Abstract
Caribbean-born African green monkeys (AGMs) were classified as Chlorocebus sabaeus by cytochrome b sequencing. Guided by these phylogenetic analyses, we developed a new model for the study of simian immunodeficiency virus (SIV) infection in natural hosts by inoculating Caribbean AGMs with their species-specific SIVagm.sab. SIVagm.sab replicated efficiently in Caribbean AGM peripheral blood mononuclear cells in vitro. During SIVagm.sab primary infection of six Caribbean AGMs, the virus replicated at high levels, with peak viral loads (VLs) of 107 to 108 copies/ml occurring by day 8 to 10 postinfection (p.i.). Set-point values of up to 2 × 105 copies/ml were reached by day 42 p.i. and maintained throughout follow-up (through day 450 p.i.). CD4+ T-cell counts in the blood showed a transient depletion at the peak of VL, and then returned to near preinfection values by day 28 p.i. and remained relatively stable during the chronic infection. Preservation of CD4 T cells was also found in lymph nodes (LNs) of chronic SIVagm.sab-infected Caribbean AGMs. No activation of CD4+ T cells was detected in the periphery in SIV-infected Caribbean AGMs. These virological and immunological profiles from peripheral blood and LNs were identical to those previously reported in African-born AGMs infected with the same viral strain (SIVagm.sab92018). Due to these similarities, we conclude that Caribbean AGMs are a useful alternative to AGMs of African origin as a model for the study of SIV infection in natural African hosts.
Simian immunodeficiency virus (SIV) infection in macaques, compared to SIV infection of African nonhuman primates (NHPs) such as African green monkeys (AGMs), sooty mangabeys (SMs), mandrills, and chimpanzees, differs in one fundamental aspect: clinical outcome. While macaques infected with SIVmac progress relatively rapidly to AIDS, African NHPs naturally or experimentally infected with their species-specific SIV rarely develop disease (4-6, 12, 26, 39, 42, 43, 47, 49, 50). Only a few cases of immunodeficiency have been reported to date in African NHPs (3, 29, 31, 44, 55). Numerous investigations have tried to explain this paradox, but no generally accepted explanation presently exists.
In the past several years three experimental “nonpathogenic” models have been developed: infection of (African-origin) AGM species with their respective variants of SIVagm (9, 26, 42), infection of sooty mangabeys with SIVsm or SIVmac (25, 49), and infection of mandrills with SIVmnd-1 and SIVmnd-2 (39, 40). The study of these models showed that in naturally and experimentally SIV-infected African monkeys, the dynamics of plasma viral load (VL) is surprisingly similar to human immunodeficiency virus-infected humans and SIVmac-infected rhesus macaques (Rh), with peaks of viral replication ranging between 3 × 105 and 3 × 109 and chronic VL levels of 1 × 103 to 1 × 107 (5, 9, 12, 13, 25, 26, 39, 40, 42, 43, 49, 50). High levels of viral replication in the absence of clinical progression were reported for SIVagm-infected AGMs (5, 12, 13, 26, 42) as well as for other African NHP hosts of SIV, such as SMs (6, 25, 30, 47, 49, 50) and mandrills (39, 40, 43, 44).
However, although these models provided key new insights into SIV biology, they suffer from significant practical limitations, such as availability of experimental animals due to difficulties in their importation, risk of transfer of various pathogenic agents from Africa, and endangered species limitations. Therefore, development of an alternative animal model for natural SIV infection would be a significant achievement in the field.
Given the endangered nature of other African NHP species currently used in AIDS research, AGMs would be the primate model of choice for investigation of viral replication and immune responses in the tissues in natural hosts infected with SIV: they are abundant and widely spread throughout sub-Saharan Africa, and they have a high prevalence of SIVagm infection in the wild (24, 45). Therefore, AGMs represent the largest reservoir of SIV. Different species of AGMs (vervet [Chlorocebus pygerythrus], grivet [C. aethiops], tantalus [C. tantalus], and sabaeus [C. sabaeus]) each carry their own SIVagm subtypes, named SIVagm.ver, SIVagm.gri, SIVagm.tan, and SIVagm.sab, respectively (1, 10, 11, 18, 23, 33). Since AGMs from Africa are currently difficult to import to the United States and may carry various pathogenic agents, a possible alternative to the AGM model based on animals from Africa exists in the colonies of AGMs established approximately 300 years ago on several Caribbean islands by the transfer of AGMs from West Africa. In contrast to their African relatives, in which SIV prevalence is up to 60 to 70% in the adult population (24, 45), Caribbean AGMs are negative for SIV (7, 16). This led to the suggestion that Caribbean AGMs might be resistant to SIV infection (20). A more plausible explanation could be that the founder animals had been captured at a young age, when they were still uninfected (36).
A second difference between the African-born and Caribbean-born AGMs seems to lie in the CD4 and CD8 expression. It is widely accepted that CD4+ T lymphocytes from most AGMs from Africa coexpress CD8 (12, 20, 32, 34, 37). However, this has been demonstrated to be due mainly to expression of the CD8α on AGM CD4+ T cells, rather than to CD8β expression (20). The same study reported that Caribbean AGMs differ from their African counterparts in that they lack the double CD4+ CD8α+ T-cell population. It was suggested that the lack of a double-positive phenotype might be considered a reversion to normal of a species that had not encountered SIVagm since separation from their African counterparts (20). The latter raised the question of whether the susceptibility of Caribbean AGMs to AIDS has changed (20).
To determine whether Caribbean AGMs represent a suitable alternative to AGMs from Africa for SIV pathogenesis studies, we investigated the dynamics of VLs and changes in the major cell populations in peripheral blood and lymph nodes (LNs) of SIVagm-infected Caribbean AGMs and compared them with previous results from SIVagm-infected AGMs from Africa.
We show here that Caribbean AGMs support productive and nonpathogenic SIV infection for up to 420 days. In SIVagm.sab92018-infected Caribbean AGMs, VLs and immunophenotypic changes are similar to those in African AGMs. No CD4 T-cell activation was detected in this new model during SIVagm infection. Thus, Caribbean AGMs infected with SIVagm.sab are appropriate models for studying the pathogenesis of SIV infection of “natural” hosts. These monkeys are plentiful and available; their use in this line of research will alleviate the previously mentioned restrictions.
MATERIALS AND METHODS
Animals.
Twelve healthy, uninfected Caribbean AGMs originating from St. Kitts island were tested for CD4, CD8α, and CD8β expression. The SIVagm.sab92018 infection study group comprised six female Caribbean AGMs (Chlorocebus sabaeus). The animals were adults (mean age, 8 years).
All animals were negative for simian T-cell lymphotropic virus (Vironostika human T-cell lymphotropic virus-I/II enzyme-linked immunosorbent assay [ELISA]; bioMérieux, Durham, NC) and SIV by both ELISA and Western blot assay using SIVmac strips (Zeptometrix, Buffalo, N.Y.).
Animals were housed at the Tulane National Primate Research Center (TNPRC), an AAALAC International-accredited facility. Housing and handling of animals were in accordance with the Guide for the Care and Use of Laboratory Animals (U.S. Public Health Service) (35) and the Animal Welfare Act. All protocols and procedures were reviewed and approved by the Tulane University Institutional Animal Care and Use Committee.
Genetic characterization of Caribbean AGMs by cytochrome b sequencing.
