Abstract
Deformed wing virus (DWV) of honeybees (Apis mellifera) is closely associated with characteristic wing deformities, abdominal bloating, paralysis, and rapid mortality of emerging adult bees. The virus was purified from diseased insects, and its genome was cloned and sequenced. The genomic RNA of DWV is 10,140 nucleotides in length and contains a single large open reading frame encoding a 328-kDa polyprotein. The coding sequence is flanked by a 1,144-nucleotide 5′ nontranslated leader sequence and a 317-nucleotide 3′ nontranslated region, followed by a poly(A) tail. The three major structural proteins, VP1 (44 kDa), VP2 (32 kDa), and VP3 (28 kDa), were identified, and their genes were mapped to the N-terminal section of the polyprotein. The C-terminal part of the polyprotein contains sequence motifs typical of well-characterized picornavirus nonstructural proteins: an RNA helicase, a chymotrypsin-like 3C protease, and an RNA-dependent RNA polymerase. The genome organization, capsid morphology, and sequence comparison data indicate that DWV is a member of the recently established genus Iflavirus.
Deformed wing virus (DWV) is the one of the main viruses associated with the collapse of honeybee (Apis mellifera L.) colonies due to infestation with the ectoparasitic mite Varroa destructor (4, 8, 12). The virus was first isolated from a sample of symptomatic honeybees from Japan in the early 1980s and is currently distributed worldwide, wherever varroa mites are found (2, 18). A recent survey of adult bee populations detected DWV in over 90% of French apiaries (66) and in 100% of mite samples. The incidence was slightly reduced when pupal samples were analyzed, especially in the spring (66). DWV has also been detected by serology in the dwarf bee A. florae Fabr. (F. R. Hunter-Fujita, M. S. Mossadegh, and B. V. Ball, Abstr. 36th “Apimondia” Int. Apic. Congr., abstr. 230, 1999) and in the Asian honeybee A. cerana Fabr. (2) and by reverse transcriptase (RT) PCR in bumblebees (29). It is serologically related to Egypt bee virus (8, 9, 13), first isolated in 1977 from infected adults from Egypt (10). Typical symptoms of deformed wing disease are vestigial and crumpled wings, bloated abdomens, paralysis, and a severely shortened adult life span for emerging worker and drone bees (44). For a long time it was believed that these symptoms were due to the feeding activity of the mites (23, 42, 72) until it was shown that, in diseased colonies, deformed bees could emerge from cells not parasitized by varroa mites (49, 55) and that the symptoms can persist in the absence of mites (13). The symptoms are perfectly correlated with the presence of large amounts of DWV, as well as with the reduced virus titers and lower prevalence found in asymptomatic bees from the same colonies (17, 19, 20, 54, 67, 68). The combination of mites and virus causes immunosuppression in the bees and increased susceptibility to other opportunistic pathogens (76), leading to a progressive reduction in colony performance and a complex disease profile at colony level that often includes other pathogens as well (12, 56, 63, 66). Symptomatic adult bees typically appear in the final stages of colony collapse, usually late summer-autumn of the second or third year of uncontrolled varroa mite infestation. Occasionally, a colony may have symptomatic bees in early spring and recover during the summer, only for symptoms to reappear at the end of the year. As is the case with acute bee paralysis virus (ABPV) and Kashmir bee virus (KBV), pupal parasitism by varroa mites is the single most important factor for both symptom development and virus titer in individual bees (17, 54, 55). An alternative route of infection is through larval contact with nurse bees, since the virus can be detected in hypopharyngeal secretions (royal jelly), broodfood, eggs, and early larval stages that are not parasitized by mites (19, 20, 77) and in uninfested pupae fed in infected colonies (54, 55). High virus levels in mites are generally associated with high virus levels in the corresponding pupae (17, 54), and there is evidence that the virus may also replicate in the mite (17, 54, 57, 77). DWV is normally a weak and insignificant virus that develops slowly, allowing the brood to develop through the pupal stage to adulthood (8, 13). It is this low virulence that has made DWV the main virus associated with varroa mite infestations, since more virulent viruses, such as chronic bee paralysis virus, ABPV, KBV, black queen cell virus (BQCV), sacbrood virus (SBV), and slow paralysis virus (SPV), kill the brood too fast for varroa mites to complete their development on the bee pupae and transmit the virus to new hosts (50, 64).
DWV produces a 30-nm icosahedral particle consisting of a single positive-stranded RNA genome and three major structural proteins (8), characteristics that are common to many insect viruses. Originally classified as picorna-like insect viruses (52, 53), these viruses now fall largely into two groups: the genus Cripavirus, family Dicistroviridae (51), with separate open reading frames for the structural and nonstructural polyproteins (in the 3′ and 5′ halves of the genome, respectively), and the genus Iflavirus, with a typical picornavirus genome organization consisting of a single open reading frame in which the structural proteins are N-terminal to the nonstructural proteins. In both genera, the viral RNA is polyadenylated and the open reading frame is flanked by nontranslated regions (NTR) containing replication and translation control elements (41, 71, 74, 75). Among the honeybee viruses, BQCV, ABPV, and KBV are cripaviruses, while SBV, Kakugo virus (KV), and Varroa destructor virus 1 (VDV-1) are iflaviruses (25, 28, 30, 35, 46, 57).
In this study, we characterize two independent DWV isolates, DWV-it, from northeastern Italy, and DWV-pa, from Pennsylvania, and provide evidence for the taxonomic assignment of DWV. The data presented here show that DWV, a honeybee virus long known to the beekeeping community, is very closely related to the recently described KV and VDV-1.
MATERIALS AND METHODS
Virus isolation and purification.
