Abstract
A highly neurovirulent murine coronavirus JHMV (wild-type [wt] JHMV) is known to spread from cells infected via the murine coronavirus mouse hepatitis virus receptor (MHVR) to cells without MHVR (MHVR-independent infection), whereas a mutant virus isolated from wt JHMV, srr7, spread only in an MHVR-dependent fashion. These observations were obtained by the overlay of JHMV-infected cells onto receptor-negative cells that are otherwise resistant to wt JHMV infection. MHVR-independent infection is hypothetically thought to be attributed to a naturally occurring fusion activation of the wt JHMV S protein, which did not occur in the case of srr7. Attachment of S protein on cells without MHVR during the S-protein activation process seems to be a key condition. Thus, in the present study, we tried to see whether wt JHMV virions that are attached on MHVR-negative cells are able to infect those cells. In order to make virions attach to the cell surface without MHVR, we have used spinoculation, namely, the centrifugation of cells together with inoculated virus at 3,000 rpm for 2 h. This procedure forces viruses to attach to the cell surface, as revealed by quantitative estimation of attached virions by real-time PCR and also facilitated wt JHMV infection to MHVR-negative cells, but failed to do so for srr7. Virions of both wt and srr7 attached on MHVR-negative cells by spinoculation were facilitated for infection in the presence of a soluble form of MHVR that induces conformational changes of both wt and srr7. It was further revealed that wt JHMV S1, but not srr7, was released from the cell surface when S protein was expressed on cells. These observations support the hypothesis that attachment of the virion to MHVR-negative cells is a critical step and that a unique feature of wt JHMV S1 to be released from S2 in a naturally occurring event is involved in an MHVR-independent infection.
The binding of virus to its specific receptor on a susceptible cell surface is an initial event in viral infection. A number of molecules classified into the immunoglobulin (Ig) superfamily are known to serve as receptors for various viruses. For example, CD4, signaling lymphocyte-activation molecule (SLAM or CD150), and a cell adhesion molecule in a carcinoembryonic antigen (CEACAM1 or MHVR) are identified as functional receptors for human immunodeficiency virus (HIV) (7, 22), measles virus (43, 46) and murine coronavirus mouse hepatitis virus (MHV), respectively (12, 45). However, additional alternative molecules are thought to be used as a receptor for HIV and measles virus, since they are known to infect cells lacking their specific receptors, CD4 or SLAM (4, 21).
MHV is classified as one of the Coronaviridae, consisting of an enveloped virus with a single, positive-stranded genomic RNA of about 31 kb (26). The characteristic spikes on the virion surface are composed of spike (S) protein. S protein is a class I fusion protein of 180 to 200 kDa in molecular mass (5, 26). It is synthesized as a large protein and cleaved by cellular protease into two subunits, N-terminal S1 and C-terminal S2 (37). The N-terminal region of S1 consisting of 330 amino acids (S1N330) is responsible for receptor binding (25, 38), and S2 is not involved in this activity (39). After receptor binding, S1 is dissociated from the membrane-anchored S2 subunit, which triggers the conformational changes of S2 to acquire fusion activity (28). The cell entry mechanism of MHV is very similar to that of HIV proposed by Chen and Kim (6). Likewise, the structural features of MHV S2 subunit are very similar to those of the membrane-anchored subunit of HIV envelope protein gp41. They both have two heptad repeats (HR) upstream of the transmembrane domain (6, 10) that play a crucial role in fusion of viral and cell membranes by forming so-called “six-helix bundles” (5, 6, 10, 14).
The major receptor for MHV, CEACAM1 or MHVR, is composed of four Ig-like ectodomains, a transmembrane domain (TM), and a cytoplasmic tail (Cy) (3, 12). There are four isoforms, two having four immunoglobulin-like domains and the other two having Ig-like domains, one of which has either a long or a short Cy (3). The N-terminal (N) domain is responsible for virus binding (13), induction of S protein conformational changes, and activation for fusion (30), although the N domain linked with TM and Cy and expressed on the cell surface is not functional, which is presumably due to inaccessibility for MHV to short molecules expressed on the cell surface (13, 30).
Although MHVR is a critical receptor for MHV, a highly neurotropic strain of JHMV was revealed by Gallagher et al. (17) to spread from DBT cells initially infected in an MHVR-mediated manner to MHVR-negative BHK cells via DBT-BHK cell fusion, when infected DBT cells were overlaid onto a BHK monolayer (called MHVR-independent fusion/infection by an infected-cell overlay test) (17). We have isolated from wild-type (wt) JHMV with this activity, a mutant virus resistant to neutralization by soluble receptor (srr7) that lacks this unique feature of infection (34, 42). Recent studies suggested that the high neurovirulence of wt JHMV was due to the MHVR-independent infection (18, 31).
The mechanism of this MHVR-independent infection and fusion is still speculative, although some unusual feature of wt JHMV S, but not srr7 S, hints that there is a possible mechanism of this unique mode of infection. wt JHMV S1 is reportedly removed from membrane-anchored S2 (23), and the conformational changes in the S2 take place (28). Both of these activities occur naturally, without binding to MHVR. These findings suggest that the spontaneous release of S1 from S2 triggers the conformational changes of the S2 in a similar fashion, since it occurs after binding its receptor. The fusion peptide in S2 could penetrate into the membrane without MHVR, if S protein is attached on the membrane when S1 is being detached from S2. Expectedly, virus inoculation onto BHK cells does not permit virus infection, whereas JHMV-infected DBT cells overlaid onto BHK cells permits the infection, since S proteins on infected DBT cells are attached onto BHK cells, whereas virion S protein fails to attach to the cell membrane because of the absence of the receptor that binds the virus and cell membrane.