In order to confirm the species of Caribbean AGMs, DNA was extracted from 300 μl of whole blood from 12 monkeys originating from St. Kitts, 5 monkeys originating from Barbados, 7 sabaeus monkeys from Senegal, 4 vervet monkeys from Tanzania, and 3 tantalus monkeys from the Central African Republic. A QIAamp blood kit (QIAGEN, Valencia, CA) was used to extract the DNA, which was eluted into 60 μl of H2O and stored at −20°C. A 267-base-pair (bp) region of cytochrome b, representing bases 14841 to 15149 of the human mitochondrial genome (2), was amplified using primers L14841 and H15149 (21). One microgram of host DNA was used in a 100-μl PCR using a PerkinElmer model PE 9700 thermocycler. The PCR conditions were as follows: 2 min at 94°C, followed by 40 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C, with a final extension of 5 min at 72°C. Following purification with a QIAquick PCR purification kit (QIAGEN), the PCR products were cycle sequenced and analyzed on an ABI-377 automated DNA sequencer.
Phylogenetic trees.
Cytochrome b sequences from different species of AGMs were aligned to that from Chlorocebus aethiops (GenBank accession no. AY863426) and to sequences from other African NHPs retrieved from GenBank or generated in our laboratory. Sequences were aligned using Clustalw version 1.8. Unrooted maximum parsimony trees were inferred using PAUP version 4.0b8 (54) with uniformly weighted unordered characters. Distance matrix-based trees were constructed with the neighbor-joining method (48) using the HKY85 model of nucleotide substitution (15). The reliability of the branching order was estimated by performing 1,000 bootstrap replicates.
In vitro infection of Caribbean PBMCs with SIVagm.sab.
To verify the capacity of SIVagm.sab to infect peripheral blood mononuclear cells (PBMCs) from Caribbean AGMs in vitro, cells from four different animals were used. In order to test the virus in both CD4 phenotypes described in Caribbean AGMs, two animals with the double-positive CD4+ CD8α+ T-cell population and two animals lacking this phenotype were examined. Cells were first stimulated with phytohemagglutinin (PHA; 1 ng/ml) for 3 days at 37°C in a 5% CO2 incubator. Cells were then washed in 10% complete RPMI, counted, resuspended at 1 × 106 cells/ml in complete medium containing 10% human interleukin-2 (IL-2), and cultured for an additional 3 days in Transwell plates (Costar, Corning, NY). In order to verify whether there was any modification in CD4 expression before and after stimulation of AGM PBMCs, CD4 expression was tested by flow cytometry at each of these steps. Before infection, the concentration of cells was again adjusted to 1 × 106/ml and 1 to 5 ml of virus stock (60 50% tissue culture infective doses of SIVagm.sab 2) was added to the culture for 24 h. The virus inoculum was washed out after 1 day, and the cell culture was maintained for 14 days. Viability of the cells was monitored daily, and fresh medium and IL-2 were added in order to maintain the culture at 1 × 106 cells/ml. Every 3 days, 1 ml of tissue culture supernatant was removed from the culture and clarified by centrifugation (1,500 rpm for 10 min). Supernatants were tested for the presence of the virus using the SIV core P27 antigen capture assay (Zeptometrix, Buffalo, NY). As a control, SIVmac251 was grown on Rh PBMCs in the same conditions.
SIVagm.sab in vivo infection.
To avoid selection of viral variants in vitro, inocula used in this study consisted of plasma obtained from an experimentally SIVagm.sab92018-infected sabaeus monkey from Senegal during primary infection, and stocks were prepared and titers were determined on SupT1 as described elsewhere (9). All six AGMs were inoculated with plasma equivalent to 300 50% tissue culture infective doses of SIVagm.sab9018. Three AGMs (EI45, EI52, and EI53) received a viral stock that was prepared in a sabaeus monkey from Senegal. The same stock had been used in our previous studies for infection of African AGMs (26, 42). The other three AGMs (EI42, EI44, and EI51) were inoculated with a stock obtained in a Caribbean AGM (EI45) experimentally infected with the virus stock mentioned above. The virus stock was named SIVagm.sab92018 (EI45). The SIVagm.sab92018 (EI45) inoculum was adjusted to a viral RNA load equivalent to the initial stock, both determined by real-time (rt)-PCR.
Specimen collection.
Five milliliters of EDTA blood was collected from the femoral vein of each monkey two times before infection (days −15 and 0), twice weekly throughout the first 3 weeks postinfection (p.i.), weekly thereafter until the set point (days 28 and 35 p.i.), and finally, every month during chronic infection (between 2 and 18 months p.i.). Whole blood was used for flow cytometry within 1 hour after collection. Plasma was aliquoted and stored at −80°C prior to the VL quantification.
Excisional biopsy specimens of axillary LNs were collected from all the animals as previously described (57). LNs were sampled before infection and at selected time points (days 0, 7, 10, 14, 28, 200, and 350 p.i.) during the acute and chronic phases of SIVagm infection.
Isolation of lymphocytes.
Lymphocytes were isolated from the axillary LNs by gently mincing and pressing tissues through nylon mesh screens and stained for flow cytometry as previously described (57).
Flow cytometry.
Whole blood was stained using a lysis technique as previously described (56). Blood cells and cell suspensions from LNs were stained for flow cytometric analysis using four-color staining combinations with monoclonal antibodies CD20-fluorescein isothiocyanate (FITC), CD3-phycoerythrin, CD8α-peridinin chlorophyll A protein, CD4-allophycocyanin, and HLA-DR (BD Biosciences Pharmingen, San Diego, CA). Cells were incubated with an excess of monoclonal antibodies at 4°C for 30 min, followed by a phosphate-buffered saline wash (400 × g, 7 min) and fixation in 2% paraformaldehyde. Data were acquired with a FACSCalibur flow cytometer and analyzed with CellQuest software (BD Bioscience Immunocytometry Systems).
CD4 and CD8 expression on T-cell subsets was determined by gating on lymphocytes and then on CD3+ T cells. To reflect the relative proportion of CD4+ and CD8+ T cells present in the LNs, we calculated an “index” of these cell populations. This index is the product of the percentage of CD3+ T cells (gated on lymphocytes) multiplied by the percentage of CD4+ or CD8+ T cells, divided by 100. This index provides a more accurate reflection of target cells, since it is a function of both total CD3+ T cells and CD4+ or CD8+ T cells.
Antibody detection.
Anti-SIVagm.sab92018 antibody dynamics were monitored by SIVagm.sab-specific enzyme immunoassay (primate immunodeficiency virus enzyme immunoassay) based on peptides mapping the conserved Gp41 immunodominant region and the highly variable V3 loop of SIVagm.sab2 (51). Serological reactivities were confirmed by Western blotting for all the monkeys included in this study (Zeptometrix Corp., Buffalo, NY).
Quantification of viral P27 antigenemia.
Plasma samples were assayed for P27 antigen by ELISA with standards provided in the SIV core p27 antigen capture assay kit (Zeptometrix Corp., Buffalo, NY). Quantification was done according to the manufacturer's instructions. This ELISA kit was initially designed to detect P27 of SIVmac but has also been shown to detect Gag antigens of SIVagm (27). In addition, in previous studies we confirmed its cross-reactivity for SIVagm.sab Gag (9, 26, 42).
Viral RNA quantification.
Viral RNA was extracted from 540 μl of plasma using the QIAamp viral RNA extraction kit (QIAGEN, Valencia, CA). Plasma RNA quantification was done using rt-PCR assays specific for SIVagm.sab as previously described (26, 42). Briefly, total RNA was retrotranscribed into cDNA using the TaqMan Gold rt-PCR kit and random hexamers (PE Applied Biosystems, Foster City, CA). PCRs were carried out in a spectrofluorometric thermal cycler (ABI PRISM 7700; PerkinElmer). The quantification was based on the amplification of a 180-bp segment located in the long terminal repeat region. This region is highly conserved within different SIVagm strains. The primers and probe were specifically designed for SIVagm.sab (J15S, 5′ J15S, and J15P) as previously described (9). Absolute viral RNA copy numbers were deduced by comparing the relative signal strength to corresponding values obtained for five 10-fold dilutions of an SIVagm.sab long terminal repeat RNA that was reverse transcribed and amplified in parallel. The detection limit of the SIVagm.sab quantification assays was 5 × 102 RNA copies/0.5 ml of plasma.
