated freezing and thawing in 10 mM Tris, pH 8.3, 1 mM EDTA. When indicated, full (DNA containing) MVMp particles were purified from empty capsids by CsCl gradient centrifugation according to their buoyant density. Recombinant vaccinia viruses were generated in CV-1 cells, plaque purified, and amplified in BSC-40 cells. Purification of recombinant vaccinia virus stocks was performed as previously described (28).
Plasmid constructs. (i) Construction of pTM1-GST-NS1x for the production of recombinant vaccinia viruses.
The construction of pTM1-GST-NS1wt was described earlier as the cloning intermediate termed pT-GST-NS1 (27). pTM1-GST-NS1:T363A and pTM1-GST-NS1:S473A were obtained by replacing the EcoRV (nucleotide [nt] 385 of the MVMp sequence)-to-BstEII (nt 1885) fragment of the wild-type NS1 sequence with the appropriate mutant sequences described by Corbau and coworkers (6) and Dettwiler and coworkers (13), respectively. pTM1-GST was constructed by transferring the entire coding sequence of the GST tag as an EcoRI-cleaved PCR fragment into similarly cleaved pTM-1 (23). Recombinant vaccinia viruses were then produced using these pTM-1 constructs as previously described by Nüesch and coworkers (28).
(ii) Isolation of mouse CKIIα cDNA.
A9 cDNA libraries were generated from mRNA preparations using a SMART PCR cDNA synthesis kit. Full-length CKIIα cDNA was isolated essentially as described previously (16) in a single PCR from the mouse fibroblast library using N-terminal (5′-ATGTCGGGACCCGTGCCAAGCAGGGCCAGAGTTTACACAG-3′) and C-terminal (5′-TTACTGCTGAGCGCCAGCGGCAGCTGGTACGGGTATCCCA-3′) primers corresponding to the published rat CKIIα sequence (NCBI L15618). PCR fragments of the appropriate length were isolated from agarose gels cloned directly into pCR2.1 (Invitrogen) and subjected to sequencing (Microsynth GmbH, Balgach, Switzerland).
(iii) Production of CKIIα mutants mATP and E81A.
Site-directed mutagenesis of the mouse CKIIα cDNA clone was performed by chimeric PCR (28) using the N- and C-terminal primers 5′-CCCGGGATATGTCGGGACCCGTGCCAAGCAGGGCCAGAGTTTACACAGA-3′ and 5′-GCGGCCGCTTACTGCTGAGCGCCAGCGGCAGCTGGTACGG-3′, together with two overlapping internal primers harboring the mutation. The N-terminal primer includes a unique SmaI site (underlined) to insert the CKIIα coding sequence (boldface) in frame with the Flag epitope in pP38-Flag (16), while the C-terminal primer contains a unique NotI cleavage site. The mutated primers used to replace the conserved lysine in the ATP-binding pocket with a serine were 5′-AAAAGTTGTTGTTAGTATTCTCAAGCCAGTAAAAAAGAAG-3′ and 5′-ACTGGCTTGACAATACTAACAACAACTTTTTCATTATTTG-3′. To replace the conserved glutamic acid at position 81 with alanine, the following primers were used: 5′-AAAAAAGAAGAAAATTAAGCGTGCTATAAAGATTTTGGAG-3′ and 5′-CTCCAAAATCTTTATAGCACGCTTAATTTTCTTCTTTTTT-3′.
(iv) Production of expression constructs to generate stably transfected cell lines.
MVM NS1-inducible expression vectors were generated from plasmid pP38-Flag (16). The mutants CKIIα:mATP and CKIIα:E81A were subcloned into pCR2.1 (Invitrogen, Groningen, The Netherlands), providing appropriate unique N-terminal SmaI and C-terminal NcoI restriction sites. pP38-Flag CKIIα:mATP and pP38-Flag CKIIα:E81A were generated by transferring the respective SmaI/NotI fragments into Sma/NotI-digested pP38-Flag.
Production and purification of recombinant proteins.
Recombinant GST, as well as wild-type and mutant GST-tagged NS1 proteins, was produced from recombinant vaccinia viruses in suspension cultures of HeLa-S3 cells (27, 29). Cultures were harvested 18 h postinfection (p.i.), and proteins were purified from whole-cell extracts after a nuclear squeeze into the cytoplasmic components using 0.6 M NaCl. After loading on glutathione-Sepharose columns and extensive washing in hypotonic buffer containing 300 mM NaCl, bound proteins were eluted using hypotonic buffer containing 150 mM NaCl and 10 mM glutathione. All protein preparations were analyzed by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and revealed by Coomassie blue staining.
Metabolic labeling and pull-down experiments using GST-NS1 affinity columns.
Metabolic labeling of cells was essentially performed as previously described (27). Asynchronously growing A9 cell cultures (108 cells) were incubated for 4 h in labeling medium (complete medium lacking cysteine and methionine [Gibco/BRL] and supplemented with 0.15 nCi/cell of Tran35S-label [ICN]). Labeled cells were washed in phosphate-buffered saline (PBS), harvested directly in hypotonic buffer (20 mM HEPES, pH 7.5, 5 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT]) containing protease inhibitors, and incubated on ice for 20 min. Cellular extracts were then prepared by cell disruption through 25 strokes in a tight-fitting Dounce homogenizer followed by a nuclear squeeze in 600 mM NaCl for an additional 30 min on ice. The soluble 35S-labeled proteins were then cleared from insoluble materials by centrifugation at 15,000 rpm in an Eppendorf centrifuge for 10 min and either used directly for pull-down experiments or further subjected to either fractionation on phosphocellulose and DE52 affinity columns (see below) or prepurification on GST-saturated glutathione-Sepharose columns. For pull-down experiments, 35S-labeled proteins corresponding to approximately 107 cells were suspended in 700 μl of coimmunoprecipitation (Co-Ip) buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.1% NP-40) containing protease inhibitors and loaded onto GST columns previously saturated with either purifed GST peptide or GST-tagged NS1 proteins. After extensive washing with Co-Ip buffer containing 150 mM NaCl, the NS1/GST-bound proteins were eluted using Co-Ip buffer containing 700 mM NaCl. NS1-associated proteins were then analyzed by SDS-PAGE and autoradiography.
Purification of NS1-interacting partner proteins. (i) Fractionation of cellular extracts on phosphocellulose and DE52 anion-exchange columns.
The procedure used has been described previously (4, 30). Extracts were prepared from 5 × 1011 A9 cells as nuclear squeezes into the cytoplasm as described above, adjusted to 150 mM NaCl, and fractionated on phosphocellulose columns to obtain fractions P1 (flow-through at 150 mM NaCl), P2 (eluate at 400 mM NaCl), and P3 (eluate at 1 M NaCl). Individual fractions were then adjusted to 200 mM NaCl by addition of 5 M NaCl, dilution or dialysis (against hypotonic buffer containing 150 mM NaCl, 20% sucrose, and 10% glycerol), respectively, and further purified on DE52 columns, resulting in fractions DE1 (flow-through at 200 mM NaCl), DE2 (eluate at 400 mM NaCl), and DE3 (eluate at 1 M NaCl). For the purification of NS1 interaction partners, proteins present in P3 were subjected to DE52 columns from which the flow-through at 200 mM NaCl (DE1) was collected and subjected to comparative NS1 affinity chromatography.