If the hypothesis described above is correct, wt JHMV can infect a cell without a receptor under conditions whereby the virion is forced to attach to the cell surface. Thus, the present study is undertaken to attach virions onto the cell surface without MHVR expression. To achieve such conditions, we have used spinoculation, which enabled or facilitated virion attachment onto the cell surface by spinning cells, as well as inoculated viruses for retroviruses (8, 15, 32). We show here that spinoculation enabled wt JHMV and srr7 attachment onto the cell membrane, while it facilitated the infection of wt JHMV, but not that of srr7, into MHV receptor-negative cells. In addition, S1 of wt JHMV but not srr7 S1 was revealed to be free from S2 when S protein was expressed on BHK cells. These results are in good agreement with the hypothesis described above.
MATERIALS AND METHODS
Cells and viruses.
The MHVR-positive cell line DBT (mouse delayed brain tumor) and the MHVR-negative BHK (baby hamster kidney), VeroE6 (African green monkey kidney), and HeLa (human cervical cancer) cell lines were maintained in Dulbecco minimum essential medium (DMEM; Nissui, Tokyo, Japan) supplemented with 5% fetal bovine serum (FBS; Tissue Culture Biologicals, California). A highly fusogenic MHV strain, MHV-JHM cl-2 (defined as wt JHMV) (40), and a soluble receptor-resistant mutant derived from wt JHMV, srr7 (34), were amplified and assayed on DBT cells as described previously (42). Viral infectivity was shown as PFU. srr7 has a single amino acid mutation at position 1114 (Leu to Phe) of the S2 subunit of the S protein relative to wt JHMV (34).
Spinoculation.
Spinoculation, with modifications, was done according to the previous reports on retroviruses (8, 15, 32). BHK and DBT cells were seeded at a concentration of 5 × 105 cells in 0.5 ml per well of a 24-well culture plate (Falcon, Franklin Lakes, NJ). After 6 h of incubation, cells were washed once with phosphate-buffered saline (pH 7.2) (PBS) and inoculated with viruses serially diluted by 10-fold in 300 μl of DMEM containing 0.5 μg of concanavalin A (ConA; Wako, Osaka, Japan)/ml. The plates were centrifuged at 3,000 rpm (1,750 × g) for 2 h at 4°C. As a control, cells in 24-well plates were inoculated with virus in 50 μl of DMEM and cultured for 1 h at 4°C without centrifugation. After infection, cells were washed three times with ice-cold PBS and incubated with DMEM supplemented with 1% FBS for further 14 h at 37°C. Cells were then fixed and stained with crystal violet, and the syncytia were counted under light microscopy.
In some experiments, a soluble form of MHVR (soMHVR) was added into the culture after spinoculation of viruses as described above. For this experiment, soMHVR1 (1) containing N domain alone of the MHVR (30), which was revealed to have a receptor functionality indistinguishable from soluble MHVR composed of two ectodomains (30), was utilized, after being expressed by recombinant baculovirus and purified by using its tag as described previously (42).
Quantitative estimation of viral RNA by real-time PCR.
BHK cells prepared in 24-well plates were inoculated with 105 PFU (corresponding to 1.1 × 107 and 1.4 × 107 copies for wt JHMV and srr7, respectively) of viruses/well and then incubated with or without centrifugation as described above. Then, cells were washed three times with PBS, and total cell-associated RNAs were extracted by using ISOGEN (Nippon Gene, Tokyo, Japan) according to the manufacturer's instruction. Five micrograms of yeast tRNA (Sigma, St. Louis, MO) was added as a carrier. The real-time reverse transcription-PCR to estimate the amount of viral RNA was performed as described previously for SARS-CoV mRNA detection (29) with slight modification. A series of reactions were performed by using LightCycler RNA Master mix (Loche Diagnostics, Mannheim, Germany). The target sequence was set at the inside of the N protein. The nucleotide sequences of the forward and reverse primers were 5′-TGTCTTTTGTTCCTGGGCA-3′ and 5′-CAAGAGTAATGGGGAACCA-3′, respectively. To detect the amplified fragments, we used hybridization probes labeled with fluorescence dye, 5′-GCTCCTCTGGAAACCGCGCTGGTAATGG-3′ (3′fluorescein isothiocyanate labeled), and 5′-ATCCTCAAGAAGACCACTTGGGCTGACCAAACC-3′ (5′LCRed640 labeled). These oligonucleotides were synthesized according to the MHV-JHM N gene sequence (36). PCR analysis was performed under the following conditions (reverse transcription, 61°C for 20 min; PCR, 95°C for 30 s; 40 cycles for 95°C for 5 s, 55°C for 15 s, and 72°C for 13 s). To obtain a calibration line, DNA plasmids encoding target sequences were serially diluted and subjected to real-time PCR analysis. The relationship between the copy number of target sequence (y axis) and cycle of real-time PCR to reach a positive level (x axis) was obtained. The amounts of virus that attached onto cells were calculated from a calibration line obtained as described above.
Expression of S proteins and evaluation of S1 dissociation.