Statistical analyses.
Flow cytometry data were analyzed for significant differences by the Wilcoxon test using StatView software.
Nucleotide sequence accession no.
The nucleotide sequences of the cytochrome b genes from different species of AGMs were deposited in GenBank (accession no., DQ451215 through DQ451245).
RESULTS
CD4, CD8α, and CD8β expression in Caribbean AGMs.
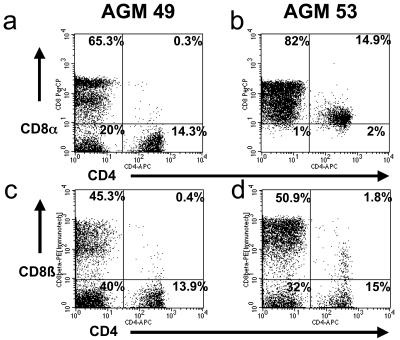
It was reported that Caribbean AGMs have a different CD4+ T-cell phenotype than their African counterparts in that they lack the double-positive CD4+ CD8+ T-cell population (20). We have analyzed the blood of 12 Caribbean AGMs originating from St. Kitts and showed that, similar to AGMs from Africa, including sabaeus monkeys originating from Senegal (20, 26), Caribbean sabaeus monkeys are actually heterogeneous in terms of CD4 and CD8α coexpression (Fig. 1a and b). Furthermore, Caribbean sabaeus monkeys showed the same pattern of CD8β expression on CD4+ T cells as AGMs from Africa (Fig. 1c and d). This indicates that there is no major difference between Caribbean monkeys and their African counterparts regarding the presence of double-positive CD4+ CD8+ T cells.
FIG. 1.
CD4 and CD8 expression in blood of Caribbean AGMs. Caribbean (St. Kitts) AGMs display two different phenotypes of CD4+ T cells; a representative example of each is shown here. In some animals, such as AGM EI49, very few CD4+ CD8α+ T cells are observed (a); AGM EI53 is representative of animals the majority of whose CD4+ T cells have a low expression of CD8α (b). When CD8+ T cells are defined with a CD8β monoclonal antibody, a similar pattern of staining is observed for all Caribbean AGMs, due to few CD4+ CD8β+ T cells (c and d).
Genetic classification of AGMs.
A cytochrome b phylogenetic tree was constructed to determine to which species of AGMs Caribbean monkeys belonged. Data were also used to determine if there were any genetic differences between the monkeys that show the double CD4+ CD8+α phenotype and the animals which lack this phenotype. The phylogenetic analysis confirms that the monkeys originating from St. Kitts are AGMs (Fig. 2). These animals tightly clustered with AGMs originating from Barbados, a different Caribbean island. All Caribbean AGMs were different from vervet monkeys originating from Tanzania, tantalus monkeys from Central Africa, and grivet monkeys (sequence retrieved from GenBank) but formed a cluster with sabaeus monkeys originating from Senegal. The 12 AGMs from our study group, originating from St. Kitts, formed two closely related groups, differentiated by one nucleotide in the studied region (Fig. 2). No correlation between the CD4+ CD8α+ phenotype or CD4+ CD8αneg phenotype and the two subgroups of Caribbean AGMs was found. Monkeys with both phenotypes clustered in both subgroups (data not shown).
FIG. 2.
Caribbean AGM speciation based on mitochondrial DNA characterization. Cytochrome b sequences (267 bp) were amplified from Caribbean AGMs and compared with those of different species of AGMs from Africa. Sequences originating from different species of AGMs of African origin are color coded: sabaeus in red, tantalus in blue, grivet in orange, and vervet in green. Sequences from Caribbean AGMs are shown in black. Monkeys coded EI are from St. Kitts island; monkeys coded A and B are from Barbados. Cytochrome b sequences from other species of African monkeys are shown in gray. The phylogenetic trees were estimated by the maximum parsimony method. The reliability was estimated from 1,000 bootstrap replicates; only bootstrap values relevant for lineage definition are shown.
Minimal CD4 down-regulation occurs after in vitro stimulation of AGM PBMCs.
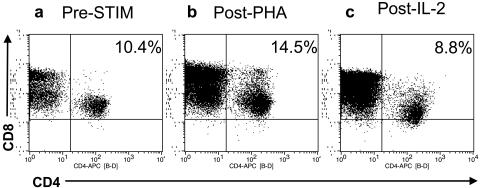
In order to study the susceptibility of Caribbean AGMs to in vitro infection by SIVagm, we first evaluated the levels of CD4 expression on activated CD4+ T cells. Early studies reported a marked down-regulation of the CD4 expression in AGM PBMCs upon stimulation and suggested that this may explain the lack of pathogenicity of SIVagm in its natural host (34). Therefore, CD4 expression was examined in unstimulated and stimulated PBMCs from four AGMs (two representative of the CD4+ CD8α+ phenotype and two that lacked CD8α on their CD4+ T cells) in order to exclude down-regulation of CD4 as a potential cause of the failure of SIVagm.sab to grow in Caribbean AGM PBMCs. The mean percentage of CD4 cells normalized to CD3 after PBMC separation was 9.7% ± 1.3% in the four animals (Fig. 3a). After 3 days of PHA stimulation, CD4 expression increased slightly (mean, 13.1% ± 2.4%; P > 0.5) (Fig. 3b) and then decreased thereafter (mean, 9.4% ± 2.6%; P > 0.5) after 3 additional days of IL-2 administration (Fig. 3c). The CD4 mean fluorescence intensity followed the same dynamics after the PHA treatment, with an increase to 153.9 ± 11.9 compared to the prestimulation level (114.2 ± 15.3) (P > 0.5). After IL-2 stimulation, the CD4 mean fluorescence intensity remained slightly but not significantly higher than the pretreatment values (141.5 ± 23) (P > 0.5). Thus, only minor modifications of CD4 expression could be observed in cell cultures of AGM PBMCs, and no significant decrease was detected, in contrast with previous reports (34).
FIG. 3.
Expression of CD4 before (a) and after stimulation with PHA (b) and IL-2 (c). An increase of CD4-positive cells was noted after 3 days of PHA stimulation (b). After 5 additional days of treatment with IL-2, a decrease of CD4 T+ cells was observed (c), but this reduction was insignificant compared with CD4 levels before stimulation (a).
Caribbean AGM PBMCs are susceptible to SIVagm.sab in vitro and in vivo.
To study whether Caribbean AGMs are susceptible to SIVagm.sab, we first verified the susceptibility of AGM PBMCs to infection with the NIH reference strain SIVagm.sab2. We tested this strain for coreceptor usage and demonstrated that it was dual tropic (data not shown), similar to SIVagm.sab92018 (42). Two different phenotypes of AGMs were tested in order to exclude a possible restriction of the virus growth to monkeys with either phenotype: PBMCs from two Caribbean AGMs in which the majority of CD4+ T cells were double positive for CD8α, and two animals that lacked this particular phenotype. As a positive control, Rh PBMCs were infected with SIVmac251. Our experiment revealed productive replication of SIVagm.sab2 in PBMCs from Caribbean AGMs (mean P27 at day 7 postinoculation = 1.73 ng/ml) without regard to CD4 phenotype. No significant differences in the P27 values could be observed between infection with SIVagm in Caribbean AGM PBMCs and SIVmac in Rh PBMCs (1.73 ± 0.03 ng/ml versus 1.15 ± 0.12 ng/ml), indicating no restriction of SIVagm.sab in the former.