Adult bees with wing deformities were collected during autumn 2000 from varroa mite-infested, diseased colonies in northeastern Italy (University of Udine) and in central Pennsylvania. Healthy bees were collected at the same time from varroa mite-free colonies with no clinical signs of disease. Virus was extracted by homogenization of deformed bees in 30% (wt/wt) 0.5 M sodium phosphate (pH 7.6)-0.02% diethylcarbamate-10% ether, filtration through gauze, and clarification with an equal volume chloroform. Virus was purified either directly from the extract (DWV-pa) or after precipitation with 1% NaCl-6% polyethylene glycol 6000 and resuspension in buffer (DWV-it) by sedimentation through a 10 to 40% sucrose gradient. The virus-containing fractions were identified by absorption at 280 nm, electron microscopy, and polyacrylamide gel electrophoresis (PAGE). The absence of SBV and ABPV, similarly shaped viruses that are common contaminants of DWV preparations, was confirmed by RT-PCR.
DWV antisera and Western blot analysis.
Antiserum was raised in rabbits against the electroeluted VP1 protein of DWV-pa and shown to be specific for DWV VP1 in Western blots, without any cross-reactivity with other bee viruses or native bee proteins (24). The immunoglobulin G fraction of the DWV VP1 antiserum was purified on protein A-agarose columns (39). Western blots were prepared by electrotransfer of proteins separated by sodium dodecyl sulfate (SDS)-PAGE to nitrocellulose (62). The blots were probed with DWV VP1 immunoglobulin G at a 1/2,000 dilution, followed by 40 ng/ml of horseradish peroxidase-conjugated protein A (Sigma P8651), and were developed with 0.6 mg/ml 3,3′-diaminobenzidine and 1 μl/ml of 30% H2O2 in 10 mM Tris-Cl (pH 7.5)-0.03% NiCl2 (39). Western blot samples were obtained by preparing extracts of weighed samples of the different bee tissues as described above, except that 1 ml of extraction buffer was used per gram of tissue. These samples included the heads, thoraxes, and abdomens of deformed and asymptomatic adult bees, varroa mite-infested and noninfested white-eyed pupae, and fifth-instar larvae from diseased colonies, plus the heads, thoraxes, and abdomens of nurse bees, guard bees, foraging bees, and drones, white-eyed pupae, and fifth-instar larvae from healthy colonies.
Electron microscopy.
Samples taken throughout the virus purification procedure were cleared by low-speed centrifugation (10 min at 8,000 × g), and 100 μl of supernatant was pelleted directly onto carbon-coated Formvar copper grids by ultracentrifugation (15 min at 82,000 × g) with a Beckman Airfuge. The grids were negatively stained with 2% sodium phosphotungstate at pH 6.8 for 90 seconds and observed with a Philips CM10 transmission electron microscope.
N-terminal protein sequencing and mass spectrometry.
For N-terminal sequence analysis, the three main structural proteins of DWV were separated by SDS-PAGE, electroeluted individually into Tris-glycine running buffer, dialyzed overnight at room temperature against sterile water, and spotted onto polyvinylidene difluoride membranes (Amersham Biosciences) (62). N-terminal sequence analysis by Edman degradation was done independently at Macromolecular Resources (Colorado State University, Boulder, Colo.) and at the Macro Core Facility (Penn State College of Medicine, Hershey, Pa.). For mass spectrometry analysis, the DWV capsid proteins were transferred to Immobilon P membranes, eluted individually, and trypsinized (6) prior to liquid chromatography coupled to mass spectrometry on a Micromass ESI-QTOF LC-MS instrument.
RNA cloning and sequencing.
Intact viral genomic RNA was extracted from the DWV-it and DWV-pa virus fractions by TRIzol-LS (Invitrogen) and retrotranscribed into cDNA (62) by using random oligonucleotides and oligo(dT)12-18 as primers. The DWV-pa cDNA fragments were ligated either directly or after PCR amplification with specific primers into plasmid cloning vectors. The DWV-it cDNA fragments were modified with synthetic EcoRI adapters, fractionated on silica columns, and ligated into pBluescript-II-SK+ (Stratagene). The ligations were transformed into Escherichia coli, and individual transformants were sequenced. The 3′ terminus was obtained by sequencing oligo(dT)-primed cDNA clones, since the virus group to which DWV belongs has a natural 3′ poly(A) tail. The 5′ terminus of the DWV-pa genome was obtained by three independent rounds of seminested 5′ rapid amplification of cDNA ends (RACE), using the SMART-Oligo protocol (Clontech) and three different cDNA primers. The PCR products were ligated into pGEM-T Easy (Promega), and a total of 17 clones from the three 5′ RACE reactions were sequenced. The 5′ terminus of the DWV-it genome was confirmed by ligating the 5′ and 3′ termini of viral RNA with T4 RNA ligase (Fermentas) in the presence of 40 U of RNasin (Promega), amplifying the resulting RNA by nested RT-PCR with primer DWV-lig2r (201ACACTAAGTGCACATATTCATAATCC176) for cDNA, primers DWV-lig2r and DWV-lig1f (10055AGTGGTACTCTAGGTTAGGTGTTACTC10081) for one round of PCR, and primers DWV-lig1f and DWV-lig3r (71ATAATGTTTGTTCATGGCTACTATCTA45) for nested PCR (6). The RT-PCR product was gel purified and sequenced directly with DWV-lig3r. The consensus 5′ end was identified by the start of the poly(A) tail of the ligated 3′ end. The sequencing strategy resulted in 3.7- and 4.7-fold coverage of the DWV-it and DWV-pa genomes, respectively.
RT-PCR.