MHV S proteins were expressed in BHK and HeLa cells by either transient expression by the transfection of expression plasmids or transient vaccinia virus expression system as previously reported (42). BHK cells were transfected by using FuGENE6 transfection reagent (Roche) with the plasmids containing wt JHMV or srr7 S gene under the control of the cytomegalovirus and T7 promoters pTarget/cl2-S or pTarget/srr7S (27). These cells were cultured for 40 h after transfection, and the expressed S protein was analyzed by flow cytometry as described below. As for the S-protein expression by vaccinia virus, BHK cells were infected with vTF7.3, a recombinant vaccinia virus harboring the T7 RNA polymerase gene (16) with a multiplication of infection of 5 and incubated at 37°C for 1 h. The cells were then treated with trypsin and transfected with plasmid pTarget/cl2-S or pTarget/srr7S (27) by electroporation using a GenePulser (Bio-Rad, Hercules, CA). Cells were then cultured in DMEM containing 5% FBS, and the culture medium was replaced with fresh medium 3 h after transfection. After an additional 12 h of incubation, the culture supernatants and cells were separately harvested. To detect the released S1 protein, culture supernatants were centrifuged at 9,000 × g for 1 min to clarify the cell debris, and a mixture of anti-S monoclonal antibodies (MAbs 2, 3, and 7) (24) and protein G-Sepharose (Amersham Bioscience, Arlington Heights, IL) was added, followed by further incubation at 4°C for 4 h with gentle rotation. After five washes with PBS, an aliquot of 2× sodium dodecyl sulfate (SDS) sample buffer (100 mM Tris-Cl [pH 6.8], 4% SDS, 200 mM dithiothreitol, 20% glycerol; volume equal to protein A-Sepharose) was added, and the lysates were subjected to Western blot analysis. To detect S proteins expressed in cells, cells infected with vTF7.3 and transfected as described above were washed twice with PBS and lysed with cell lysis buffer (PBS containing 10% glycerol and 1% Triton X-100). After centrifugation at 9,000× g for 1 min, soluble fractions were mixed with 2× SDS sample buffer as described above.
Western blot analysis.
S proteins of wt JHMV and srr7 were prepared as described above and analyzed by Western blotting as described previously (27, 34). Briefly, each sample was separated by SDS-7.5% polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA). S proteins on the membrane were reacted with anti-S1 (11F) or anti-S2 (10G) MAb (33) and subsequently with anti-mouse immunoglobulin G (IgG) serum labeled with horseradish peroxidase as described elsewhere (27, 34). The bands were then visualized by using Supersignal West Dura (Pierce, Rockford, IL) with LAS-1000PLUS (Fujifilm, Tokyo, Japan).
Indirect immunofluorescence assay (IFA).
To examine viral antigens in cells infected by spinoculation, cells spinoculated as described above were fixed with acetone-methanol (1:1) for 2 min. After three washes with PBS, cells were incubated with anti-S MAbs (a mixture of MAbs 2, 3, and 7) (24) for 1 h at 37°C. After three washes with PBS, cells were further incubated with fluorescein isothiocyanate-conjugated anti-mouse IgG (Zymed Laboratories, California).
Flow cytometry.
BHK cells transfected with pTarget/cl2-S or pTarget/srr7S as described above were examined for the S protein expression on cell surface by flow cytometric analysis with cytofluorometer FACSCalibur (Becton Dickinson, San Jose, CA). Transfected cells were treated with PBS containing 0.05% EDTA to prepare a single cell suspension and collected into microtubes. The cells were fixed with 4% paraformaldehyde at 4°C for 10 min; reacted with an aliquot of 1:200-diluted anti-S MAbs 2, 3, and 7 for 1 h on ice; and washed with PBS containing 1% FBS. The cells were then incubated with 1:200-diluted anti-mouse IgG (H+L) conjugated with phycoerythrin (Jackson Immunoresearch, West Grove, PA) for 1 h on ice. After three washes with PBS, the fluorescence intensity was analyzed with FACSCalibur.
RESULTS
Enhancement of viral attachment onto MHVR-negative cells by spinoculation.
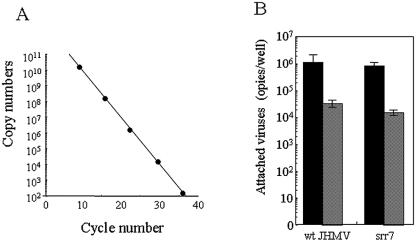
Spinoculation has been reported to force virions attached onto cell membrane for retroviruses (8, 15, 32). We first tested whether the spinoculation applied to those viruses can enhance the attachment of MHV onto MHVR-negative cells. To enhance the adsorption efficiency, Polybrene or DEAE-dextran was included in the medium, which was shown to not enhance JHMV infection (data not shown). Instead, ConA was revealed to enhance the adsorption efficiency. Finally, we spinoculated wt JHMV and srr7 onto BHK cells prepared in 24-well plates in 300 μl of DMEM containing 0.5 μg of ConA/ml/well and spun the sample at 3,000 rpm for 2 h at 4°C. After the cells were washed, the total RNA was extracted from them, and virus genome RNA copies were evaluated by real-time PCR according to the calibration line obtained using cloned DNA of target sequence (Fig. 1A). As shown in Fig. 1B, spinoculation under the above conditions augmented the virus attachment to MHVR-negative BHK by ∼100-fold. There was no significant difference between wt JHMV and srr7 in the virus copies attached to MHVR-negative BHK by the spinoculation used in the present study. Unexpectedly, considerable amounts of both wt and srr7 attached onto BHK cells after ordinary infection, although the molecule(s) participating into this binding has not yet been identified.