To test the in vivo susceptibility of Caribbean AGMs to SIVagm.sab infection, six animals were inoculated with SIVagm.sab92018. Animals were again selected to display the different phenotypes CD4+ CD8α+ and CD4+ CD8αneg (four versus two) in order to determine if these cell subsets play a particular role in SIVagm pathogenesis in Caribbean AGMs, as has been suggested (20).
All animals became infected and seroconverted between 21 and 45 days p.i. (Fig. 4a), similar to AGMs from Africa infected with the same SIVagm strain (9). None of the monkeys had clinical signs related to the SIV infection during the follow-up (up to day 420 p.i.). No weight loss, fever, opportunistic infections, or neoplasia was noted. One monkey died at 132 days postinfection due to causes unrelated to SIV infection.
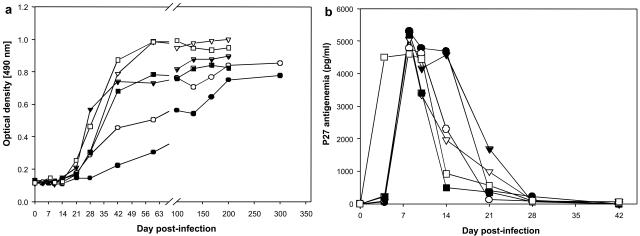
FIG. 4.
Antibody responses (a) and p27 antigenemia (b) in Caribbean AGMs infected with SIVagm.sab. Anti-gp41 antibodies against SIVagm.sab were detected by an in-house peptide ELISA using specific peptides (51); all animals seroconverted between days 21 and 45 p.i. Positive P27 antigenemia was detected only during the acute infection (up to day 28 p.i.). Symbols for animals: no. EI45 (•), no. EI53 (○), no. EI52 (▾), no. EI42 (▿), no. EI51 (▪), and no. EI44 (□).
The P27 antigenemia levels in SIVagm.sab92018-infected Caribbean AGMs are similar to those observed in their African counterparts.
To evaluate systemic viral replication, two viral parameters were measured: P27 antigenemia and plasma RNA VL. P27 antigenemia quantification showed that all animals were positive for P27 during primary infection at day 4 p.i. The P27 values peaked at up to 4.6 to 5.2 ng/ml of plasma between days 8 and 10 p.i. (Fig. 4b). These values are very high, and in the same range as those recorded for SIVagm.sab92018-infected AGMs from Africa (9, 14, 42) and for SIVmac-infected Rh (17, 28, 38, 53, 58). After primary infection, P27 antigenemia became undetectable in SIVagm.sab-infected Caribbean AGMs by day 28 p.i., similar to what was described for the asymptomatic phase of chronic SIVmac, HIV-1, SIVagm, SIVsm, and SIVmnd-1/2 infections (9, 17, 39, 40, 42, 43). No differences in P27 antigenemia could be detected between monkeys with high numbers of CD4+ CD8α+ T cells and the animals that lacked this phenotype. These results demonstrate that SIVagm.sab replicates well in Caribbean AGMs.
SIVagm.sab92018-infected Caribbean and African AGMs show the same range of plasma RNA VLs.
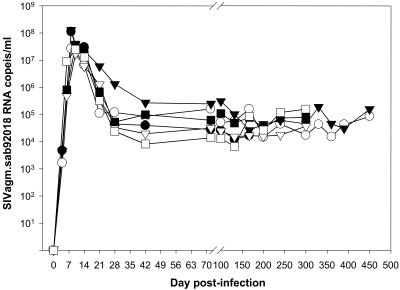
The dynamics of plasma RNA VL of SIVagm.sab in Caribbean AGMs is shown in Fig. 5. Plasma viremia peaked between days 8 and 10 and the values ranged between 2.7 × 107 and 1.3 × 108 copies/ml. This is very similar to what has been described in 11 sabaeus monkeys from Senegal experimentally infected with the same SIVagm.sab92018 strain (42). Thus, plasma RNA quantification confirmed the high antigenemia levels detected in SIVagm.sab-infected Caribbean AGMs. No difference in plasma viremia was observed between animals infected with viral stocks of isolate 92018 produced in African or Caribbean AGMs. As reported earlier, these VL values are high and are comparable to levels previously described for pathogenic SIVmac infections (17, 28, 38, 52, 58).
FIG. 5.
Viral RNA copy numbers in plasma of SIVagm.sab92018-infected Caribbean AGMs. The detection limit of the assay was 103 copies/ml. The range of plasma VLs in Caribbean AGMs is in the same range as values observed in African AGMs infected with the same viral strain (42). Symbols for animals: no. EI45 (•), no. EI53 (○), no. EI52 (▾), no. EI42 (▿), no. EI51 (▪), and no. EI44 (□).
The set-point levels in Caribbean AGMs ranged between 104 and 105 copies/ml (Fig. 5). The plasma VLs during the postacute and chronic phases were in the same range as those we and others reported previously in experimentally or naturally SIVagm.sab-infected AGMs from Africa (5, 9, 12, 13, 19, 26, 42).
Altogether, our results show that SIVagm.sab replicates at the same level and following the same pattern in Caribbean AGMs as in those from Africa (9, 14, 26, 42). Therefore, we concluded that Caribbean AGMs may be used as an alternative to AGMs from Africa for the study of immunological factors of protection against AIDS.
SIVagm.sab-infected Caribbean AGMs showed transient CD4+ T-cell declines in peripheral blood and LNs.
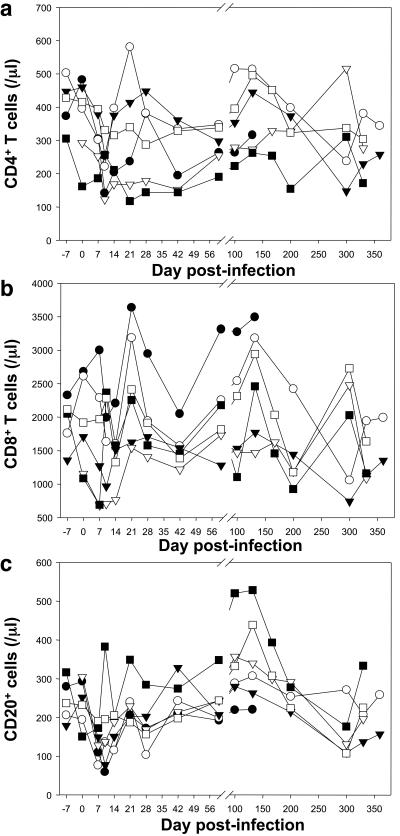
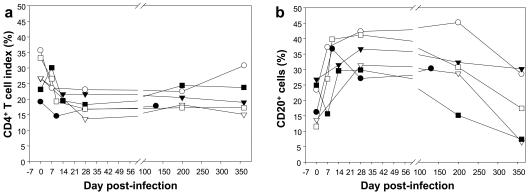
The dynamics of CD3+, CD4+, and CD8+ T cells and of CD20+ B cells were measured in SIVagm.sab92018-infected Caribbean AGMs, in blood and LNs (Fig. 6). Caribbean AGMs had a mean of 383 CD4+ T cells/μl (range: 292 to 502 cells/μl) prior to SIVagm.sab infection. After inoculation, similar to SIVagm.sab-infected African sabaeus monkeys, Caribbean AGMs demonstrated a significant drop in CD4+ T-cell counts, reaching the nadir at day 10 p.i. (P < 0.01) (Fig. 6a). After the viral peak, the CD4+ T-cell values rebounded close to preinfection values (reached by day 28 for most of the animals) and remained at relatively the same levels during chronic SIV infection. A transient decrease in CD4+ T cells was also observed in the LNs that reached a significant degree at day 28 (P < 0.03), similar to what has been previously reported in SIVmnd-1-infected mandrills (39). By day 200 the CD4 values were slightly (but not significantly) decreased compared with preinfection values (Fig. 7a). A small but insignificant decrease was also observed in peripheral CD8+ T cells at the peak of the VL, but this was readily recovered thereafter (Fig. 6b). No significant changes were noted in the dynamics of CD8+ T cells in the LNs of the SIVagm.sab-infected AGMs. Thus, the CD4+ and CD8+ T-cell dynamics in LNs was similar to those described in AGMs of African origin (9, 26, 42). The numbers of CD20 cells transiently decreased (P < 0.05) in peripheral blood during primary infection, with the lowest values occurring at days 7 to 10 p.i (Fig. 6c). At day 21, however, numbers of CD20+ cells returned to the preinfection values. During the chronic phase (between days 100 and 175), B lymphocytes transiently increased (P < 0.05), and thereafter they returned to normal at around day 200. In the LNs, the percentage of CD20+ cells increased temporarily at day 28 (P = 0.03) and returned to normal by day 200, concomitant with CD4+ T-cell normalization (Fig. 7b).