RNA was extracted with RNeasy (QIAGEN) from purified virus, from extracts of different bee tissues, and from type strains of DWV, SBV, and ABPV (kindly provided by B. V. Ball). The RNAs were converted to cDNA and amplified by PCR for 30 cycles, with annealing at 50 to 60°C for 30 seconds and elongation at 72°C for 50 to 100 seconds, depending on the primer sets used. The two primer sets used for DWV were D (8714GGATGTTATCTCTTGCGTGGA8734 and 9125CGATAATAATTTCGAACGCTGA9104) and D′ (5689TGATTTTCAGCCTGTTTTGTG5709 and 5967CCTCGCTTCTTCTCTTCACTC5947). Other virus-specific primer pairs were primer set S (2351GTGGCAGTGTCAGATAATCC2370 and 3185GTCAGAGAATGCGTAGTTCC3166) for SBV (36) and primer set A (7847GGTGATTTGAGAGATGTATTTCCT7870 and 8570ACACTTGCGTTGGTCCTGAA8551) for ABPV.
Northern blot analysis.
Bee tissue was ground in a mortar in the presence of liquid nitrogen, and total RNA was extracted with RNeasy (QIAGEN) by the protocol for tissues rich in proteins. Poly(A) RNA was purified with FastTrack 2.0 (Invitrogen). Poly(A) RNA (800 ng) from deformed bees and total RNA (20 μg) from deformed and healthy bees were resolved in 1.2% denaturing agarose gels containing 2.2 M formaldehyde, transferred to Hybond N+ (Amersham Biosciences) membranes (62), and hybridized with a DNA probe (nucleotides 1118 to 3496) labeled with the ECL random prime labeling kit (Amersham Biosciences).
Sequence and phylogenetic analyses.
Nucleotide sequences were assembled with Lasergene (DNASTAR) and DNA Club (freeware). Primary nucleotide and protein sequence analyses were done online at http://www.ncbi.nlm.nih.gov/ and at http://us.expasy.org/. The amino acid sequences of all unique iflaviruses were aligned with CLUSTAL W (69) at http://www.ebi.ac.uk/clustalw/ by using three picornaviruses—poliovirus (PV), hepatitis A virus (HAV), and encephalomyocarditis virus (EMCV)—as the outgroup. The alignments were edited by hand where necessary, with the conserved protein domains used as a guide. The available partial sequences from other DWV isolates were manually inserted into this alignment. Phylogenetic analysis adhered to maximum parsimony criteria as implemented by PAUP (4.0b10) (65). Gaps were treated as a separate character state. Bootstrap analyses were based on 1,000 replicates, and nodes with less than 60% support were collapsed.
Nucleotide sequence accession numbers.
The following sequences from the GenBank database were used in the protein domain and phylogenetic analyses. Iflaviruses: DWV from Italy (DWV-it), AJ489744; DWV from Pennsylvania (DWV-pa), AY292384; DWV from France (DWV-fr), AY224602; KV, AB070959; VDV-1, AY251269; an uncharacterized sequence isolated from a human tracheal cDNA library, AK128556; Venturia canescens virus (VeCV), AY534885; SBV from the United Kingdom (SBV-uk), AF092924; SBV from China (SBV-cn), AF469603; infectious flacherie virus (IFV), AB000906; Ectropis obliqua picorna-like virus (EoPV), AY365064; Perina nuda picorna-like virus (PnPV), AF323747. Marnavirus: Heterosigma akashiwo virus (HaRNAV), AY337486. Cripaviruses: ABPV, AF150629; BQCV, AF183905; KBV, AY275710; cricket paralysis virus (CrPV), AF218039; Drosophila C virus (DCV), AF014388. Picornaviruses: PV, CAA24465; HAV, BAA35107; EMCV, M81861.
The consensus sequences reported here have been deposited in the EMBL, GenBank, and DDBJ sequence databases under accession numbers AJ489744 (DWV-it) and AY292384 (DWV-pa).
RESULTS
Virus identification and molecular characterization.
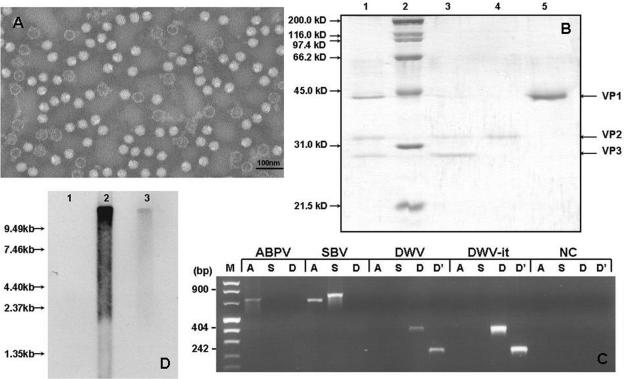
Electron microscopy revealed the presence of large amounts of empty and filled icosahedral virus particles of about 30 nm in diameter in virus preparations from bees with wing deformities (Fig. 1A) that were absent in control preparations from healthy bees. The virus was purified and shown by RT-PCR to contain DWV RNA, with no evidence of SBV or ABPV, the two most common copurifying viruses (Fig. 1C). DWV, SBV, and ABPV reference strains served as positive controls for the RT-PCRs and to confirm the identity of the virus. The sizes of the RT-PCR products for ABPV (723 nucleotides), SBV (815 nucleotides), and DWV (411 and 278 nucleotides) correspond to their respective primer sets. The reactions also showed that the ABPV preparation was free of SBV or DWV RNA, while the SBV preparation was free of DWV but contained detectable amounts of ABPV.
FIG. 1.