FIG. 1.
Quantitative estimation by real-time PCR of virus attached onto cells after spinoculation: (A) Calibration in real-time PCR. Control DNA encoding the target sequence was subjected to real-time PCR. The relationship between the copy numbers of target sequence (y axis) and the cycles of real-time PCR to reach a positive level (x axis) is shown. (B) Effect of spinoculation on virus binding. BHK cells were spinoculated (black column) or inoculated by ordinary methods (shaded column) with 105 PFU of wt and srr7 as described in Materials and Methods. Cell-attached virus copies were estimated by real-time PCR using a total RNA extracted from infected cells. They were calculated from the calibration line shown in panel A. Error bars represent the standard deviations of the results of three independent experiments.
Enhancement of MHVR-independent infection of wt JHMV by spinoculation.
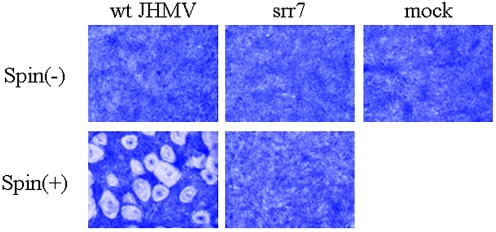
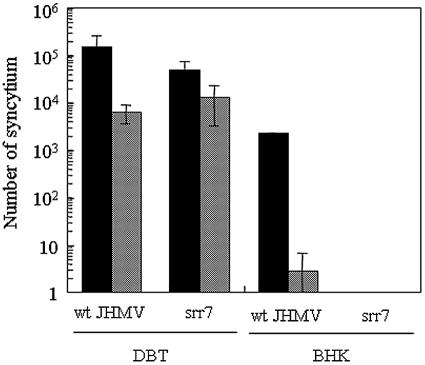
Since spinoculation was revealed to enhance the virus attachment onto cells for both wt JHMV and srr7, we then examined whether it enhances virus infection or not. BHK cells were spinoculated with either wt JHMV or srr7, and syncytium formation was examined at 14 h after incubation. As shown in Fig. 2, a large number of syncytia were produced after infection with wt JHMV but not at all after infection with srr7. To quantitatively examine the effect of spinoculation, we inoculated about 104 PFU/well of either wt JHMV or srr7, and the number of syncytia formed was determined. As shown in Fig. 3, spinoculation of wt JHMV augmented the infection on MHVR-negative BHK cells by 200- to 1,000-fold (with a mean value of ca. 800-fold), while it failed to facilitate the infection of srr7. wt JHMV infected BHK cells very inefficiently (2.75 syncytia per well after infection with 104 PFU) by standard procedures of infection (nonspinoculated and incubated at 4°C for 1 h). Although the enhancement was not so remarkable, spinoculation of both wt and srr7 enhanced their infection on MHVR-positive DBT cells by ca. 10-fold (Fig. 3).
FIG. 2.
Syncytium formation of BHK cells with wt JHMV infected by spinoculation: wt and srr7 (104 PFU) were spinoculated [spin (+)] or inoculated by an ordinary method [spin (−)] and cultured in DMEM containing 5% FBS for 14 h. Cells were fixed and stained with crystal violet. Mock-infected cells are shown as a control (mock).
FIG. 3.
Quantitative estimation of syncytium formation after spinoculation: DBT or BHK cells were infected by spinoculation (black column) or by ordinary methods (shaded column) with 104 PFU of wt JHMV or srr7. Cells were further incubated in DMEM containing 5% FBS for 14 h, and the number of syncytia was determined after staining with crystal violet. Error bars the represent standard deviations of the results of three independent experiments.
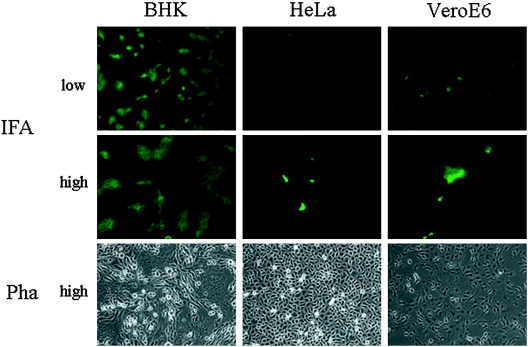
We then examined whether spinoculation facilitates the wt JHMV infection in other MHVR-negative cells. We have tested HeLa cells and VeroE6 cells, as well as BHK cells, all of which are MHVR negative and resistant to infection to wt JHMV. All of these cell lines were infected by spinoculation with wt JHMV, as described for the infection to BHK cells, and syncytium formation was observed. As shown in Fig. 4, after phase-contrast microscopy no syncytium formation was found in two cell lines, HeLa and VeroE6 cells, whereas syncytia were clearly visible for infection of BHK cells. We then examined these cells by IFA to determine whether a small number of cells unrecognized by microscopy were infected or not. As shown in Fig. 4, tiny syncytia were detected by IFA on both HeLa and VeroE6 cells, showing that wt JHMV infected various cells without MHVR, when they are forced to attach on these cells by spinoculation. After the spinoculation of 105 PFU of wt JHMV, ca. 4 × 103, 3 × 103, and 8 × 102 syncytia were detected on BHK, HeLa, and VeroE6 cells, respectively. Srr7 failed to produce syncytium, and no virus antigen was detected by IFA after srr7 spinoculation in MHVR-negative cell lines (data not shown).