FIG. 6.
T- and B-cell dynamics in blood of SIVagm.sab-infected Caribbean AGMs. CD4+ T cells transiently decreased in the blood (a) of SIVagm.sab-infected AGMs at the peak of VLs. The levels rebounded to levels close to preinfection values during chronic SIVagm.sab infection. No significant modification was observed in the dynamics of CD8+ T cells in the peripheral blood. (b) A transient decrease in the numbers of CD20+ B cells was present in the blood of SIVagm.sab-infected AGMs (c). Symbols for animals: no. EI45 (•), no. EI53 (○), no. EI52 (▾), no. EI42 (▿), no. EI51 (▪), and no. EI44 (□).
FIG. 7.
CD4+ T-cell and B-cell dynamics in LNs in SIVagm.sab-infected Caribbean AGMs. A significant (P < 0.05) decrease in CD4+ T-cell percentages was observed in the LNs at day 28 post-SIVagm.sab inoculation. The levels of CD4 T+ cells remained decreased at day 200, but the difference was not significant compared with preinfection values (a). Concomitant relative increase and return to normal levels of CD20 cells in the same compartment (b). Symbols for animals: no. EI45 (•), no. EI53 (○), no. EI52 (▾), no. EI42 (▿), no. EI51 (▪), and no. EI44 (□).
The comparison of CD4+ T-cell dynamics in blood and LNs of SIVagm.sab-infected Caribbean and African AGMs clearly shows similarities. In both species, similar to SIVmac251-infected Rh, we observed an early decline in the blood CD4+ T cells. However, in contrast to the pathogenic SIV model, there is better preservation of the CD4+ T cells in both Caribbean AGMs and those from Africa during the chronic phase of SIV infection.
Caribbean AGMs and AGMs from Africa respond to SIVsm infection with a similar pattern of T-cell activation.
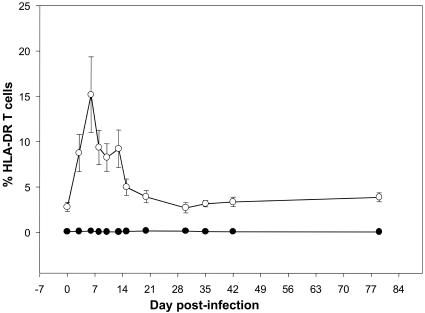
The dynamics of HLA-DR on CD4+ and CD8+ T cells was determined by flow cytometry in order to determine whether, similarly to AGMs from Africa, Caribbean-born AGMs infected with SIVagm show low levels of T-cell activation. SIVagm.sab92018 infection of Caribbean AGMs was associated with only a transitory increase in the activation status of CD8+ T cells. DR+ CD8+ T cells significantly increased at as early as day 3 p.i. and reached a peak at day 7 p.i. (15.19% ± 4.17%) (Fig. 8). By day 20 p.i., these cells returned to preinoculation values (3.9% ± 0.67% versus 2.8% ± 0.5%) (Fig. 8). No significant change was observed in peripheral blood for DR+ CD4+ T cells in SIV-infected Caribbean AGMs (Fig. 8). This pattern of dynamics is very similar to that previously reported in sabaeus monkeys of African origin, in which T-cell activation occurs very early and only transiently during SIVagm.sab infection (26).
FIG. 8.
Kinetic expression of percent HLA-DR upon CD4+ (•) and CD8+ (○) T lymphocytes in peripheral blood of SIVagm.sab92018-infected Caribbean AGMs. A significant but transitory activation of CD8 T cells can be observed during the primary infection. No modification of the activation status was observed for the CD4+ T cells in SIVagm.sab-infected Caribbean AGMs.
DISCUSSION
In this study we show that AGMs from the Caribbean are C. sabaeus and that they apparently respond identically to SIVagm.sab92018 infection as do African-born AGMs. This is a significant result, since it offers alternatives and expands the avenues for pathogenesis studies in NHP hosts of SIVs. In these monkeys, SIVs generally do not induce disease, in spite of active viral replication (5, 6, 12, 13, 25, 26, 30, 39, 40, 42-44, 47, 49, 50). Therefore, the study of these models allowed the investigation of the reasons for reduced pathogenicity. Such models for pathogenesis studies are needed in order to identify factors associated with protection against lentiviral disease. However, extensive studies are hampered because most currently available animal models for SIV infection in natural hosts are either highly endangered species (such as SMs, mandrills, and chimpanzees) or difficult to obtain (AGMs from Africa). A Caribbean AGM model for SIV infection is therefore a significant new resource, due to the availability of these monkeys and to the fact that Caribbean AGM colonies are currently present in the United States.
Caribbean AGMs are an appropriate model for studying SIVagm infection.
Our data clearly show that Caribbean AGMs are susceptible to SIVagm.sab infection in vitro and in vivo. SIVagm.sab infection in Caribbean AGMs faithfully reproduces the viral parameters of SIVagm infection in AGMs from Africa. This contradicts anecdotal reports that Caribbean AGMs are resistant to SIV, which were based on several lines of evidence. (i) First, there is the absence of SIV infection in Caribbean AGM colonies despite repeated large-scale serological surveys (7). Our results support the hypothesis that the founder Caribbean AGMs were probably SIVagm negative when separated from AGMs in Africa and brought to the Caribbean islands, rather than being naturally resistant to SIV (8, 32). Their young age at the time of separation may be the key to their SIV negativity, as it has been demonstrated that AGMs become SIVagm positive at sexual maturity (45), and no vertical transmission of SIVagm has been described to date (41). (ii) Second, previous attempts to infect Caribbean monkeys with SIV resulted in significantly less efficient viremia compared with AGMs from Africa, and some of the animals failed to become persistently infected (13). The reason for these failures may be the use of SIVagm strains belonging to different subtypes. Prior studies used SIVagm.ver, which naturally infects a different AGM subspecies, the vervets (Chlorocebus pygerythrus). (iii) Finally, differences in CD4 and CD8 expression reported by Holznagel et al. suggested that a modification of the immune system of Caribbean AGMs may have occurred as the result of them being separated from their African relatives for 200 to 400 years (20). However, we did not find any difference in CD4+ T-cell phenotype regarding CD8 coexpression between Caribbean and African sabaeus monkeys. Monkeys harboring double-positive CD4/CD8 T cells were present in both populations. A possible explanation for the difference in results may rely on the use of inbred animals in the previous study (20). Also, in our study, only animals originating in St. Kitts were included. It is possible that monkeys born in Barbados might be more homogeneous in terms of CD4/CD8 expression. Regardless, no difference was found in the pattern of viral replication after SIVagm in vitro and in vivo infection of Caribbean AGMs harboring CD4+ CD8α+ or CD4+ CD8αneg, indicating that these phenotypes do not play a role in their susceptibility to infection.