(ii) Preparation of NS1 affinity columns.
Hi-Trap 1-ml glutathione-Sepharose columns (Pharmacia-Amersham) were loaded with GST-tagged wild-type or mutant (T363A and S473A) NS1 proteins from recombinant vaccinia virus-infected HeLa-NE.6 cell extracts and washed extensively with hypotonic buffer containing 300 mM NaCl and 0.1% NP-40. Columns were used for comparative affinity chromatography after equilibration in hypotonic buffer containing 150 mM NaCl and 0.1% NP-40.
(iii) Comparative NS1 affinity chromatography.
Proteins extracted from approximately 1010 cells and segregating in the P3-DE1 fraction were first loaded onto columns carrying either GST-NS1:T363A or GST-NS1:S473A. The individual flow-through from each column was collected and then loaded onto a second column carrying GST-NS1wt. The latter columns were washed extensively with Co-Ip buffer containing 150 mM NaCl, and the bound proteins were eluted with Co-Ip buffer containing 700 mM NaCl. Proteins present in the individual eluates were dialyzed and frozen at −80°C. Analysis and/or additional purification for tandem mass spectrometry (MS/MS) analyses was performed by 12% SDS-PAGE and Coomassie blue staining. Protein bands present in significantly different amounts between the two eluates were excised from the gel and subjected to MS/MS analyses (Keck facility, Yale University, New Haven, Conn.).
Western blotting analyses.
Protein extracts were fractionated by discontinuous SDS-PAGE and blotted onto nitrocellulose membranes. Proteins of interest were detected by incubation with appropriate primary antibodies in 10% dry milk-PBS for 18 h and staining with horseradish peroxidase-conjugated secondary antibodies for 1 h followed by chemiluminescence detection (Amersham).
In vitro kinase reactions.
In vitro kinase reactions were performed as described previously (30) using, besides the various forms of NS1/CKIIα-complexes, recombinant CKIIαβ (Roche), PDK-1 (Upstate Biochemicals), and the various PKC isoforms derived from recombinant vaccinia virus expression in HeLa cells (13, 16, 31). Purified MVMp capsids were used as substrates. Assays were performed using 30 μCi of [γ-32P]ATP (3,000 mCi/mmol) in 50 μl of 20 mM HEPES-KOH (pH 7.5), 7 mM MgCl2, 5 mM KCl, and 1 mM DTT in the presence of the appropriate cofactors for 40 min at 37°C. Reactions were stopped by adding the same volume of 20 mM Tris [pH 7.5], 5 mM EDTA, and 0.2% SDS and heating for 30 min at 70°C. The reaction products were analyzed either directly or after immunoprecipitations with antibody EIIF3 (recognizes intact MVM capsids) by 10% SDS-PAGE and semidry transfer onto polyvinylidene difluoride membranes (Millipore).
Generation of stably transfected A9 cell lines.
Stable transfectants were generated as previously described by Lachmann and coworkers (16). In brief, 105 A9 cells were cotransfected with 25 μg of the appropriate pP38-X construct together with pSV2neo in a molar ratio of 25:1, using 25 μl of Lipofectamine (Invitrogen) according to the manufacturer's protocol. Two days posttransfection, cultures were split 1:10 and transfected cells were selected using 400 μg/ml G418 (Sigma). Colonies were pooled after growth for approximately 4 weeks under selection, and frozen stocks were prepared. All experiments were performed after additional cell growth for several passages in the absence of selection in order to avoid physiological side effects of G418. To obtain optimal reproducibility, all transfectants were kept in culture for limited times only (less than 25 passages).
Immunofluorescence microscopy.
For examination by immunofluorescence microscopy, cells were grown on spot slides, mock or MVMp infected, and further incubated for the appropriate times (33). Cultures were then fixed at indicated times postinfection using 3% paraformaldehyde in PBS, pH 7.4, for 30 min at room temperature, neutralized with PBS containing 50 mM NH4Cl for 6 min, and permeabilized by treatment with PBS containing 0.1% Triton X-100 for 10 min. All solutions were supplemented with 1 mM MgCl2 and 0.5 mM CaCl2. After extensive washing, cells were blocked with 10% goat serum for 30 min, incubated with primary antibodies for 2 h at room temperature, and stained with fluorescein isothiocyanate (FITC)-, Cy2-, Cy3-, or rhodamine-conjugated species-specific antibodies (Dianova). After mounting using Elvanol, cells were analyzed by conventional epifluorescence microscopy (Leica; ×63 lens with immersion oil). Phase-contrast microscopy of living cells was performed using an Olympus IMT-2 reverse-angle microscope.
MVM DNA replication in infected cells.
The accumulation of MVM DNA replicative forms was analyzed by Southern blotting as described by Corbau et al. (7). A total of 3 × 105 A9 cells (or derivatives) were infected with MVMp (30 PFU/cell). Cells were harvested in TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) at 4 h, 24 h, and 48 h postinfection and digested with proteinase K in 0.1% SDS for 18 h at 46°C. Total DNA was then sheared by passing through a syringe, fractionated by 0.8% agarose gel electrophoresis, and blotted on a nitrocellulose membrane. Viral replicative intermediates were then detected using a 32P-labeled probe corresponding to nt 385 to 1885 of the NS1-coding region of MVMp.
Production of infectious MVM viruses.
To determine formation and release of progeny virions, cultures were infected with MVMp as described above. At indicated times p.i., medium was removed and kept separately. Attached cells were washed with medium, harvested in DMEM without serum by scraping from the surface, and collected by centrifugation. The amounts of medium- and cell-associated virions were then measured after repeated freezing and thawing steps using standard plaque assays.
The number and morphology of the lysis plaques induced by MVMp in A9 indicator cells or derivatives thereof were essentially determined as described by Daeffler and coworkers (12). Briefly, cell monolayer cultures were seeded at a concentration of 105 cells per 60-mm2 dish and infected 24 h later with serial dilutions of a CsCl-purified MVMp stock in DMEM without FCS. The inoculum was replaced 2 h p.i. with a Bacto-agar overlay (1.8% in MEM containing 5% FCS). After incubation for 6 days, cultures were stained for 18 h by addition of neutral red (0.2 mg/ml)-containing Bacto-agar. Stained cells were fixed on the plates with formaldehyde after removing the agar overlay.
Biochemical fractionation of cellular extracts.
Cell fractionation according to solubility was performed essentially as described by Nüesch and coworkers (32). Briefly, cells were harvested by scraping into the medium and washed repeatedly in PBS. Cell pellets were suspended in proportional amounts to the cell number in hypotonic buffer (20 mM HEPES-KOH, pH 7.5, 5 mM MgCl2, 5 mM KCl, 1 mM DTT), and proteins were extracted by repeated cycles of freezing and thawing. After incubation for 10 min on ice, soluble proteins (S1) were separated from insoluble material by centrifugation at 15,000 × g for 3 min. S2 was generated by extraction of the insoluble pellet for 5 min at room temperature in the same amount of CHAPS buffer—50 mM Tris, pH 8.0, 5 mM EDTA, 100 mM NaCl, 0.5% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}—followed by centrifugation to recover the solubilized components in the supernatant. S3 and S4 fractions were similarly obtained through successive extraction of the insoluble pellet with CHAPS-DOC buffer (CHAPS buffer supplemented with 0.2% Na-deoxycholate) and CHAPS-DOC-SDS-buffer (containing 0.1% SDS), respectively. The remaining insoluble pellet was then heated at 100°C in loading buffer. All fractions were analyzed by SDS-PAGE and Western blotting. It should be stated that the fractions were adjusted to the same volume, allowing their direct comparison for the determination of the distribution of selected proteins.