FIG. 4.
MHVR-independent infection of wt JHMV in various cell lines by spinoculation: wt JHMV of 105 PFU was spinoculated onto cells without MHVR expression, BHK cells, HeLa cells, or VeroE6 cells, and incubated for 14 h. Syncytium formation was microscopically observed by phase-contrast microscopy (Pha) or by immunofluorescence (IFA) with anti-spike MAbs under low and high magnifications (high magnification = 2× low magnification).
These results collectively suggest that wt JHMV infects MHVR-negative cells if it is forced to attach the cell surface. However, srr7 fails to infect cells in an MHVR-independent fashion, being consistent with our previous results obtained by the infected-cell overlay test.
Facilitation of infection by spinoculation with soMHVR.
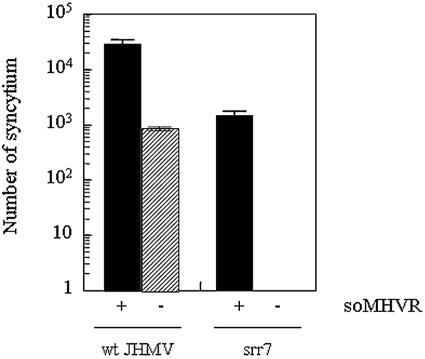
Previous studies on the infected-cell overlay test have shown that MHVR-independent infection was facilitated by soluble MHVR (42). DBT cells infected with srr7 did not induce fusion on BHK cell monolayers. However, they induced fusion when cells were cultured in the presence of soMHVR. wt JHMV infection was also augmented by soMHVR in terms of the size of syncytia produced in an MHVR-independent fashion. These findings suggested that S protein adjacent to MHVR-negative target cells can be activated by the soMHVR (42). In the present study, we also addressed whether or not soMHVR enhances MHVR-independent infection. wt JHMV or srr7 infected by spinoculation, as described above, were incubated in the presence of 50 nM soMHVR (1), consisting of the N domain alone of MHVR (30). As shown in Fig. 5, soMHVR enhanced the spinoculated wt JHMV by about 30-fold. Although srr7 attached by spinoculation failed to infect cells without MHVR, soMHVR potentiated the srr7 infection, which shows that srr7 infection is solely dependent upon the presence of MHVR. These results with virions are similar to those found with MHVR-independent infection, when examined by the infected-cell overlay test.
FIG. 5.
Effect of soMHVR on the infection after spinoculation: BHK cells were infected by spinoculation with 103 PFU of wt and srr7, and infected cells were cultured for 14 h in DMEM containing 5% FBS in the presence (+) or absence (−) of soMHVR (50 nM final concentration). The number of syncytia was counted after staining with crystal violet. Error bars represent the standard deviations of the results of three independent samples.
Naturally occurring release of S1 of wt JHMV from the cell surface.
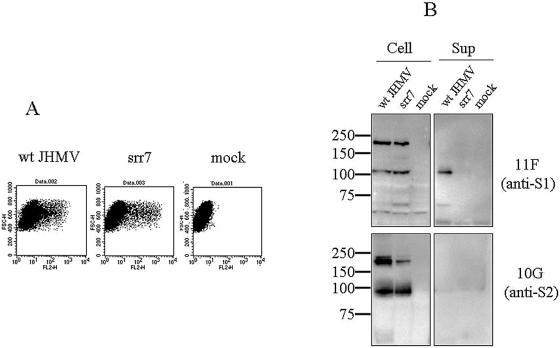
S1 of JHMV with MHVR-independent infection activity has been described as being released in a naturally occurring fashion from the cell surface, but this result was not seen in JHMV-derived mutant viruses without this activity (23). Thus, we also examined wt JHMV and srr7, with and without MHVR-independent infection activity, respectively, to assess whether their S1 is released from S2 when expressed on cells. S protein of wt JHMV and srr7 was expressed in BHK cells by transfecting the plasmids harboring wt or srr7 S gene. The S proteins of both viruses were expressed in similar amounts and transported equally onto the cell surface, as revealed by flow cytometric analysis (Fig. 6A). However, S1 proteins were not clearly detected in the culture fluids of cells expressing these S proteins. We then expressed S protein with the T7 vaccinia virus expression system, which normally permits higher expression of the protein. S1 or S2 subunits released in the culture fluids were detected by immunoprecipitation, followed by Western blotting with subunit specific MAbs, as described in Materials and Methods. MAbs used to precipitate S protein in the supernatant recognize epitopes located in the S1 and were shown to coimmunoprecipitate S2 that interacts with S1 in our previous study (24). It was revealed that large amounts of the S1 subunit of wt JHMV were released into culture supernatants of cells transfected with wt S expression plasmid (Fig. 6B). On the other hand, there was no release of S1 subunit in the supernatants of cells introduced with srr7 S expression plasmid. The S2 subunit of each strain was not released into the supernatant. The amounts of wt and srr7 S proteins expressed were not different, as demonstrated by Western blotting (Fig. 6B). These results agree with the observations by Krueger et al. (23) that S1 of MHV showing MHVR-independent infection is released in a naturally occurring event, whereas this phenomenon was not observed for those without such activity.