In order to match the virus with the host species, the species of our Caribbean AGMs was confirmed by genotyping. Based on cytochrome b typing, we demonstrated for the first time that Caribbean AGMs originating from St. Kitts cluster with sabaeus monkeys from Senegal. All Caribbean AGMs showed clear differences from three other species of AGMs, such as vervet, tantalus, and grivet monkeys. Surprisingly, despite their putatively more diverse origin (8), Caribbean AGMs from Barbados showed a high homogeneity in their cytochrome b structure, which was identical with sabaeus species of AGM from St. Kitts and Senegal. Our study therefore genetically traced the origins of all Caribbean AGMs to West Africa (Senegal) (Fig. 1) and confirmed that the use of an SIVagm.sab strain is the best approach for SIV pathogenesis studies in this AGM species.
Furthermore, there were no significant differences detected in the dynamics of blood and LN CD4+ T cells between SIVagm.sab-infected Caribbean and African AGMs. Moreover, Caribbean AGMs and AGMs from Africa respond to SIVsm infection with similar patterns of T-cell activation, as showed by the dynamics of the HLA-DR on their T cells. Finally, no difference in the availability of target cells (CD4+ CCR5+ T cells) was observed between Caribbean and African AGMs (I. Pandrea et al., submitted for publication). A longer follow-up of the animals in our study group will be done in order to verify potential differences in viral replication, immune cell dynamics, and clinical outcomes that can occur later. However, due to the previously mentioned similarities between Caribbean-born and African-born AGMs, our study strongly supports the usefulness of the Caribbean model to study SIVagm pathogenesis.
SIVagm.sab92018 as a reference strain for pathogenesis studies on AGM.
This study was based on the hypothesis that SIVagm.sab92018 would replicate at high levels in Caribbean AGMs and therefore may be used as a tool to study the SIVagm infection in the natural host. In addition to the reasons above regarding matching the species of animal and the viral strain, the selection of SIVagm.sab92018 was prompted by the following rationales. (i) We previously used this strain to infect sabaeus and vervet African monkeys and showed that SIVagm.sab92018 replicates at similar levels in both AGM species, although at slightly lower levels in vervets. These two species separated millennia ago (32). Therefore, we hypothesized that Caribbean AGMs that diverged from their African relatives only 300 years ago would support high levels of viral replication when infected with SIVagm.sab. (ii) This strain was previously used to infect a large number of AGMs from Africa, and we had sufficient data to compare the dynamics of SIVagm.sab92018 infection (9, 14, 26, 42, 46) in order to assess the efficacy of viral replication in Caribbean AGMs and validate our new model. (iii) All necessary experimental tools to investigate SIVagm.sab92018 VLs and CD4+ T-cell dynamics had already been developed in our previous studies (9, 14, 26, 42, 46), and overlapping peptides mapping gag, env, and nef genes of SIVagm.sab92018 recently became available for cellular immunology studies through the NIH. (iv) Finally, SIVagm.sab92018 is a wild-type virus. All the other SIVagm.sab strains available through the NIH AIDS Research Reference Reagent Program are laboratory-adapted strains grown on human cell lines (1, 22). Most of these strains are either not infectious in vivo, have not yet been tested in vivo, or have a truncated gp41 (SIVagm.sab1c). Recently the use of another primary isolate of SIVagm.sab from the central nervous system was reported (SIVagm.sab4br), but the in vivo replication of this virus was lower than that reported for SIVagm.sab92018 (J. S. Allan, B. K. Wilson, R. G. White, L. M. Parodi, V. L. Hodara, and L. D. Giaverdoni, 23rd Annu. Symp. Nonhuman Primate Models AIDS, abstr. 13, 2005). Combined with the above rationales, our results on VLs in vitro and in vivo demonstrate that primary SIVagm.sab92018 is an appropriate strain for pathogenesis studies using Caribbean AGMs.
In conclusion, by matching the Caribbean AGMs with their species-specific virus, we have developed a model for pathogenesis studies of SIV infection in natural African NHP hosts. This new model may be extensively employed to determine the factors associated with the low pathogenicity of these viruses and with the mechanisms of immune protection against disease progression. In the context of an expanding pandemic and of repeated failures of current vaccines to induce protective immune responses, our model, developed in a nonendangered species, is an invaluable tool for the study of host factors associated with the control of lentiviral infection.
Acknowledgments
We have no conflicting financial interests.
We thank O. Diop for the SIVagm.sab92018 (00008) virus stock preparation and M. J. Dodd, J. Leblanc, K. Rasmussen, J. Bruhn, E. Benes, and C. Lanclos for excellent technical help.
This study was supported by grants R01 AI064066 (I.P.), R01 AI49080 (R.V.), P20 RR020159 (C.A.), and P51 RR000164 (TNPRC) from the National Institutes of Health and by a grant from the Pasteur Institute (M.M.T. and F.B.S.).
REFERENCES
- 1.Allan, J. S., M. Short, M. E. Taylor, S. Su, V. M. Hirsch, P. R. Johnson, G. M. Shaw, and B. H. Hahn. 1991. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J. Virol. 65:2816-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, S., A. T. Bankier, B. G. Barrell, M. H. de Bruijn, A. R. Coulson, J. Drouin, I. C. Eperon, D. P. Nierlich, B. A. Roe, F. Sanger, P. H. Schreier, A. J. Smith, R. Staden, and I. G. Young. 1981. Sequence and organization of the human mitochondrial genome. Nature 290:457-465. [DOI] [PubMed] [Google Scholar]
- 3.Apetrei, C., B. Gormus, I. Pandrea, M. Metzger, P. ten Haaft, L. N. Martin, R. Bohm, X. Alvarez, G. Koopman, M. Murphey-Corb, R. S. Veazey, A. A. Lackner, G. Baskin, J. Heeney, and P. A. Marx. 2004. Direct inoculation of simian immunodeficiency virus from sooty mangabeys in black mangabeys (Lophocebus aterrimus): first evidence of AIDS in a heterologous African species and different pathologic outcomes of experimental infection. J. Virol. 78:11506-11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer, B., J. Denner, C. R. Brown, S. Norley, J. zur Megede, C. Coulibaly, R. Plesker, S. Holzammer, M. Baier, V. M. Hirsch, and R. Kurth. 1998. Simian immunodeficiency virus of African green monkeys is apathogenic in the newborn natural host. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 18:210-220. [DOI] [PubMed] [Google Scholar]
- 5.Broussard, S. R., S. I. Staprans, R. White, E. M. Whitehead, M. B. Feinberg, and J. S. Allan. 2001. Simian immunodeficiency virus replicates to high levels in naturally infected African green monkeys without inducing immunologic or neurologic disease. J. Virol. 75:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakrabarti, L. A., S. R. Lewin, L. Zhang, A. Gettie, A. Luckay, L. N. Martin, E. Skulsky, D. D. Ho, C. Cheng-Mayer, and P. A. Marx. 2000. Normal T-cell turnover in sooty mangabeys harboring active simian immunodeficiency virus infection. J. Virol. 74:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniel, M. D., N. L. Letvin, P. K. Sehgal, D. K. Schmidt, D. P. Silva, K. R. Solomon, F. S. Hodi, Jr., D. J. Ringler, R. D. Hunt, N. W. King, et al. 1988. Prevalence of antibodies to 3 retroviruses in a captive colony of macaque monkeys. Int. J. Cancer 41:601-608. [DOI] [PubMed] [Google Scholar]