RESULTS
NS1 interaction partners related to MVM-induced CPE.
A hallmark of NS1 cytotoxic activity consists of the ability of the viral product to induce morphological alterations to the host cell (3, 6). To determine properties and enzymatic functions of NS1 involved in cytopathogenicity, a correlation was sought between biochemical features of NS1 mutants and their potency to induce CPE. These analyses led to the conclusion that the NS1 helicase and nickase activities involved in viral DNA amplification or trans regulation of the P38 promoter are dispensable for cytotoxicity, while the ability of NS1 to bind and/or hydrolyze ATP and to self-assemble into (higher order) oligomers appear to be essential (6). In addition, as indicated in Fig. 1A, mutations modulating the cytotoxic activity of NS1 were found to cluster in distinct areas of the viral polypeptide and, at least in some cases, to affect potential or known PKC phosphorylation sites (6, 12, 26, 31). Altogether, these findings led us to hypothesize that NS1 may induce CPE in target cells through its interactions with host cell factors rather than its enzymatic functions involved in progeny particle production.
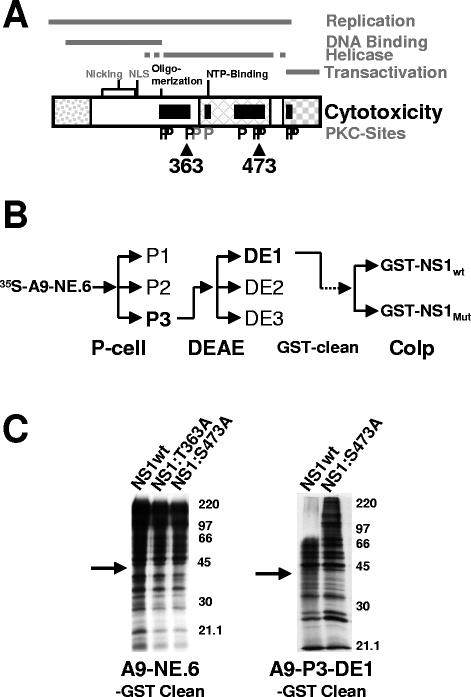
FIG. 1.
Cellular interaction partners of NS1 lacking affinity to cytotoxicity mutants. (A) Domain structure of NS1 (25). The common N terminus of NS1 and NS2 is shown as a dotted box. The crosshatched box represents the homology region with SV40 large-T antigen, and the checkered box denotes the transactivation domain. NS1 domains involved in the DNA binding, helicase, and overall replication functions of the viral product are delineated on top. The nucleotide-binding site (NTP-Binding), oligomerization region, nuclear localization signal (NLS), and DNA-nicking motifs (metal coordination site and linkage tyrosine) are indicated. NS1 regions harboring mutations affecting CPE induction are represented as black boxes. Consensus PKC phosphorylation sites (P) are indicated in black if the corresponding mutant was impaired in its ability to induce CPE and in gray if no significant change was detected compared to the wild-type protein. Positions of NS1 mutants T363 and S473 used to search for NS1 interaction partners are indicated. (B) Cell fractionation procedure used to enrich for “rare” NS1 interaction partners. P-cell, phosphocellulose column with P1, flow-through at 150 mM NaCl concentration; P2, eluate at 400 mM NaCl; P3, eluate at 1 M NaCl. DEAE, DE52 anion-exchange column with DE1, flow-through at 200 mM NaCl; DE2, eluate at 400 mM NaCl; DE3, eluate at 1 M NaCl. Cellular proteins in P3-DE1 with affinity for GST were purified on GST-coated glutathione Sepharose beads; residual polypeptides were then coimmunoprecipitated with either wild-type or mutant GST-tagged NS1. (C) Coimmunoprecipitation of 35S-labeled cellular proteins with GST-NS1 derived from vaccinia virus expression in HeLa cells. The left panel shows a comparison between wild-type NS1, NS1:T363A, and NS1:S473A using total nuclear squeeze from A9 cells, while the right panel presents a comparison between wild-type NS1 and NS1:S473A using partially purified proteins after phosphocellulose and DE52 column chromatography. Proteins with affinity for the GST tag have been eliminated prior to the immunoprecipitation with GST-NS1. The arrow points at a ∼40-kDa polypeptide specifically interacting with wild-type NS1, while lacking affinity to the cytotoxic NS1 mutant S473A.
A number of NS1 interaction partners were reported to serve as cofactors for viral DNA amplification (5), transcriptional regulation (15, 20), or up-to-date unknown functions (10). To specifically select for NS1 interaction partners involved in cytotoxicity and to isolate sufficient amounts of these polypeptides for identification, we developed the following strategy. (i) To reveal protein interactions potentially involved in cytotoxicity, cellular partners were sought that bind to wild-type toxic NS1 but lack affinity to mutant NS1 proteins specifically debilitated for CPE induction. (ii) To produce NS1 proteins serving as baits, GST-tagged NS1 proteins were expressed by means of recombinant vaccinia viruses in mammalian cells. The N-terminal GST fusion peptide gives rise to stable NS1 dimers (27), a prerequisite for cytotoxicity (6). Furthermore, NS1 expression driven by recombinant vaccinia viruses in HeLa cells results in large amounts of proteins that are suitable for purification (29) and endowed with a phosphorylation pattern highly similar to the one generated during the replicative phase of an MVMp infection in A9 cells (7). (iii) To facilitate the isolation of “rare” host factors, cellular proteins were prepurified by consecutive exchange chromatography steps as previously performed to identify NS1 modulating PKs (Fig. 1B) (30), prior to the NS1 affinity selection. This prepurification procedure allows the isolation of even low-abundance proteins in sufficient amounts for the identification by mass spectrum analyses. Moreover, it may avoid NS1 interactions with rare partners becoming masked by other more abundant partners of similar size.
To evaluate the feasibility of this approach, we first performed a small-scale study using metabolically 35S-labeled proteins extracted from A9 cells and partially purified by exchange chromatography according to the scheme depicted in Fig. 1B. Whole-A9-cell extracts or partially purified proteins were subjected to affinity chromatography using columns coupled with either wild-type or nontoxic mutant NS1 proteins. Bound fractions were eluted with 0.7 M NaCl and further analyzed by SDS-PAGE and autoradiography. As shown in Fig. 1C (left panel), a large number of polypeptides from whole-A9-cell extracts interacted with phosphorylated GST-tagged wild-type NS1 protein. Among these polypeptides, only a minor protein band migrating around 40 kDa (arrow) was specifically absent from the eluate derived from the affinity column coupled with nontoxic mutant NS1. Interestingly, this protein band became enriched through fractionation of A9 cell extracts by phosphocellulose and anion-exchange chromatography (right panel), demonstrating the feasibility of our approach.