Molecular characterization. (A) Electron microscopy image showing empty and filled DWV particles purified from an extract from deformed adult bees. (B) SDS-PAGE of purified DWV. Lane 1, purified virus; lane 2, molecular weight marker; lanes 3 to 5, gel-purified structural proteins. (C) Verification of the purity of DWV preparations by RT-PCR amplification of purified SBV, ABPV, DWV, DWV-it, and negative controls (NC) with primers specific for ABPV (A), SBV (S), and DWV (D and D′). (D) Northern blot of DWV RNA. Total RNA extracted from healthy bees (lane 1) was compared to mRNA (lane 2) and total RNA (lane 3) from infected bees.
The purified virus particle has three major proteins, with apparent masses of 44 kDa, 32 kDa, and 28 kDa (Fig. 1B), corresponding to the structural proteins VP1, VP2, and VP3 in the Picornaviridae nomenclature. No low-molecular-weight protein corresponding to the picornavirus VP4 was detected, despite repeated attempts.
Northern blot analysis with a DWV-specific probe revealed a signal of up to about 10 kb in both total RNA and poly(A)-enriched RNA from deformed bees, while no signal was detected in total RNA from healthy bees (Fig. 1D). This marks the upper size limit of DWV RNA and indicates that DWV has a naturally polyadenylated tail. There is no evidence of discreet subgenomic RNA molecules, suggesting that DWV does not use this strategy for translation of its genome. Furthermore, purified viral RNA was invariably partially degraded, in contrast to the internal control (bee ATPase mRNA; data not shown), supporting previous reports of the lability of the DWV capsid and genomic RNA (8).
Viral loads of DWV in infected bees.
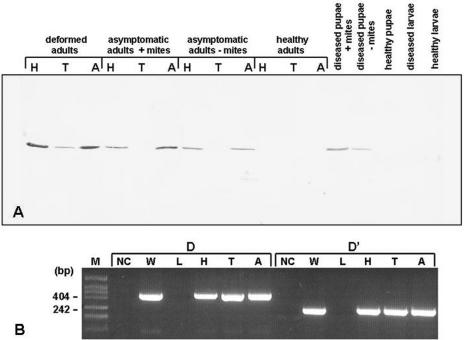
Western blot analysis of weight-adjusted extracts of different bee tissues showed that deformed adult bees have the highest DWV titers, followed by mite-infested asymptomatic adults and mite-free asymptomatic adults from severely diseased colonies, with the greatest concentration of signal in the head and abdomen and considerably less in the thorax (Fig. 2A). Significant titers were also found for mite-infested pupae, with reduced levels for noninfested pupae and nondetectable levels for fifth-instar larvae from diseased colonies. DWV VP1 was not detected in healthy worker bees, pupae, or larvae (Fig. 2A) or in the heads, thoraxes, or abdomens of healthy nurse, forager, or aggressive or passive guard bees or drones (data not shown). DWV RNA was readily detected by RT-PCR in the heads, thoraxes, abdomens, and wings of deformed bees but not in the legs (Fig. 2B), while no DWV RNA was detected in these tissues from healthy bees (data not shown). These data concur largely with similar observations made elsewhere (77). We conclude that DWV infection, at least in later stages when the symptoms become obvious, is systemic.
FIG. 2.
Virus distribution. (A) Distribution of DWV protein in adult worker bees, white-eyed pupae, and fifth-instar larvae of healthy and diseased colonies, as detected by Western blotting using DWV VP1 antiserum. All extracts were normalized by tissue weight. H, head; T, thorax; A, abdomen. (B) Distribution of DWV RNA in deformed worker bees as detected by RT-PCR using primer sets D and D′. M, molecular weight markers; NC, negative control; W, wings; L, legs; H, head; T, thorax; A, abdomen.
Nucleotide sequence analysis.
The complete viral genome sequence is 10,140 nucleotides long, excluding the poly(A) tail, and is enriched in A (29.5%) and U (32.3%) compared to G (22.4%) and C (15.8%). Analysis of codon usage suggests a preference for U (39.5%) versus A (26.8%) as the codon's third base. Analysis of natural variation within each isolate showed that single-nucleotide polymorphisms are spread evenly throughout the genome, with a frequency of about 0.7% for DWV-it and 0.8% for DWV-pa. The vast majority (82%) of these variants were transitions (C-U and A-G changes), which are common for RNA viruses and a consequence of the relative stability of the G-U pairing in RNA molecules during replication (60). Twenty-eight percent of the nucleotide variations within the coding region resulted in amino acid changes. The DWV genome contains a single long open reading frame flanked by 5′ and 3′ NTR of 1,140 to 1,144 nucleotides and 317 nucleotides, respectively. The Italian and Pennsylvanian DWV nucleotide sequences are 98% identical to each other and to the (partial) sequences of other DWV isolates. DWV is also 97% identical to KV, which was isolated in Japan from the mushroom bodies of phenotypically normal honeybees (28), and 84% identical (95% amino acid identity) to VDV-1, which was isolated from mites infesting A. mellifera colonies in The Netherlands (57). There is significant variability between these isolates at the 5′ terminus, where DWV-it is 5 nucleotides longer than DWV-pa, 13 nucleotides longer than VDV-1, and 11 nucleotides shorter than KV. These differences may be genuine, since for all viruses multiple clones with the same 5′ terminus were obtained in specific experiments designed to identify the 5′-most nucleotide (seminested 5′ RACE for DWV-pa, KV, and VDV-1; RNA ligation-PCR for DWV-it). There are also other minor insertion/deletion variations between the viruses elsewhere in the 5′ NTR, including a major 11-nucleotide deletion in VDV-1 immediately prior to the start of the open reading frame (57). The 3′ NTR is much less variable than the 5′ NTR, as observed in many positive-strand RNA viruses.
Mapping of DWV capsid protein genes.