- 8.Denham, W. W. 1981. History of green monkeys in the West Indies. Part I. Migration from Africa. J. Barbados Museum Historical Soc. 36:210-229. [Google Scholar]
- 9.Diop, O. M., A. Gueye, M. Dias-Tavares, C. Kornfeld, A. Faye, P. Ave, M. Huerre, S. Corbet, F. Barre-Sinoussi, and M. C. Muller-Trutwin. 2000. High levels of viral replication during primary simian immunodeficiency virus SIVagm infection are rapidly and strongly controlled in African green monkeys. J. Virol. 74:7538-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fomsgaard, A., J. Allan, M. Gravell, W. T. London, V. M. Hirsch, and P. R. Johnson. 1990. Molecular characterization of simian lentiviruses from East African green monkeys. J. Med. Primatol. 19:295-303. [PubMed] [Google Scholar]
- 11.Fukasawa, M., T. Miura, A. Hasegawa, S. Morikawa, H. Tsujimoto, K. Miki, T. Kitamura, and M. Hayami. 1988. Sequence of simian immunodeficiency virus from African green monkey, a new member of the HIV/SIV group. Nature 333:457-461. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein, S., I. Ourmanov, C. R. Brown, B. E. Beer, W. R. Elkins, R. Plishka, A. Buckler-White, and V. M. Hirsch. 2000. Wide range of viral load in healthy African green monkeys naturally infected with simian immunodeficiency virus. J. Virol. 74:11744-11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein, S., C. R. Brown, I. Ourmanov, I. Pandrea, A. Buckler-White, C. Erb, J. S. Nandi, G. J. Foster, P. Autissier, J. E. Schmitz, and V. M. Hirsch. 2006. Comparison of simian immunodeficiency virus SIVagmVer replication and CD4+ T-cell dynamics in vervet and sabaeus African green monkeys. J. Virol. 80:4868-4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gueye, A., O. M. Diop, M. J. Ploquin, C. Kornfeld, A. Faye, M. C. Cumont, B. Hurtrel, F. Barre-Sinoussi, and M. C. Muller-Trutwin. 2004. Viral load in tissues during the early and chronic phase of non-pathogenic SIVagm infection. J. Med. Primatol. 33:83-97. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 16.Hendry, R. M., M. A. Wells, M. A. Phelan, A. L. Schneider, J. S. Epstein, and G. V. Quinnan. 1986. Antibodies to simian immunodeficiency virus in African green monkeys in Africa in 1957-62. Lancet ii:455. [DOI] [PubMed] [Google Scholar]
- 17.Hirsch, V. M., T. R. Fuerst, G. Sutter, M. W. Carroll, L. C. Yang, S. Goldstein, M. Piatak, Jr., W. R. Elkins, W. G. Alvord, D. C. Montefiori, B. Moss, and J. D. Lifson. 1996. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J. Virol. 70:3741-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch, V. M., C. McGann, G. Dapolito, S. Goldstein, A. Ogen-Odoi, B. Biryawaho, T. Lakwo, and P. R. Johnson. 1993. Identification of a new subgroup of SIVagm in tantalus monkeys. Virology 197:426-430. [DOI] [PubMed] [Google Scholar]
- 19.Holzammer, S., E. Holznagel, A. Kaul, R. Kurth, and S. Norley. 2001. High virus loads in naturally and experimentally SIVagm-infected African green monkeys. Virology 283:324-331. [DOI] [PubMed] [Google Scholar]
- 20.Holznagel, E., S. Norley, S. Holzammer, C. Coulibaly, and R. Kurth. 2002. Immunological changes in simian immunodeficiency virus SIVagm-infected African green monkeys (AGM): expanded cytotoxic T lymphocyte, natural killer and B cell subsets in the natural host of SIVagm. J. Gen. Virol. 83:631-640. [DOI] [PubMed] [Google Scholar]
- 21.Irwin, D. M., T. D. Kocher, and A. C. Wilson. 1991. Evolution of the cytochrome b gene of mammals. J. Mol. Evol. 32:128-144. [DOI] [PubMed] [Google Scholar]
- 22.Jin, M. J., H. Hui, D. L. Robertson, M. C. Muller, F. Barre-Sinoussi, V. M. Hirsch, J. S. Allan, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J. 13:2935-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, P. R., M. Gravell, J. Allan, S. Goldstein, R. A. Olmsted, G. Dapolito, C. McGann, W. T. London, R. H. Purcell, and V. M. Hirsch. 1989. Genetic diversity among simian immunodeficiency virus isolates from African green monkeys. J. Med. Primatol. 18:271-277. [PubMed] [Google Scholar]
- 24.Jolly, C., J. E. Phillips-Conroy, T. R. Turner, S. Broussard, and J. S. Allan. 1996. SIVagm incidence over two decades in a natural population of Ethiopian grivet monkeys (Cercopithecus aethiops aethiops). J. Med. Primatol. 25:78-83. [DOI] [PubMed] [Google Scholar]
- 25.Kaur, A., R. M. Grant, R. E. Means, H. McClure, M. Feinberg, and R. P. Johnson. 1998. Diverse host responses and outcomes following simian immunodeficiency virus SIVmac239 infection in sooty mangabeys and rhesus macaques. J. Virol. 72:9597-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornfeld, C., M. J. Ploquin, I. Pandrea, A. Faye, R. Onanga, C. Apetrei, V. Poaty-Mavoungou, P. Rouquet, J. Estaquier, L. Mortara, J. F. Desoutter, C. Butor, R. Le Grand, P. Roques, F. Simon, F. Barre-Sinoussi, O. M. Diop, and M. C. Muller-Trutwin. 2005. Antiinflammatory profiles during primary SIV infection in African green monkeys are associated with protection against AIDS. J. Clin. Investig. 115:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lairmore, M. D., D. E. Hofheinz, N. L. Letvin, C. S. Stoner, S. Pearlman, and G. P. Toedter. 1993. Detection of simian immunodeficiency virus and human immunodeficiency virus type 2 capsid antigens by a monoclonal antibody-based antigen capture assay. AIDS Res. Hum. Retrovir. 9:565-571. [DOI] [PubMed] [Google Scholar]
- 28.Lifson, J. D., M. A. Nowak, S. Goldstein, J. L. Rossio, A. Kinter, G. Vasquez, T. A. Wiltrout, C. Brown, D. Schneider, L. Wahl, A. L. Lloyd, J. Williams, W. R. Elkins, A. S. Fauci, and V. M. Hirsch. 1997. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J. Virol. 71:9508-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ling, B., C. Apetrei, I. Pandrea, R. S. Veazey, A. A. Lackner, B. Gormus, and P. A. Marx. 2004. Classic AIDS in a sooty mangabey after an 18-year natural infection. J. Virol. 78:8902-8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ling, B., M. L. Santiago, S. Meleth, B. Gormus, H. M. McClure, C. Apetrei, B. H. Hahn, and P. A. Marx. 2003. Noninvasive detection of new simian immunodeficiency virus lineages in captive sooty mangabeys: ability to amplify virion RNA from fecal samples correlates with viral load in plasma. J. Virol. 77:2214-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClure, H. M., D. C. Anderson, T. P. Gordon, A. A. Ansari, P. N. Fultz, S. A. Klumpp, P. Emau, and M. Isahakia. 1992. Natural simian immunodeficiency virus infection in nonhuman primates. Top. Primatol. 3:425-438. [Google Scholar]
- 32.Muller, M. C., and F. Barre-Sinoussi. 2003. SIVagm: genetic and biological features associated with replication. Front. Biosci. 8:d1170-d1185. [DOI] [PubMed] [Google Scholar]
- 33.Muller, M. C., N. K. Saksena, E. Nerrienet, C. Chappey, V. M. Herve, J. P. Durand, P. Legal-Campodonico, M. C. Lang, J. P. Digoutte, A. J. Georges, et al. 1993. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J. Virol. 67:1227-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murayama, Y., A. Amano, R. Mukai, H. Shibata, S. Matsunaga, H. Takahashi, Y. Yoshikawa, M. Hayami, and A. Noguchi. 1997. CD4 and CD8 expressions in African green monkey helper T lymphocytes: implication for resistance to SIV infection. Int. Immunol. 9:843-851. [DOI] [PubMed] [Google Scholar]
- 35.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, D.C.