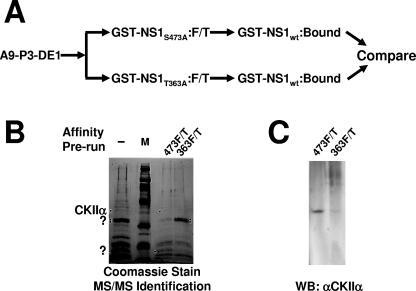
The detection of a host cellular protein that specifically interacts with wild-type (toxic) NS1, while showing little or no affinity to a nontoxic mutant, prompted us to purify sufficient amounts of this species for identification by MS analyses. To this end, 5 × 1011 A9 cells grown in suspension were first fractionated on phosphocellulose and anion-exchange chromatography columns. The A9-P3-DE1 was then precleaned, eliminating, through affinity chromatography, the proteins that were able to bind to either GST-NS1:S473A, a nontoxic NS1 variant harboring a mutation within the helicase domain, or to GST-NS1:T363A, a less toxic NS1 variant harboring a mutation with the spacer region between nickase and helicase domain (for details see Fig. 1A). The flow-through samples from these affinity columns were loaded separately on columns coupled with wild-type GST-NS1, and after an extensive washing procedure, the bound polypeptides were eluted using 0.7 M NaCl. These final eluates were then further subjected to 12% SDS-PAGE and stained with Coomassie brilliant blue (Fig. 2B). Polypeptides, which were found in either but not both of the eluates, were cut from the gel and subjected to MS/MS analyses (Keck facility, Yale University, New Haven, CT). Interestingly, among the proteins found to interact with wild-type NS1 after cleaning on mutant NS1:S473A, only two polypeptides were found to be absent or present in significantly smaller amounts after purification on NS1-T363A. Therefore, a polypeptide with an approximate size of 40 kDa could be assigned by MS/MS to CKIIα, the catalytic subunit of casein kinase II, while a doublet migrating around 20 kDa could not be identified so far. Conversely, prepurification by NS1:T363A also revealed an as-yet-unidentified wild-type NS1-interacting doublet migrating around 32 kDa, which was not present in the eluate of NS1:S473. To confirm MS identification of the 40-kDa polypeptide, individual fractions were characterized by Western blotting. As shown in Fig. 2C, CKIIα was readily detected as an NS1-interacting protein lacking affinity to NS1:S473A. Altogether, these findings suggested that CKIIα interacts with NS1 within the helicase domain, most likely under control of NS1 phosphorylation by PKCλ at residue S473 (13, 26, 31).
FIG. 2.
Isolation and identification of cellular proteins interacting with wild-type but not nontoxic mutant NS1. (A) Purification of cytotoxicity-related NS1 partner proteins by sequential NS1 affinity chromatography. The P3-DE1 prepurified protein fractions were run first over a column carrying either cytotoxicity mutant of NS1 (T363A or S473A). Unbound proteins (flow-through; F/T) failing to interact with the nontoxic NS1 derivative were then run over a second affinity column coupled with wild-type GST-NS1 to capture cytotoxicity-dependent NS1 partners. After extensive washing, eluates were collected, containing the cellular proteins which bind to wild-type but not cytotoxic mutant NS1 proteins. (B) SDS-PAGE separation and Coomassie blue staining of proteins retained by GST-NS1wt affinity columns and subsequently eluted at 700 mM NaCl. A9 cell proteins from the P3-DE1 fraction (see Fig. 1B) were run over the GST-NS1wt column without prior additional purification (−, total NS1-binding protein) or after prior elimination of proteins binding to either mutant NS1 affinity column (473F/T; proteins lacking affinity to GST-NS1:S473A and binding to wild-type NS1; 363F/T, proteins lacking affinity to GST-NS1:T363A and binding to wild-type NS1). NS1wt partner proteins overrepresented in 474F/T versus 363F/T, and conversely, are marked with dots. Marked bands were cut from the gel and submitted to MS/MS analyses. The 40-kDa species overrepresented in 473F/T (i.e., with a reduced affinity for NS1:S473A) was identified as CKIIα. M, molecular weight markers (Amersham-Pharmacia). (C) This identification was confirmed by Western blotting (WB) of the two protein fractions obtained after differential affinity chromatography with NS1 variants using antibodies specific for CKIIα.
NS1-dependent capacity of CKIIα for phosphorylating MVMp capsids in vitro.
The identification of CKIIα as a cellular interaction partner of the viral NS1 protein is of particular interest. The generation of a complex between NS1 and the catalytic subunit of a cellular kinase raises the possibility that MVM could interfere directly with intracellular signaling by retargeting the enzyme on new viral or cellular substrates or by modulating its activity. MVM capsids have been shown to become phosphorylated during infection, a posttranslational modification that seems to be essential for the export of newly synthesized infectious virions from the host cell (21, 22). To investigate whether NS1 is indeed able to modulate CKIIα, we tested the isolated GST-NS1/CKIIα complex for its kinase activity on purified viral capsids as substrates in comparison with recombinant cellular PKs. In vitro kinase assays were carried out using [γ-32P]ATP. Radiolabeled substrates were isolated by immunoprecipitation and analyzed by SDS-PAGE and autoradiography.
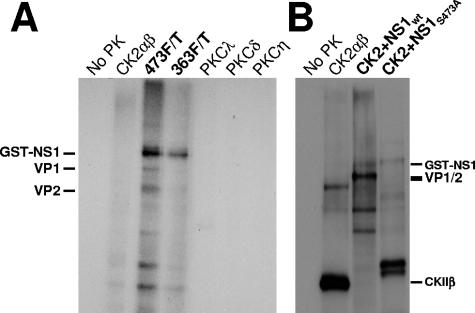
As shown in Fig. 3A, NS1-associated CKIIα purified from A9 cells by affinity chromatography was able to phosphorylate both the VP1 and VP2 polypeptides present in MVMp virions. In contrast, members of the PKC family, the genuine recombinant CKIIαβ, and the NS1 eluate derived after precleaning by GST-NS1:T363A (serving as a negative control) were unable to phosphorylate isolated MVMp capsids. This finding is remarkable since it implies that NS1 either targets CKIIα onto novel substrates or alters the substrate specificity of the PK, at least in vitro. It cannot be ruled out, however, that mouse and human CKIIαs differ in their specificities. To address the latter possibility, we performed a second approach mixing recombinant CKIIαβ with purified wild-type or mutant GST-NS1. As shown in Fig. 3B, purified wild-type GST-NS1 was able to modify human recombinant CKII activity, mediating the phosphorylation of MVMp capsids and inhibiting autophosphorylation of the regulatory CKIIβ subunit. In contrast, CKIIαβ alone or in combination with GST-NS1:S473A was unable to phosphorylate MVM capsids. It should be stated here that besides MVM capsids, GST-NS1 itself appeared to be a target for CKII and NS1-CKIIα kinase activity, in agreement with our previous findings that NS1 is a substrate for casein kinase II in vitro (27).
FIG. 3.