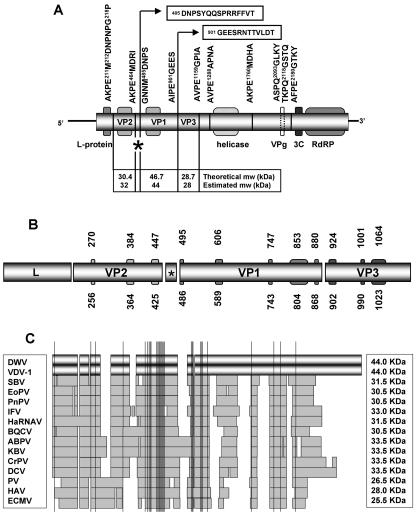
The DWV capsid proteins were individually purified (Fig. 1B) for N-terminal sequence determination and mass spectrometry analysis. The N-terminal sequences of VP1 (DNPSYQQSPRRF) and VP3 (GEESRNTTVLDT) map to amino acids 486 to 497 and 902 to 913, respectively, of the deduced polyprotein sequence (Fig. 3A). No reliable sequence data were obtained for VP2. Such failure is not uncommon and is most likely due to secondary modifications of the N terminus of the protein (47). The mass spectrometry data confirm and extend these results (Fig. 3B). Peptides identified from VP1 collectively match five sequence blocks of the predicted polyprotein, spanning amino acids 486 to 880. The first VP1 peptide corresponds exactly to sequence identified by N-terminal degradation, while the last peptide is only 21 amino acids upstream of the confirmed N terminus of VP3 (amino acid 902). These data imply a VP1 with a theoretical mass of 46.7 kDa, a figure close to the 44 kDa estimated by SDS-PAGE (Fig. 1B). The VP2 peptides correspond to three sequence blocks between amino acids 256 and 448 of the polyprotein. This places VP2 upstream of VP1 on the DWV genome, although the exact boundaries of the VP2 gene can only be hypothesized (see below). Finally, peptides from VP3 correspond to four blocks on the polyprotein sequence between the VP3 N terminus and amino acid 1064. The nearest putative 3C protease cleavage site downstream from amino acid 1064 is AVPE1159/GPIA. If this defines the correct VP3 C terminus, then VP3 has a theoretical mass of 28.7 kDa, close to the 28 kDa estimated by SDS-PAGE (Fig. 1B).
FIG. 3.
Genome mapping. (A) Map of the DWV genome. The long shaded box represents the single open reading frame, with various recognizable protein domains shown. The short vertical lines (dashed and solid) represent predicted protease cleavage sites: their positions and surrounding amino acid sequences are also shown. The long vertical lines lead to the experimentally determined N-terminal amino acid sequences of VP1 and VP3 (boxed). The figure also shows, for each of the main structural proteins, the size estimated by PAGE and the theoretical calculations based on putative protease cleavage sites. The asterisk marks the position of the putative VP4 capsid protein (2.3 kDa). (B) Genome location and ranges of the peptides identified by mass spectrometry analysis of purified DWV VP2, VP1, and VP3 capsid proteins. (C) Schematic representation of the optimal alignment of the DWV/KV VP1 structural protein, with its homologues in other iflaviruses, cripaviruses, and picornaviruses. Shaded areas indicate blocks of aligned amino acids, and vertical lines represent highly conserved amino acids, based on the following amino acid groups: aliphatic (M, I, L, V), aromatic (F, W, Y), charged positive (H, K, R), charged negative (D, E), polar (N, Q), small nonpolar (A, G), small polar (S, T), cysteine (C), and proline (P).
The large size of VP1 is unusual among iflaviruses and related picorna-like viruses, whose homologues of DWV VP1 are no larger than 35 kDa and are often much smaller (48). Comparative analysis of these proteins shows that the additional amino acids in DWV VP1 are mostly located in the C-terminal part of the protein (Fig. 3C), with the most strongly conserved protein domains in the middle of the N-terminal half of the protein. Since VP1 is an integral component of the virus particle, it remains to be seen how these extra amino acids are incorporated into the shell to produce the typical 30-nm icosahedral shape shown in Fig. 1A.
Protease site analysis.
The N-terminal amino acid sequences of VP1 and VP3 define bona fide cleavage sites of the polyprotein and provide some insight into the type of protease activity associated with processing of the DWV polyprotein. The VP3 protease site (AIPE901GEES) conforms to the classic pattern for viral 3C proteases that cut after either glutamine (Q) or glutamic acid (E) (37, 58). The flanking amino acids are usually also conserved for individual 3C proteases (37, 41, 58), with the glycine (G), proline (P), and alanine (A) residues common in the shown positions (16, 58, 59). Using the positions of these residues as a guide, we found several putative recognition sites for the DWV 3C protease in the polyprotein (Fig. 3A). All sites are suitably located for the separation of functional viral proteins from the polyprotein, with excellent agreement between the estimated and predicted molecular weights for the structural proteins. The most intriguing sites, which occur at amino acids 211, 464, and 1760, are identical over six consecutive amino acids (AKPE/MD), a coincidence too great not to be of significance. Furthermore, they fall in optimal positions for the production of several viral proteins. Site 211 could separate a leader polypeptide (L protein) from the polyprotein and is consistent with the mass spectrometry data for VP2. Site 464 would, together with the VP1 N terminus, generate a small, 2.3-kDa VP4, which is a product of particle maturation in many picorna-like viruses, including certain iflaviruses (21, 40, 58), and is usually part of a precursor protein (VP0) together with VP1 (14, 22, 40). A VP4 protein has thus far not been detected in purified DWV (8), although this is not surprising considering its small predicted size. Site 1760 could separate the helicase from the polyprotein. Sites 1159, 1288, and 2180 are very similar to the confirmed VP3 protease site and would perfectly process the C terminus of VP3, the N terminus of the helicase, and the N terminus of the 3CD protein comprising the 3C protease and RNA-dependent RNA polymerase (RdRp) (40). There are two further sites of interest (at positions 2093 and 2118) that have glutamine (Q) as the canonical site rather than glutamate (E) and that are perfectly positioned to release the putative DWV VPg from the polyprotein. Both of these sites are conservatively altered in the closely related VDV-1, as are sites 464, 901, and 1288. The remaining putative 3C protease sites between DWV and VDV-1 are identical.