FIG. 6.
Analysis of wt and srr7 S proteins expressed in BHK cells: (A) Expression of S protein on cell surface after transfection with expression plasmid containing S gene of wt or srr7. Cells were transfected with plasmid containing wt (wt) or srr7 (srr7) S gene. They were also transfected with plasmid containing no S gene (mock). The cells were harvested at 8 h after transfection and stained with anti-S MAbs and anti-mouse IgG antibody conjugated with phycoerythrin. Flow cytometric analysis was done with FACSCalibur, and the fluorescence intensity was analyzed by dot plot analysis. (B) Western blot analysis of the S protein in culture fluid of BHK cells transfected with S gene. Expression plasmids harboring either wt (wt) or srr7 (srr7) S gene, as well as plasmid alone (mock), were transfected into BHK cells previously infected with vTF7.3 for the S protein expression. S protein in culture supernatants (Sup) was immunoprecipitated by MAbs against S protein and analyzed by Western blotting with MAbs 11F and 10G specific for S1 and S2, respectively. S protein in cells (Cell) was analyzed by using total cell lysates.
DISCUSSION
Viral infection is generally restricted to cells that express the receptor molecule, with which the virus binds specifically and invades cells. Some viruses are known to infect cells on which a specific receptor molecule is not expressed, and these infections are generally not as efficient as the infection via an identified receptor. For example, HIV infects cells that do not express its receptor CD4 (4) and measles virus can infect cells lacking both SLAM and CD46, molecules identified as measles virus receptor (21). These infections are supposedly attributed to another cellular receptor (4, 21). MHV mutants derived from cells persistently infected with MHV-A59 strain or cells doubly infected with JHMV and MHV-A59 were reported to infect a variety of MHVR-negative cells derived from different species (1, 2, 35). In these cases, infections were induced by using standard protocols and did not require an infected cell overlay or spinoculation, suggesting that their infections are mediated by molecule(s) that function as an alternative receptor. Some of these MHV mutants were revealed to utilize human CEA as a receptor, although some other molecules were supposed to work as a receptor, since these mutants infected cells derived from nonhuman species as well (1). MHV mutant isolated from the above-described persistent infection was recently reported to obtain the ability to use heparan sulfate for entry receptor (11). In contrast, some strains of MHV are reported to spread in an MHVR-independent manner (41). For example, highly neurovirulent MHV-JHMV is able to spread from cells initially infected via its receptor CEACAM1 to CEACAM1-negative cells (17, 31). Although it is clear that MHVR is not involved in this MHVR-independent infection, no other specific molecules are supposedly involved, since this infection differs from those that use alternative, less-functional receptors, such as HIV, measles virus, or MHV mutant isolated from persistent infection (4, 11, 21) in that the infection is not accomplished after standard protocol of infection.
MHVR-independent infection had been restricted from cells initially infected via MHVR to MHVR-negative cells (infected-cell overlay test), whereas viruses failed to directly infect MHVR-negative cells when ordinary infection procedures were used. The proposed mechanism of MHVR-independent infection is that S protein adjacent to the cell membrane is activated for fusion in a naturally occurring manner, which allows it to induce a fusion of the viral envelope and cell membrane without receptor involvement. However, an infected-cell overlay test could not completely rule out the possible contribution of some cellular factors derived from initially infected MHVR-positive cells. Even the possibility is not thoroughly excluded that MHVR expressed on MHV-infected DBT cells plays a part in this infection. To explore the hypothesized mechanism of MHVR-independent infection described above and to determine whether there are possible cellular factors, we have performed spinoculation that was reported by retrovirologists to force virions to attach onto the cell surface (8, 15, 32). For spinoculation in the present study, cells were spun together with inoculated viruses in medium containing 0.5 μg of ConA/ml at 1,750 × g for 2 h at 4°C. This procedure enhanced virus attachment by ∼100-fold, irrespective of the virus strains used, i.e., wt JHMV and srr7, with and without MHVR-independent infection activity, respectively. It also enhanced the infection of wt JHMV by more than 100-fold but not at all for srr7 infection, which displayed for the first time an MHVR-independent infection by using wt JHMV virions. Since our virus materials used for spinoculation could contain some components of infected cells, we excluded those materials by the purification of MHV with sucrose gradient centrifugation. These viruses also showed the similar enhanced infection by spinoculation (data not shown). These findings collectively exclude the possible participation of some specific cellular factors into the MHVR-independent infection.
Although the data now available suggest the mechanism of MHVR-independent infection as described above, they fail to exclude the possibility that another receptor molecule is responsible for this infection. Cellular factors such as heparan sulfate or several chemical reagents were reported to function as a receptor (11, 19, 20). However, we think the possibility is quite low for the following reasons. A possible receptor, if there is one, has very weak receptor functionality; it is more than 104 and 105 less functional compared to CEACAM1 for wt JHMV and srr7, respectively. Actually, it does not function at all as a receptor for srr7. Moreover, such a receptor would permit the binding of wt and srr7 to almost the same extent (Fig. 1B) and be expressed ubiquitously in various species, since not only BHK cells derived from hamster but also HeLa and VeroE6 cells derived from human and the monkey are target cells for MHVR-independent infection. To finally exclude such a molecule to work as a receptor, liposome-binding of wt JHMV but not srr7 in the absence of soMHVR would be a convincing evidence.