In vitro kinase activity of the NS1/CKIIα complex. (A) CsCl-purified MVMp capsids were subjected to phosphorylation using [γ-32P]ATP and indicated recombinant protein kinases or the NS1wt complexes with 473F/T or 363F/T proteins. Treated capsids were immunoprecipitated with the monoclonal antibody EIIF3 and analyzed by 10% SDS-PAGE and autoradiography. The migrations of GST-NS1 (∼110 kDa), VP1, and VP2 are indicated. (B) Phosphorylation of MVMp capsids with recombinant CKIIαβ either alone or in the presence of wild-type or mutant (S473A) GST-tagged NS1 proteins produced by recombinant vaccinia virus expression. Labeled proteins were analyzed directly by 12% SDS-PAGE and autoradiography. The migration of GST-NS1, VP1, and VP2, as well as the regulatory subunit of CKII, CKIIβ (which is a target for CKII phosphorylation), is indicated.
Impact of CKII activity on virus replication.
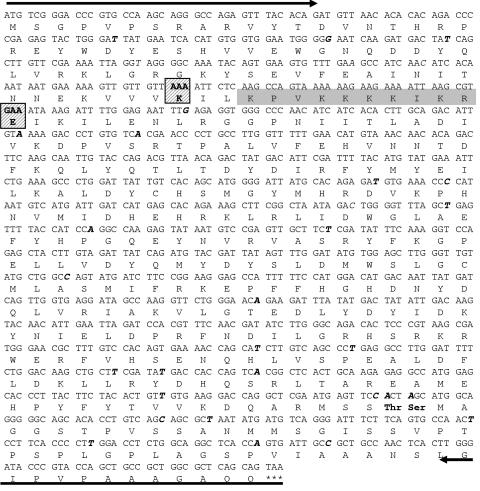
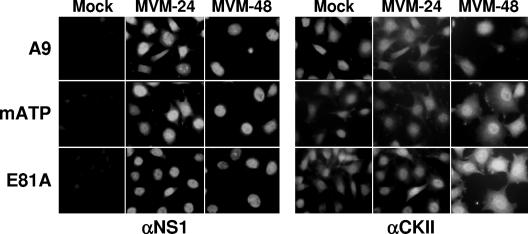
To further evaluate the impact of CKII on the virus life cycle, we generated cell lines stably expressing dominant-negative mutant forms of CKIIα under control of the NS1-inducible P38 promoter. This approach proved useful in the past to study the role of PKCη-dependent phosphorylation in MVM DNA replication in vivo (16) and to determine the impact of PKCλ-driven phosphorylation on the microtubule dynamics in MVM-infected cells (32). Mouse fibroblast CKIIα cDNA was isolated by PCR amplification from an A9 cDNA bank (16) using N- and C-terminal primers corresponding to the rat CKIIα sequence (NCBI L15618). As indicated in Fig. 4, sequence comparison between the published rat and present mouse cDNAs revealed only 20 nucleotide differences, of which only 2 led to amino acid substitutions, i.e., A337 to threonine and G338 to serine. Since overexpression of catalytically inactive protein kinase mutants has been shown to exert a specific dominant-negative effect on the appropriate endogenous PK for a variety of kinases, including PKC (16, 38), we generated two such mutant forms of the cloned mouse CKIIα cDNA. In analogy to the usually used PKC mutants, we decided to inactivate the nucleotide-binding domain by replacing the α-phosphate-interacting lysine to a serine (K68S). Alternatively, we replaced the invariant glutamic acid at position 81 (E81) with alanine (37). The whole Flag-tagged CKIIα coding sequence harboring these individual mutations was then stably transfected under the control of the parvoviral P38 promoter into A9 cells as previously described (16), generating the A9-P38:CKII(mATP) and A9-P38:CKII(E81A) cell lines. As shown by double-immunofluorescence staining for NS1 and CKIIα proteins, both transfectants could be readily infected with MVMp, which induced them to accumulate enhanced levels of CKIIα lines in over 90% of the cell population at 2 days postinfection as compared to the parental A9 cell line or noninfected transfectants (Fig. 5). This suggested that overproduction of catalytically inactive CKII mutants could actively interfere with the endogenous CKII activity in the presence of NS1.
FIG. 4.
Nucleotide and amino acid sequence of mouse CKIIα cDNA derived from A9 cells. The cDNA was isolated from an A9 cDNA bank using the primers indicated by arrows. Differences between the nucleotide sequence obtained and the published rat CKIIα sequence (NCBI L15618) are indicated in boldface and italics, with corresponding amino acid changes shown in boldface and by the triple-letter code. Amino acids targeted for mutagenesis to generate dominant-negative mutants are indicated by hatched boxes, while the nuclear targeting sequence is shown in light gray.
FIG. 5.
Immunofluorescence detection of CKIIα expressed from the endogenous gene and transfected mutant clones. A9 cells or derivatives thereof stably transfected with indicated mutants of CKIIα expression constructs were grown on spot slides, infected (or not) with MVMp (30 PFU/cell), and fixed 24 or 48 h p.i. CKIIα was detected using a polyclonal goat antiserum (αCKII) and FITC-conjugated anti-goat IgGs. NS1 was revealed using a polyclonal rabbit anti-NS1C antiserum (αNS1) and rhodamine-conjugated anti-rabbit IgGs.
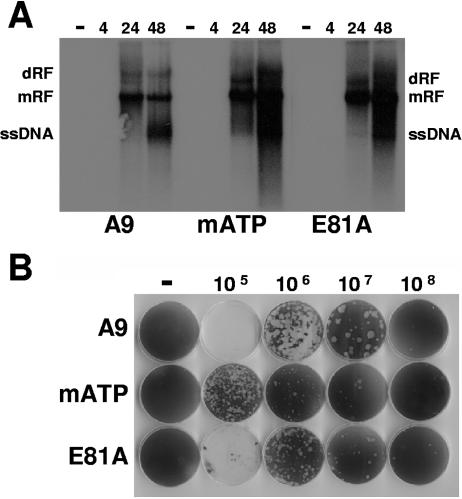
To further investigate the impact of CKII on MVM replication, we performed Southern blotting experiments, analyzing the accumulation of viral DNA replication intermediates and the production of progeny particle production (as revealed by the presence of displaced single-stranded virion DNA). A9 cells or derivatives thereof were infected (or not) with 30 PFU/cell MVMp. This infectivity ratio was sufficient to infect over 90% of the cell population as apparent from the amount of NS1-positive cells detected already 24 h p.i. (Fig. 5). Cells were then harvested at intervals postinfection and analyzed for their content in viral DNA species by agarose gel electrophoresis and Southern blotting using a hybridization probe corresponding to nt 385 to 1885 of the MVMp genome. As shown in Fig. 6A, over the whole time period monitored, the induction of dominant-negative CKIIα expression did not impair viral DNA amplification, leading to the accumulation of at least as much replicative-form intermediates (mRF and dRF) and single-stranded virion DNA as seen in parental A9 cells.
FIG. 6.
MVMp replication in A9 cells and derivatives expressing dominant-negative CKIIα. (A) Asynchronously growing A9 cell derivatives (A9-P38:CKIImATP and A9-P38:CKIIE81A) expressing CKIIα variants under control of the viral P38 promoter were infected (or not) with MVMp (30 PFU/cell). The accumulation of double-stranded replicative forms and single-stranded virion DNA (ssDNA) was determined by Southern blotting at 4 h, 24 h, and 48 h p.i. dRF, dimer replicative form; mRF, monomer replicative form. Prolonged exposure showed similar amounts of input ssDNA in infected A9 cells and derivatives at 4 h p.i. (data not shown), indicating that modulation of the casein kinase II pathway did not affect virus entry. (B) Determination of MVMp plaquing efficiency and plaque morphology in A9 cells and derivatives expressing dominant-negative CKIIα according to standard plaque assays.