The VP1 N-terminal junction (NM485DNPS) is generated by an unknown protease activity, which is also a feature of most picorna-like viruses (14, 22, 40). Comparison with the homologous VP1 termini of the other iflaviruses shows that, in particular, the D and P residues in positions P1′ and P3′ are highly conserved (Table 1). There is only one similar sequence in the DWV polyprotein, EM212DNPN, which falls in the region of the VP2 N terminus. The same region also contains an autocatalytic NPG218P tetrad, or 2A-like protease site (58). With potentially three different protease targets between amino acids 211 and 218, it is likely that DWV, like IFV (41), PnPV (75), and the cardio- and aphthoviruses (58), produces a leader polypeptide (L protein) prior to the three structural proteins.
TABLE 1.
VP4/VP1 protease cleavage sites
| Virus | Amino acid at indicated processing sitea
|
|||||||
|---|---|---|---|---|---|---|---|---|
| P4 | P3 | P2 | P1 | P1′ | P2′ | P3′ | P4′ | |
| DWV (L/VP2) | K | P | E | M212 | D | N | P | N |
| DWV | G | N | N | M485 | D | N | P | S |
| SBVb | S | N | N | Q428 | D | K | P | K |
| IFV | M | R | N | K465 | D | K | P | V |
| PnPVc | K | K | D | M637 | D | R | P | Q |
| EoPVc | K | K | D | M639 | D | R | P | Q |
Comparison of the VP4/VP1 processing sites of different iflaviruses. In the case of DWV, the putative L/VP2 site is also shown for comparison (first row). P1/P1′ correspond to the positions of scissile-bond amino acids. Highly conserved residues are indicated in bold. Other residues are conserved among groups of viruses.
The SBV site was deduced from amino acid sequence comparisons.
Amino acid sequence analysis.
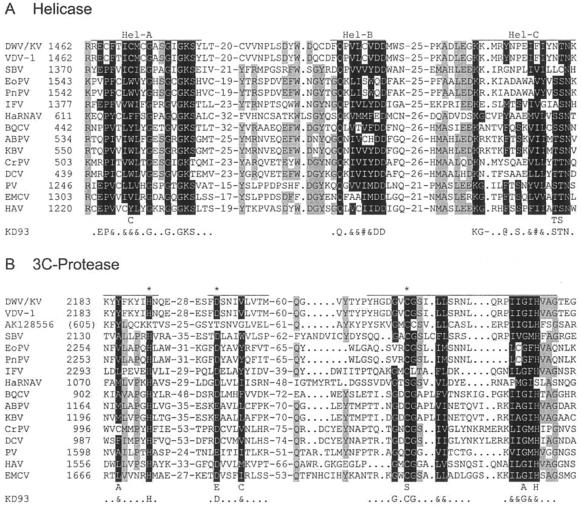
Domain analysis of the polyprotein shows the order of the viral proteins to be identical to that of mammalian picornaviruses (37), with the structural proteins in the N-terminal portion and the nonstructural proteins in the C-terminal portion of the polyprotein (Fig. 3A). The helicase domains A, B, and C (43) are found between amino acids 1460 and 1575 (Fig. 4A), including the perfectly conserved putative nucleoside triphosphate binding residues 1472GxxGxGKS1479 and 1518Qx5DD1525 in domains A and B, respectively. The C domain (1561KKx4Px5NTN1575) is slightly divergent from the consensus, KGx4Sx5STN (33).
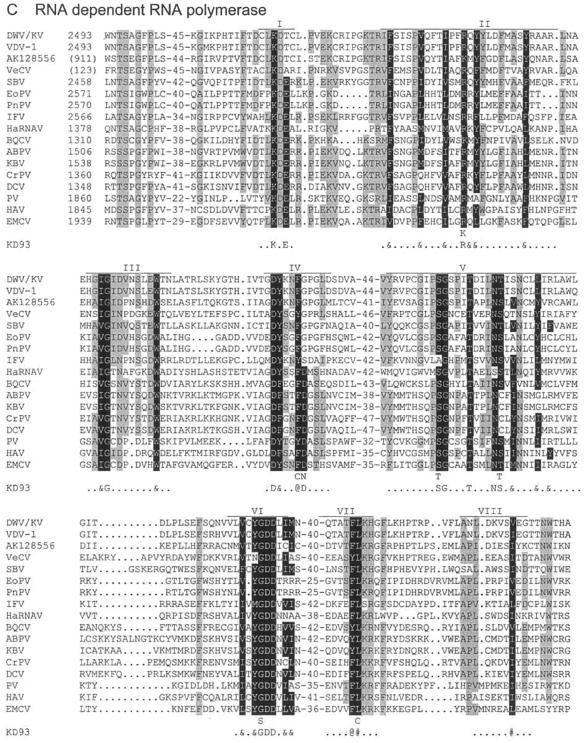
FIG.4.