The receptor molecule for enveloped viruses has at least two distinct, critical functions, namely, its ability to bind to the virus surface protein and its ability to activate fusion of the protein, which would result in the fusion of envelope and cell membrane, thereby permitting virus entry into the cell. Spinoculation can replace one of these functions, the binding of envelope protein to cell surface. After binding to the cell surface, virus surface protein is activated for fusion if a soluble form of the receptor interacts with the protein. This is evident for the S protein of srr7 that does not show MHVR-independent fusion activity. This feature is usually observed in other viruses, such as the retrovirus avian sarcoma leukosis virus (ASLV), which is able to infect cells lacking its receptor, if ASLV is allowed to attach those cells by spinoculation and soluble ASLV receptors are mixed in the culture (8).
To show that the MHVR-independent infection is not restricted in BHK cells, two other cell lines were used for spinoculation. Although remarkable syncytia were not produced, as seen in BHK cells, infection by wt JHMV of HeLa and VeroE6 cells was detected by IFA with anti-S antibodies. There are two possibilities to explain this difference in cytopathic changes caused by wt JHMV. One is that there are differences in the degree of replication among these cells after spinoculation. wt infection in BHK cells could produce more abundant S protein than in VeroE6 or HeLa cells. The other possibility is that the components of plasma membrane of these cell lines, especially the lipid composition, are varied. Cellular cholesterol was reported to influence replication efficiency and fusion formation ability of MHV (9, 44).
In conclusion, we showed in the present study that the virus with MHVR-independent infection activity is able to infect cells when it is attached to the target cell surface. However, the virus without this activity fails to infect cells, even if it is attached to the cell surface. It is also confirmed that S1 release from S2 in a naturally occurring event is one of the important features for the virus with MHVR-independent infection activity. These observations support the proposed mechanism of MHVR-independent infection; S protein attached to the MHVR-negative cell surface is activated for fusion of the virus-cell membrane, which is mediated by a naturally occurring dissociation of S1 from S2. However, the possibility that the cellular molecule is a receptor with a weak function for wt JHMV, but not for srr7, cannot be thoroughly excluded.
Acknowledgments
We thank Keiko Nakagaki and Kazumitsu Suzuki for valuable discussions and comments.
This study was financially supported by a grant from the Ministry of Education, Culture, Sports, Science, and Technology (grant 16017308) and a grant from the Ministry of Health, Labor, and Welfare (H16-Shinkoh-9).
REFERENCES
- 1.Baric, R., E. Sullivan, L. Hensley, B. Yount, and W. Chen. 1999. Persistent infection promotes cross-species transmissibility of mouse hepatitis virus. J. Virol. 73:638-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baric, R., B. Yount, L. Hensley, S. A. Peel, and W. Chen. 1997. Episodic evolution mediates interspecies transfer of a murine coronavirus. J. Virol. 71:1964-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beauchemin, N., P. Draber, G. Dveksler, P. Gold, S. Gray-Owen, F. Grunert, S. Hammarstrom, K. V. Holmes, A. Karlsson, M. Kuroki, S. H. Lin, L. Lucka, S. M. Najjar, M. Neumaier, B. Obrink, J. E. Shively, K. M. Skubitz, C. P. Stanners, P. Thomas, J. A. Thompson, M. Virji, S. von-Kleist, C. Wagener, S. Watt, and W. Zimmermann. 1999. Redefined nomenclature for members of the carcinoembryonic antigen family. Exp. Cell Res. 252:243-249. [DOI] [PubMed] [Google Scholar]
- 4.Bhat, S., S. L. Spitalnik, F. Gonzalez-Scarano, and D. H. Silberberg. 1991. Galactosyl ceramide or a derivative is an essential component of the neural receptor for human immunodeficiency virus type 1 envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 88:7131-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 7.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 8.Damico, R., and P. Bates. 2000. Soluble receptor-induced retroviral infection of receptor deficient cells. J. Virol. 74:6469-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daya, M., M. Cervin, and R. Anderson. 1988. Cholesterol enhances mouse hepatitis virus-mediated cell fusion. Virology 163:276-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Groot, R. J., W. Luytjes, M. C. Horzinek, B. A. van der Zeijst, W. J. Spaan, and J. A. Lenstra. 1987. Evidence for a coiled-coil structure in the spike of coronaviruses. J. Mol. Biol. 196:963-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Haan, C. A., Z. Li, E. te Lintelo, B. J. Bosch, B. J. Haijema, and P. J. Rottier. 2005. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J. Virol. 79:14451-14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dveksler, G. S., M. N. Pensiero, C. B. Cardellichio, R. K. Williams, G.-S. Jiang, K. V. Holmes, and C. W. Dieffenbach. 1991. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 65:6881-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dveksler, G. S., M. N. Pensiero, C. W. Dieffenbach, C. B. Cardellichio, A. A. Basile, P. E. Elia, and K. V. Holmes. 1993. Mouse hepatitis virus strain A59 and blocking antireceptor monoclonal antibody bind to the N-terminal domain of cellular receptor. Proc. Natl. Acad. Sci. USA 90:1716-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 15.Forestell, S. P., J. S. Dando, E. Bohnlein, and R. J. Rigg. 1996. Improved detection of replication-competent retrovirus. J. Virol. Methods 60:171-178. [DOI] [PubMed] [Google Scholar]