In addition, the release and spread of infectious progeny virus were analyzed by standard plaque assays (39). To this end, virus titrations were performed by infecting in parallel A9, A9-P38:CKII(mATP), and A9-P38:CKII(E81A) cell monolayers at different multiplicities. As shown in Fig. 6B, the plaque numbers were comparable between these cell lines: despite similar growth rates, the sizes of the plaques differed markedly with both transfectants, presenting a small-to-pinpoint plaque phenotype compared to parental A9 cells. Together with the viral DNA accumulation data, this result suggested that CKII may control the release of the virus burst (through cell lysis and/or virus export), thereby facilitating the cell-to-cell spreading of progeny particles.
Involvement of CKII in MVM-induced cell alterations.
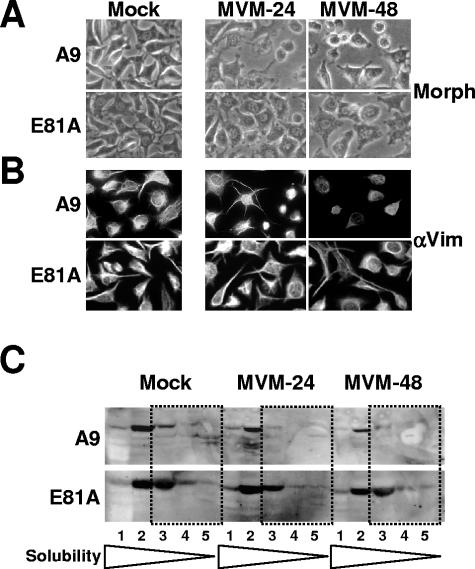
Due to the reduced affinity for the nontoxic NS1 mutant S473A, CKIIα is a cellular candidate for the mediation of NS1-dependent CPE induction. This prompted us to investigate whether functional CKII is involved in MVM-induced CPE. To monitor the fate of infected cells and particular cytoskeletal filaments in situ in the presence or absence of functional CKII, parental A9 cells or cells of the derivative cell line A9-P38:CKII(E81A) were infected (or not) with MVMp (30 PFU/cell) and examined at 24 and 48 h p.i. by phase-contrast microscopy of living cells (Fig. 7A), as well as immunofluorescence staining (Fig. 7B) and biochemical fractionation of vimentin (Fig. 7C), an intermediate filament structure, whose solubility and intracellular structure are strongly affected upon MVM infection (32). As shown in Fig. 7A, in contrast to the parental A9 cells, which displayed severe CPE in the majority of infected cells, most cells in cultures overexpressing the dominant-negative mutant form, CKII:E81A, kept a “normal” morphology over the first 48 h p.i. In addition to the overall cell morphology, we were interested in investigating the fate of the cytoskeleton network in both parental A9 cells and the transfectant expressing CKII:E81A. In agreement with previously reported observations (32), upon MVMp infection of A9 cells the intermediate filament vimentin is rearranged and becomes degraded upon infection until the remaining filaments collapse around the nucleus. In contrast, A9-P38:CKII(E81A) cells were protected against this CPE. No significant change in the vimentin structure could be detected in most of the infected versus control cells during the 48-h p.i. interval studied by both immunofluorescence staining of single cells (Fig. 7B) and biochemical fractionations of vimentin according to the solubility of the protein (Fig. 7C). This suggested that CKII plays a major role in the rearrangement of the cell morphology and particularly the cytoskeleton network during infection. Moreover, considering that the mutant NS1:S473A (lacking affinity for CKIIα) is debilitated for this function (6), it seems likely that CKII-mediated CPE induction can be triggered through interaction with the viral NS1 protein.
FIG. 7.
MVMp-induced CPE in the presence of a dominant-negative CKIIα mutants (E81A). (A) Asynchronously growing A9 cells were infected (or not) with MVMp (30 PFU/cell) and examined for morphological alterations (Morph) by phase-contrast microscopy. (B) Asynchronously growing A9 cells grown on spot slides were infected (or not) with MVMp (30 PFU/cell) and fixed with paraformaldehyde at indicated times postinfection. Preparations were analyzed by immunofluorescence for the organization of vimentin filaments using goat polyclonal antiserum raised against vimentin (αVim) and FITC-coupled anti-goat IgGs. (C) Biochemical determination of MVMp-induced alterations to the cytoskeleton. Asynchronously growing A9 cells were infected with MVMp (30 PFU/cell), harvested 24 h and 48 h p.i., and analyzed for the solubility of the intermediate filament vimentin. Cell extracts were treated with increasing amounts and strengths of detergents, and the individual fractions (S1 to S5) were analyzed for their vimentin content by Western blotting. The most insoluble filament proteins are indicated by dotted frames.
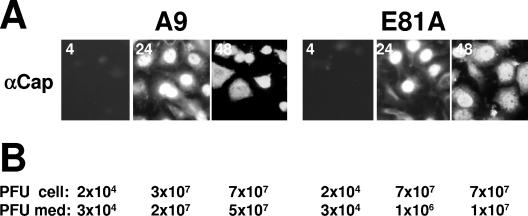
The resistance to CPE induction and the small-to-pinpoint plaque phenotype of wild-type MVMp in transfectants expressing dominant-negative CKIIα mutants raised the question of whether the functional inactivation of CKII and the resultant preservation of the cytoskeleton network had an effect on the intracellular distribution and release of progeny particles. To investigate this question, we monitored A9 versus A9-P38:CKII(E81A) for their production and distribution of capsids by indirect immunofluorescence staining release of infectious progeny particles from the cells into the culture medium by standard plaque assays. As shown in Fig. 8A, progeny particle production, nuclear accumulation, and leakage to the cytoplasm took place to similar extents in the presence (A9) or absence [A9-P38:CKII(E81A)] of functional CKIIα, as determined by capsid staining. Yet an intriguing difference was observed in the late cytoplasmic distribution of capsids, which was diffuse in A9 cells but had a vesicular appearance in A9-P38:CKII(E81A) (Fig. 8A, 48h p.i.) and A9-P38:CKII(mATP) cells (data not shown). This difference may be indicative of an impairment of the active transport of progeny virions to the cell surface in the absence of functional CKIIα. This possibility was supported by the determination of the distribution of progeny virions between cells and culture medium. The ratio of free (medium associated) versus bound (cell associated) virions was indeed 20-fold (24 h p.i.) to 5-fold (48 h p.i.) lower in the A9-P38:CKII(E81A) transfectant than in the parental A9 cells. Altogether, our data suggest that though dispensable for progeny particle production, CKII takes part in the control of progeny particles' release. It remains to be determined whether this control depends on the above-mentioned ability of the CKIIα-NS1 complex to drive the destruction of the cytoskeleton network and/or to modify assembled capsids.
FIG. 8.
Release of progeny virus particles from A9 cells and derivatives expressing dominant-negative CKIIα mutants. (A) Cells grown on spot slides were fixed with paraformaldehyde at 4 h, 24 h, and 48 h p.i. and analyzed by immunofluorescence using a mixture of two mouse monoclonal anticapsid (αCap) antibodies (EIIF3 and B7) and Cy2-conjugated anti-mouse IgGs. (B) Cell-associated infectious virions (PFU cell) and progeny particles released into the medium (PFU med) were determined by standard plaque assays.