Protein domain alignments. Shown are alignments of the helicase (A), 3C proteinase (B), and RdRp (C) domains of DWV/KV with those of the other iflaviruses (VDV-1, SBV, VeCV, EoPV, PnPV, and IFV), a human tracheal cDNA clone (AK128556), an ABPV marnavirus (HaRNAV), several (honeybee) cripaviruses (BQCV, KBV, CrPV, and DCV), and three picornaviruses (PV, ECMV, and HAV). The genome location of the first amino acid of each alignment is shown, with those of partial sequences shown in brackets. The lengths of the domains are indicated by the lines above each segment; the conserved amino acids, as identified by Koonin and Dolja (43), are indicated in reverse type and in the line KD93, where “@” refers to an aromatic residue (F, Y, or W); “#” refers to a bulky aliphatic residue (M, I, L, or V); and “&” refers to any bulky hydrophobic residue, aliphatic or aromatic. Other residues that are also conserved among these sequences are shaded in gray.
The 3C protease domains (Fig. 4B) span amino acids 2183 to 2327 and conserve the cysteine protease motif 2305GxCG2308 and the putative substrate binding residues 2322GxHxxG2327 (32). C2307 is the third residue of the protease catalytic triad that also involves a histidine residue and either an aspartate or glutamate residue (15, 61). H2190 and D2225 are the most likely candidates between the processing site at position 2180 and the cysteine protease motif to complete the catalytic triad with C2307. All other possibilities are either too close to the other domains or are in regions where VDV-1 is variant. Neither the H nor D residue is conserved in sequence AK128556, which is most closely related to DWV/KV/VDV-1, but this sequence also has a seriously disrupted cysteine protease motif and so may not possess a functional 3C protease at all. This sequence was extracted from a Homo sapiens tracheal cDNA library, which raises all sorts of interesting questions, but no further characterization has been reported.
Prior to DWV VP2 is a putative leader polypeptide (L protein), also found in other iflaviruses (41, 75) and cardio- and aphthopicornaviruses (37, 58). The DWV L protein has two motifs (123CIFx7V133 and 172HL173) that weakly resemble the catalytic sites of papain-like cysteine proteases found in some aphthoviruses (34, 38, 40, 43). The L protein also appears to be a hotspot for microvariation. Despite occupying only 7.3% of the polyprotein, it bears 26.2% to 33.3% of the amino acid differences found among DWV, KV, and VDV-1. Combining data from all comparisons, the L protein is 5.4 times more variable than the rest of the polyprotein and 3.3 times more variable than the next most variable region (between the helicase and 3C protease domains). This variability stops abruptly right after the postulated L-VP2 cleavage sites, providing further validation for the existence of these sites. In the light of the proposed biological differences between these very closely related viruses (28, 57), it is interesting to note that the L proteins of other picornaviruses are also highly variable and are implicated in disease pathology through the inhibition of host cap-dependent mRNA translation (31) and stimulation of viral internal ribosome entry site activity (40).
VPg, a small protein common to many RNA viruses, is responsible for stabilizing the 5′ end of the genomic RNA for replication and translation. In picornaviruses, VPg is about 23 amino acids long, with an early tyrosine (Y) residue as the physical link to the RNA (73), and is located immediately prior to the 3C protease domains. We identified a weak VPg motif for DWV between the putative 3C protease sites at positions 2093 and 2118 that includes a Y residue at position 2097.
Finally, all eight RdRp domains are located between amino acids 2493 and 2828 (Fig. 4C), including the “core” RdRp motifs IV, V, and VI, thought to be involved in catalysis and nucleoside triphosphate binding (43). A highly conserved domain prior to the first RdRp domain (2495TSxGxP2500) was also identified.
Phylogenetic analysis.
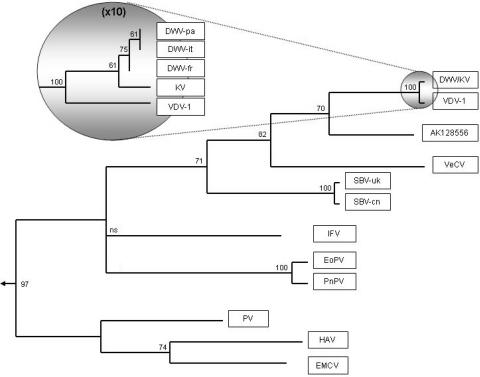
The relationships between DWV, KV, VDV-1, and the other iflaviruses are depicted in a phylogram (Fig. 5) obtained by comparing the RdRp regions of the viruses. The phylogram shown is based on maximum parsimony criteria, but identical trees were obtained with neighbor-joining and maximum likelihood criteria. There is clear separation between the picornavirus outgroup and the iflaviruses. The lack of resolution at the base of the Iflavirus clade, particularly between IFV and EoPV/PnPV, has been described before (45) and is also found when the other regions of the genome are analyzed. Ultimately, it will be resolved only by the addition of other deep-rooting iflaviruses. All of the other partitions in the tree are well supported, with support weakening within the DWV/KV/VDV-1 clade due to a dearth of differentiating characters. It is clear that DWV, KV, and VDV-1 are variants of a single major virus, with little geographic differentiation (the European, American, and Japanese [KV] versions are practically identical); only VDV-1 is truly distinct. Similar internal identity is found for EoPV and PnPV, where the virus is also given different names depending on host origin, and for SBV, where a single name is retained. As stated above, the presence of a purported human tracheal cDNA sequence (AK128556), solidly entrenched between DWV from honeybees and VeCV from a parasitic wasp (Venturia canescens), is a major curiosity still to be resolved.
FIG. 5.
Phylogram of the genus Iflavirus, with three picornaviruses (PV, HAV, and EMCV) used as the outgroup. The phylogram is based on the RdRp sequence and was constructed by maximum parsimony criteria. The percentage of bootstrap support for each branch is indicated. Branches with less than 60% bootstrap support were collapsed.