- 16.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher, T. M., M. J. Buchmeier, and S. Perlman. 1992. Cell receptor-independent infection by a neurotropic murine coronavirus. Virology 191:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guibinga, G. H., A. Miyanohara, J. D. Esko, and T. Friedmann. 2002. Cell surface heparan sulfate is a receptor for attachment of envelope protein-free retrovirus-like particles and VSV-G pseudotyped MLV-derived retrovirus vectors to target cells. Mol. Ther. 5:538-546. [DOI] [PubMed] [Google Scholar]
- 20.Harada, S., and Y. Maeda. 1999. Chemically induced infection of CD4-negative HeLa cells with HIV-1. Microbiol. Immunol. 43:1077-1086. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto, K., N. Ono, H. Tatsuo, H. Minagawa, M. Takeda, K. Takeuchi, and Y. Yanagi. 2002. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J. Virol. 76:6743-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klazmann, D., E. Champagne, S. Chameret, J. Gruest, D. Guetard, T. Hercend, J. C. Gluckman, and L. Montagnier. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767-768. [DOI] [PubMed] [Google Scholar]
- 23.Krueger, D. K., S. M. Kelly, D. N. Lewicki, R. Ruffolo, and T. M. Gallagher. 2001. Variations in disparate regions of the murine coronavirus spike protein impact the initiation of membrane fusion. J. Virol. 75:2792-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubo, H., S. Y. Takase, and F. Taguchi,. 1993. Neutralization and fusion inhibition activities of monoclonal antibodies specific for the S1 subunit of the spike protein of neurovirulent murine coronavirus JHMV cl-2 variant. J. Gen. Virol. 74:1421-1425. [DOI] [PubMed] [Google Scholar]
- 25.Kubo, H., Y. K. Yamada, and F. Taguchi. 1994. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J. Virol. 68:5403-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus. Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuyama, S., and F. Taguchi. 2000. Impaired entry of soluble receptor-resistant mutants of mouse hepatitis virus into cells expressing MHVR2 receptor. Virology 273:80-89. [DOI] [PubMed] [Google Scholar]
- 28.Matsuyama, S., and F. Taguchi. 2002. Receptor-induced conformational changes of murine coronavirus spike protein. J. Virol. 76:11819-11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuyama, S., M. Ujike, S. Morikawa, M. Tashiro, and F. Taguchi. 2005. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. USA 102:12543-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miura, S. H., K. Nakagaki, and F. Taguchi. 2004. N-terminal domain of the murine coronavirus receptor CEACAM1 is responsible for fusogenic activation and conformational changes of the spike protein. J. Virol. 78:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakagaki, K., K. Nakagaki, and F. Taguchi. 2005. Receptor-independent spread of a highly neurotropic murine coronavirus JHMV strain from initially infected microglial cells in mixed neural cultures. J. Virol. 79:6102-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Dherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Routledge, E., R. Stauber, M. Pfleiderer, and S. G. Siddell. 1991. Analysis of murine coronavirus surface glycoprotein functions by using monoclonal antibodies. J. Virol. 65:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saeki, K., N. Ohtsuka, and F. Taguchi. 1997. Identification of spike protein residues of murine coronavirus responsible for receptor-binding activity by use of soluble receptor-resistant mutants. J. Virol. 71:9024-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schickli, J. H., B. D. Zelus, D. E. Wentworth, S. G. Sawicki, and K. V. Holmes. 1997. The murine coronavirus mouse hepatitis virus strain A59 from persistently infected murine cells exhibits an extended host range. J. Virol. 71:9499-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skinner, M. A., and S. G. Siddell. 1983. Coronavirus JHM: nucleotide sequence of the mRNA that encodes nucleocapsid protein. Nucleic Acids Res. 11:5045-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturman, L. S., C. A. Ricard, and K. V. Holmes. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: activation of cell-fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J. Virol. 56:904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, H., and F. Taguchi. 1996. Analysis of the receptor-binding site of murine coronavirus spike protein. J. Virol. 70:2632-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taguchi, F. 1995. The S2 subunit of the murine coronavirus spike protein is not involved in receptor binding. J. Virol. 69:7260-7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taguchi, F., S. G. Siddell, H. Wege, and V. ter Meulen. 1985. Characterization of a variant virus selected in rat brains after infection by coronavirus mouse hepatitis virus JHM. J. Virol. 54:429-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taguchi, F., S. Matsuyama, and K. Saeki. 1999. Difference in Bgp-independent fusion activity among mouse hepatitis viruses. Arch. Virol. 1444:2041-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taguchi, F., and S. Matsuyama. 2002. Soluble receptor potentiates receptor-independent infection by murine coronavirus. J. Virol. 76:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatsuo, H., N. Ono, K., Tanaka, and Y. Yanagi. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406:893-897. [DOI] [PubMed] [Google Scholar]
- 44.Thorp, E. B., and T. M. Gallagher. 2004. Requirements for CEACAMs and cholesterol during murine coronavirus cell entry. J. Virol. 78:2682-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams, R. K., G.-S. Jiang, and K. V. Holmes. 1991. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. USA 88:5533-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanagi, Y., N. Ono, H. Tatsuo, K. Hashimoto, and H. Minagawa. 2002. Measles virus receptor SLAM (CD150). Virology 299:155-161. [DOI] [PubMed] [Google Scholar]