DISCUSSION
Due to their specificity for oncogene-transformed cells (24), NS1 cytotoxic functions have drawn attention for a long time. Although NS1 is endowed with a variety of enzymatic and regulatory functions that may contribute to cytotoxicity (1, 6, 17, 18, 34-36), the viral trigger(s) leading to alterations of the cell morphology, collapse of the cytoplasm, and eventual cell death (3) remains elusive. The clustering of mutations affecting NS1 cytotoxicity in defined regions of the viral polypeptides (6, 12, 17) raised the possibility that NS1 exerts its cytotoxic activity by interacting with distinct host cell partner proteins rather than its own enzymatic activities. Indeed, the present study led to the identification of a new NS1 interaction partner, whose interaction with the viral polypeptide correlated with its cytotoxicity. Evidence was obtained to suggest that this partner, CKIIα, is directly involved in the induction of morphological alterations upon infection of host cells with MVMp and particularly the cytoskeleton filament network. Thereby, the interaction of NS1 with CKIIα, the catalytic subunit of a cellular protein kinase, points to a possible interference of the viral regulatory protein with intracellular signaling, which in consequence could modify the cytoskeleton dynamics. Indeed, stably transfected A9 cell lines expressing catalytically inactive mutant forms of CKIIα proved to be protected from MVM-induced CPE and more particularly from the destruction of vimentin filaments due to interference of these CKIIα variants with endogenous CKII activity. Although CKIIα (probably in the form of a complex with NS1) appeared to control the fate of infected cells and the release of progeny viruses, functional CKII was dispensable for intracellular virus multiplication. This result led us to conclude that retargeting of the CKII phosphorylation activity represents a distinct “late” function of NS1, aimed at poisoning host cells and facilitating efficient virus release and spread.
The NS1/CKIIα complex proved to differ from genuine CKII by its substrate specificity. Indeed, purified MVMp capsids were efficiently phosphorylated by NS1/CKIIα while being poor targets for recombinant CKIIαβ. The mechanisms underlying this difference in substrate specificity remain to be unraveled. The interaction with NS1 may indeed facilitate the interaction with novel substrate proteins and/or cause a structural alteration of the substrate interaction pocket of CKIIα, which could modify the affinity to the substrate recognition site, i.e., interaction with the adjacent amino acids of the substrate serine/threonines. Another, nonexclusive possibility would be that the intracellular distribution of the enzyme gets modified due to its binding to NS1. Irrespective of the substrate specificity of the kinase, this interference of NS1 with CKIIα in vivo would result in a modulation of CKIIα activity or a change of the availability to its (cellular) substrates and in consequence could trigger the observed changes of the cytoskeleton. NS1 may thus act by affecting CKII functioning in a direct way and/or indirectly by targeting the enzyme to novel substrates usually not reached or rarely reached. Complex formation between NS1 and the catalytic subunit of a cellular PK may thus serve multiple purposes, at the level of both the host cell physiology and the fate of progeny virions. The hereby described involvement of CKII in the facilitation of virus release may thus result from cytoskeletal alterations and/or capsid modifications. The latter possibility is in keeping with recent reports showing that the export of newly produced viruses is dependent on the phosphorylation of capsids (21, 22). The relative contributions of cytoskeleton and capsid modification to the NS1/CKIIα-mediated stimulation of progeny virus release remain to be determined. This is a complex issue since MVMp infection is known to affect the phosphorylation patterns of several cellular proteins (1, 32) and to modulate interconnected PK cascades (16, 31). NS1/CKIIα is therefore a piece of an intricate puzzle which may also include feedback loops such as NS1 itself, regulated by differential phosphorylation through the PDK/PKC pathway during infection (7, 12).
Besides CKIIα, other cellular interaction partners (such as, for instance, the 30/32-kDa doublet lacking affinity to NS1:T363) and particularly multiple interaction domains of NS1 appear to be involved in the cytotoxic functions of the viral product. In particular, the C-terminal-end part of NS1 becomes phosphorylated late during infection and amino acid substitutions for some of these (putative) phosphorylation sites impair the capacity of NS1 to induce cell morphological alterations and death (12). This leads us to speculate that proteins still to be identified may interact with this and possibly other regions of NS1 and further contribute to jeopardize the survival of infected cells. The NS1:S473A mutant (which failed to bind CKIIα) had no detectable cytotoxicity: i.e., its expression was well tolerated by target cells for longer than a week (6). This suggests that the interaction of NS1 with CKIIα is essential to trigger CPE, in agreement with the ability of CKIIα dominant-negative mutant forms to fully protect cells from MVMp-induced cell death and lysis. On the other hand, the NS1:T363A selectively binding to the 30/32-kDa doublet or NS1:T585A (12) located in the C terminus of NS1 only showed a delay in the CPE that eventually appeared 4 days after transfection. The fact that NS1:363A and NS1:T585A kept a significant residual cytotoxicity may tentatively be assigned to NS1's ability to interact with several (multiple) cellular targets, which could become modified by the catalytic activity of the NS1/CKIIα complex. Our current investigations aim to identify additional cellular NS1 interaction partners involved in cytotoxicity (particularly the 30/32-kDa doublet lacking affinity to NS1:T363A) and to assess the role of NS1/CKIIα in the modulation of intracellular signaling.
Acknowledgments
Particular thanks are granted to Claudia Plotzky for excellent technical assistance. In addition, we are grateful to Peter Tattersall, Susan Cotmore, Colin Parrish, and José Almendral providing us with plasmid constructs and antisera.