- 36.Norley, S., and R. Kurth. 2004. The role of the immune response during SIVagm infection of the African green monkey natural host. Front. Biosci. 9:550-564. [DOI] [PubMed] [Google Scholar]
- 37.Norley, S. G., G. Kraus, J. Ennen, J. Bonilla, H. Konig, and R. Kurth. 1990. Immunological studies of the basis for the apathogenicity of simian immunodeficiency virus from African green monkeys. Proc. Natl. Acad. Sci. USA 87:9067-9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowak, M. A., A. L. Lloyd, G. M. Vasquez, T. A. Wiltrout, L. M. Wahl, N. Bischofberger, J. Williams, A. Kinter, A. S. Fauci, V. M. Hirsch, and J. D. Lifson. 1997. Viral dynamics of primary viremia and antiretroviral therapy in simian immunodeficiency virus infection. J. Virol. 71:7518-7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onanga, R., C. Kornfeld, I. Pandrea, J. Estaquier, S. Souquiere, P. Rouquet, V. P. Mavoungou, O. Bourry, S. M'Boup, F. Barre-Sinoussi, F. Simon, C. Apetrei, P. Roques, and M. C. Muller-Trutwin. 2002. High levels of viral replication contrast with only transient changes in CD4+ and CD8+ cell numbers during the early phase of experimental infection with simian immunodeficiency virus SIVmnd-1 in Mandrillus sphinx. J. Virol. 76:10256-10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onanga, R., S. Souquiere, M. Makuwa, A. Mouinga-Ondeme, F. Simon, C. Apetrei, and P. Roques. 2006. Primary simian immunodeficiency virus SIVmnd-2 infection in mandrills (Mandrillus sphinx). J. Virol. 80:3301-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otsyula, M. G., A. Gettie, M. Suleman, R. Tarara, I. Mohamed, and P. Marx. 1995. Apparent lack of vertical transmission of simian immunodeficiency virus (SIV) in naturally infected African green monkeys, Cercopithecus aethiops. Ann. Trop. Med. Parasitol. 89:573-576. [DOI] [PubMed] [Google Scholar]
- 42.Pandrea, I., C. Kornfeld, M. J.-Y. Ploquin, C. Apetrei, A. Faye, P. Rouquet, P. Roques, F. Simon, F. Barré-Sinoussi, M. C. Müller-Trutwin, and O. M. Diop. 2005. Impact of viral factors on very early in vivo replication profiles in simian immunodeficiency virus SIVagm-infected African green monkeys. J. Virol. 79:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandrea, I., R. Onanga, C. Kornfeld, P. Rouquet, O. Bourry, S. Clifford, P. T. Telfer, K. Abernethy, L. T. White, P. Ngari, M. Muller-Trutwin, P. Roques, P. A. Marx, F. Simon, and C. Apetrei. 2003. High levels of SIVmnd-1 replication in chronically infected Mandrillus sphinx. Virology 317:119-127. [DOI] [PubMed] [Google Scholar]
- 44.Pandrea, I., R. Onanga, P. Rouquet, O. Bourry, P. Ngari, E. J. Wickings, P. Roques, and C. Apetrei. 2001. Chronic SIV infection ultimately causes immunodeficiency in African non-human primates. AIDS 15:2461-2462. [DOI] [PubMed] [Google Scholar]
- 45.Phillips-Conroy, J. E., C. J. Jolly, B. Petros, J. S. Allan, and R. C. Desrosiers. 1994. Sexual transmission of SIVagm in wild grivet monkeys. J. Med. Primatol. 23:1-7. [DOI] [PubMed] [Google Scholar]
- 46.Ploquin, M. J., O. M. Diop, N. Sol-Foulon, L. Mortara, A. Faye, M. A. Soares, E. Nerrienet, R. Le Grand, Y. Van Kooyk, A. Amara, O. Schwartz, F. Barre-Sinoussi, and M. C. Muller-Trutwin. 2004. DC-SIGN from African green monkeys is expressed in lymph nodes and mediates infection in trans of simian immunodeficiency virus SIVagm. J. Virol. 78:798-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rey-Cuille, M. A., J. L. Berthier, M. C. Bomsel-Demontoy, Y. Chaduc, L. Montagnier, A. G. Hovanessian, and L. A. Chakrabarti. 1998. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J. Virol. 72:3872-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 49.Silvestri, G., A. Fedanov, S. Germon, N. Kozyr, W. J. Kaiser, D. A. Garber, H. McClure, M. B. Feinberg, and S. I. Staprans. 2005. Divergent host responses during primary simian immunodeficiency virus SIVsm infection of natural sooty mangabey and nonnatural rhesus macaque hosts. J. Virol. 79:4043-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silvestri, G., D. L. Sodora, R. A. Koup, M. Paiardini, S. P. O'Neil, H. M. McClure, S. I. Staprans, and M. B. Feinberg. 2003. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18:441-452. [DOI] [PubMed] [Google Scholar]
- 51.Simon, F., S. Souquiere, F. Damond, A. Kfutwah, M. Makuwa, E. Leroy, P. Rouquet, J. L. Berthier, J. Rigoulet, A. Lecu, P. T. Telfer, I. Pandrea, J. C. Plantier, F. Barre-Sinoussi, P. Roques, M. C. Muller-Trutwin, and C. Apetrei. 2001. Synthetic peptide strategy for the detection of and discrimination among highly divergent primate lentiviruses. AIDS Res. Hum. Retrovir. 17:937-952. [DOI] [PubMed] [Google Scholar]
- 52.Smith, S. M., B. Holland, C. Russo, P. J. Dailey, P. A. Marx, and R. I. Connor. 1999. Retrospective analysis of viral load and SIV antibody responses in rhesus macaques infected with pathogenic SIV: predictive value for disease progression. AIDS Res. Hum. Retrovir. 15:1691-1701. [DOI] [PubMed] [Google Scholar]
- 53.Staprans, S. I., P. J. Dailey, A. Rosenthal, C. Horton, R. M. Grant, N. Lerche, and M. B. Feinberg. 1999. Simian immunodeficiency virus disease course is predicted by the extent of virus replication during primary infection. J. Virol. 73:4829-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swofford, D. 1999. PAUP* v 4: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Massachusetts.
- 55.Traina-Dorge, V., J. Blanchard, L. Martin, and M. Murphey-Corb. 1992. Immunodeficiency and lymphoproliferative disease in an African green monkey dually infected with SIV and STLV-I. AIDS Res. Hum. Retrovir. 8:97-100. [DOI] [PubMed] [Google Scholar]
- 56.Veazey, R. S., R. J. Shattock, M. Pope, J. C. Kirijan, J. Jones, Q. Hu, T. Ketas, P. A. Marx, P. J. Klasse, D. R. Burton, and J. P. Moore. 2003. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat. Med. 9:343-346. [DOI] [PubMed] [Google Scholar]
- 57.Veazey, R. S., I. C. Tham, K. G. Mansfield, M. DeMaria, A. E. Forand, D. E. Shvetz, L. V. Chalifoux, P. K. Sehgal, and A. A. Lackner. 2000. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4+ T cells are rapidly eliminated in early SIV infection in vivo. J. Virol. 74:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watson, A., J. Ranchalis, B. Travis, J. McClure, W. Sutton, P. R. Johnson, S. L. Hu, and N. L. Haigwood. 1997. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J. Virol. 71:284-290. [DOI] [PMC free article] [PubMed] [Google Scholar]