REFERENCES
- 1.Anouja, F., R. Wattiez, S. Mousset, and P. Caillet-Fauquet. 1997. The cytotoxicity of the parvovirus minute virus of mice nonstructural protein NS1 is related to changes in the synthesis and phosphorylation of cell proteins. J. Virol. 71:4671-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bashir, T., J. Rommelaere, and C. Cziepluch. 2001. In vivo accumulation of cyclin A and cellular replication factors in autonomous parvovirus minute virus of mice-associated replication bodies. J. Virol. 75:4394-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caillet-Fauquet, P., M. Perros, A. Brandenburger, P. Spegelaere, and J. Rommelaere. 1990. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding non-structural proteins. EMBO J. 9:2989-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen, J., S. F. Cotmore, and P. Tattersall. 1997. Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J. Virol. 71:5733-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, J., and P. Tattersall. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbau, R., V. Duverger, J. Rommelaere, and J. P. Nüesch. 2000. Regulation of MVM NS1 by protein kinase C: impact of mutagenesis at consensus phosphorylation sites on replicative functions and cytopathic effects. Virology 278:151-167. [DOI] [PubMed] [Google Scholar]
- 7.Corbau, R., N. Salomé, J. Rommelaere, and J. P. Nüesch. 1999. Phosphorylation of the viral nonstructural protein NS1 during MVMp infection of A9 cells. Virology 259:402-415. [DOI] [PubMed] [Google Scholar]
- 8.Cotmore, S. F., and P. Tattersall. 1987. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 33:91-174. [DOI] [PubMed] [Google Scholar]
- 9.Cotmore, S. F., and P. Tattersall. 1988. The NS-1 polypeptide of minute virus of mice is covalently attached to the 5′ termini of duplex replicative-form DNA and progeny single strands. J. Virol. 62:851-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cziepluch, C., E. Kordes, R. Poirey, A. Grewenig, J. Rommelaere, and J.-C. Jauniaux. 1998. Identification of a novel cellular TPR-containing protein, SGT, that interacts with the nonstructural protein NS1 of parvovirus H-1. J. Virol. 72:4149-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cziepluch, C., S. Lampel, A. Grewenig, C. Grund, P. Lichter, and J. Rommelaere. 2000. H-1 parvovirus-associated replication bodies: a distinct virus-induced nuclear structure. J. Virol. 74:4807-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daeffler, L., R. Hörlein, J. Rommelaere, and J. P. F. Nüesch. 2003. Modulation of minute virus of mice cytotoxic activities through site-directed mutagenesis within the NS coding region. J. Virol. 77:12466-12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dettwiler, S., J. Rommelaere, and J. P. F. Nüesch. 1999. DNA unwinding functions of minute virus of mice NS1 protein are modulated specifically by the lambda isoform of protein kinase C. J. Virol. 73:7410-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichwald, V., L. Daeffler, M. Klein, J. Rommelaere, and N. Salomé. 2002. The NS2 proteins of parvovirus minute virus of mice are required for efficient nuclear egress of progeny virions in mouse cells. J. Virol. 76:10307-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krady, J. K., and D. C. Ward. 1995. Transcriptional activation by the parvoviral nonstructural protein NS-1 is mediated via a direct interaction with Sp1. Mol. Cell. Biol. 15:524-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lachmann, S., J. Rommeleare, and J. P. F. Nüesch. 2003. Novel PKCη is required to activate replicative functions of the major nonstructural protein NS1 of minute virus of mice. J. Virol. 77:8048-8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legendre, D., and J. Rommelaere. 1992. Terminal regions of the NS-1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans inhibition. J. Virol. 66:5705-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, X., and S. L. Rhode III. 1990. Mutation of lysine 405 to serine in the parvovirus H-1 NS1 abolishes its functions for viral DNA replication, late promoter trans activation, and cytotoxicity. J. Virol. 64:4654-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, J., Z. H. Ran, S. D. Xiao, and J. Rommelaere. 2005. Changes in gene expression profiles induced by parvovirus H-1 in human gastric cancer cells. Chin. J. Dig Dis. 6:72-81. [DOI] [PubMed] [Google Scholar]
- 20.Lorson, C., J. Pearson, L. Burger, and D. J. Pintel. 1998. An Sp1-binding site and TATA element are sufficient to support full transactivation by proximally bound NS1 protein of minute virus of mice. Virology 240:326-337. [DOI] [PubMed] [Google Scholar]
- 21.Maroto, B., J. C. Ramirez, and J. M. Almendral. 2000. Phosphorylation status of the parvovirus minute virus of mice particle: mapping and biological relevance of the major phosphorylation sites. J. Virol. 74:10892-10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maroto, B., N. Valle, R. Saffrich, and J. M. Almendral. 2004. Nuclear export of the nonenveloped parvovirus virion is directed by an unordered protein signal exposed on the capsid surface. J. Virol. 78:10685-10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. Product review. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 24.Mousset, S., Y. Ouadrhiri, P. Caillet-Fauquet, and J. Rommelaere. 1994. The cytotoxicity of the autonomous parvovirus minute virus of mice nonstructural proteins in FR3T3 rat cells depends on oncogene expression. J. Virol. 68:6446-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nüesch, J. 2005. Regulation of non-structural protein functions by differential synthesis, modification and trafficking, p. 275-289. In M. E. Bloom, S. F. Cotmore, R. M. Linden, C. R. Parrish, and J. R. Kerr (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 26.Nüesch, J. P. F., J. Christensen, and J. Rommelaere. 2001. Initiation of minute virus of mice DNA replication is regulated at the level of origin unwinding by atypical protein kinase C phosphorylation of NS1. J. Virol. 75:5730-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nüesch, J. P. F., R. Corbau, P. Tattersall, and J. Rommelaere. 1998. Biochemical activities of minute virus of mice nonstructural protein NS1 are modulated in vitro by the phosphorylation state of the polypeptide. J. Virol. 72:8002-8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nüesch, J. P. F., S. F. Cotmore, and P. Tattersall. 1992. Expression of functional parvoviral NS1 from recombinant vaccinia virus: effects of mutations in the nucleotide-binding motif. Virology 191:406-416. [DOI] [PubMed] [Google Scholar]
- 29.Nüesch, J. P. F., S. F. Cotmore, and P. Tattersall. 1995. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology 209:122-135. [DOI] [PubMed] [Google Scholar]
- 30.Nüesch, J. P. F., S. Dettwiler, R. Corbau, and J. Rommelaere. 1998. Replicative functions of minute virus of mice NS1 protein are regulated in vitro by phosphorylation through protein kinase C. J. Virol. 72:9966-9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nüesch, J. P. F., S. Lachmann, R. Corbau, and J. Rommelaere. 2003. Regulation of minute virus of mice NS1 replicative functions by atypical PKCλ in vivo. J. Virol. 77:433-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nüesch, J. P. F., S. Lachmann, and J. Rommelaere. 2005. Selective alterations of the host cell architecture upon infection with parvovirus minute virus of mice. Virology 331:159-174. [DOI] [PubMed] [Google Scholar]
- 33.Nüesch, J. P. F., and P. Tattersall. 1993. Nuclear targeting of the parvoviral replicator molecule NS1: evidence for self-association prior to nuclear transport. Virology 196:637-651. [DOI] [PubMed] [Google Scholar]
- 34.Op De Beeck, A., F. Anouja, S. Mousset, J. Rommelaere, and P. Caillet-Fauquet. 1995. The nonstructural proteins of the autonomous parvovirus minute virus of mice interfere with the cell cycle, inducing accumulation in G2. Cell Growth Differ. 6:781-787. [PubMed] [Google Scholar]
- 35.Op De Beeck, A., and P. Caillet-Fauquet. 1997. The NS1 protein of the autonomous parvovirus minute virus of mice blocks cellular DNA replication: a consequence of lesions to the chromatin? J. Virol. 71:5323-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Op De Beeck, A., J. Sobczak-Thepot, H. Sirma, F. Bourgain, C. Brechot, and P. Caillet-Fauquet. 2001. NS1- and minute virus of mice-induced cell cycle arrest: involvement of p53 and p21cip1. J. Virol. 75:11071-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pyerin, W., K. Ackermann, and P. Lorenz. 1996. Casein kinases, p. 117-147. In F. Marks (ed.), Protein phosphorylation. VCH Verlagsgesellschaft GmbH, Weinheim, Germany.
- 38.Suzuki, A., T. Yamanaka, T. Hirose, N. Manabe, K. Mizuno, M. Shimizu, K. Akimoto, Y. Izumi, T. Ohnishi, and S. Ohno. 2001. Atypical protein kinase C is involved in the evolutionarily conserved PAR protein complex and plays a critical role in establishing epithelia-specific junctional structures. J. Cell Biol. 152:1183-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tattersall, P., and J. Bratton. 1983. Reciprocal productive and restrictive virus-cell interactions of immunosuppressive and prototype strains of minute virus of mice. J. Virol. 46:944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]