DISCUSSION
Wing deformities in honeybees have long been associated with a virus, appropriately named deformed wing virus, transmitted by varroa mites and symptomatic of the final stages of colony collapse due to uncontrolled varroa mite infestation (8, 9, 12, 13, 17, 50, 54, 56). Examination of the first samples of deformed bees in the early 1980s revealed very high titers of a rather unstable icosahedral virus with a single RNA genome (8). Subsequent studies using a variety of techniques confirmed a nearly 100% association of the wing deformities with highly elevated titers of the virus and reduced titers in phenotypically normal bees from the same colonies (17, 20, 29, 54, 55, 67, 68); the detection of the virus in eggs, larvae, pupae, and broodfood (19, 20, 77); a strong association of individual virus titers with pupal infestation by varroa mites (54, 55); the transmission by varroa mites of both symptoms and virus titers (17); and both indirect and direct evidence of viral replication in the mites, as well as in bees (17, 54, 57, 77). Nevertheless, a direct causal link between the virus and the disease has not yet been established, mostly due to the difficulty of excluding other pathogens as being (partly) responsible for the symptoms. This study may constitute a first step toward the precise definition and extent of DWV involvement in causing the disease. The data presented here confirm that DWV presence is strongly associated with typical pathological symptoms and provide information about the molecular features of the virus. DWV has an icosahedral particle structure, with a considerable proportion (25%) of empty particle shells visible after purification, possibly due to particle instability. Western blotting with a monospecific antiserum against the DWV VP1 protein shows that the virus is concentrated in the heads and abdomens of infected adult bees, with significantly reduced titers in the thorax. DWV RNA was detected by RT-PCR in the head, thorax, abdomen, and wings of infected bees, with only the legs being devoid of virus. Reduced virus titers in the thorax have also been observed for ABPV and SPV, two other viruses associated with varroa mite infestation. ABPV accumulates almost exclusively in the hypopharyngeal, mandibular, and salivary glands, with minor amounts in the crop, midgut, and hind legs, while SPV has a slightly wider distribution that includes the brain and fat bodies (26). DWV was also detected in mite-infested and noninfested pupae of severely diseased colonies. Western blotting could not detect the virus in fifth-instar larvae from diseased colonies, nor in any bees (adults of various ages and functions, pupae and larvae, of both sexes) from healthy colonies. More-sensitive techniques, such as RT-PCR, have been used successfully to detect DWV in larvae and eggs from diseased colonies (as well as in adults and pupae) but not in those from healthy colonies (19, 20, 68, 77). The VP1 distribution and our RT-PCR experiments clearly show that, despite their extensive sequence identity, the mode of DWV infection is profoundly different from the striking organ tropism of KV (28), which was isolated from the mushroom bodies of adult honeybees from healthy hives and shown by RT-PCR to occur at elevated levels only in the brains of “aggressive” guard bees, not those of “passive” guard bees, nurse bees, or foraging bees or in other bee tissues. Only upon artificial inoculation did KV accumulate to high levels in other tissues and in bees of different functions.
Analysis of the DWV nucleotide and predicted amino acid sequences revealed a perfect match with the N-terminal amino acid sequences and with mass spectrometry data of the purified structural proteins, proving that the nucleotide and protein data (including the Western blots with DWV VP1 antiserum) belong to the same virus. The data also provide direct mapping of the VP1, VP2, and VP3 genes and reliable sizing of the structural proteins that matches the estimated molecular weights obtained by PAGE. It is noteworthy that the apparent molecular weight of VP1 is indeed due to its large size and is not an artifact of secondary modifications or abnormal gel migration. This makes DWV VP1 the largest protein of its kind in the picornavirus supergroup. Since the sizes of both VP2 and VP3 are within the normal range, it would be interesting to see how VP1 fits into the typical 30-nm virus particle. The N-terminal sequences of VP1 and VP3 define two important polyprotein cleavage sites. Combining these data with the sizes of the structural proteins determined by gel and mass spectrometry and the known specificity of the 3C protease leads to a fairly good prediction of additional polyprotein cleavage sites. Analysis of the conserved protein domains and the proteolytic processing sites identified a leader protein (L protein) preceding VP2, followed by a putative VP4, VP1, and VP3, a helicase, VPg, 3C protease, and RdRp, all with recognizable and conserved protease sites in suitable locations for processing. The VP1 N terminus does not fit with the conventional 3C protease targets; as in other picornaviruses, VP1 may be produced as a precursor protein, VP0, made of VP1 plus VP4, to be processed in the final stages of particle maturation by an unconventional protease. Due to the very low predicted molecular size of the putative VP4, it is not surprising that it may go undetected by SDS-PAGE analysis. At a mere 2.3 kDa, the DWV VP4 is one of the smallest VP4 proteins in the picornavirus supergroup. As seen in many positive-strand RNA viruses, the single open reading frame of DWV is flanked by a long 5′ NTR, presumably involved in translation initiation, and a short, conserved 3′ NTR.
The combination of direct mapping with sequence and phylogenetic analyses of the nonstructural region shows that the DWV genome organization is typical of iflaviruses, a group of insect-infecting viruses with many similarities to the picornaviruses. The Italian and Pennsylvanian DWV sequences are 98% identical to each other and to the (partial) sequences of other DWV isolates. DWV is also 97% identical to KV, which was isolated from the mushroom bodies of phenotypically normal honeybees (28), and 84% identical to VDV-1, a virus coexisting with DWV in varroa mites infesting honeybee colonies in The Netherlands (57) and likewise without any overt disease pathology in mites or bees. Positive- and negative-strand RNAs of both VDV-1 and DWV were detected in the same mites by strand-specific RT-PCR (57). It is possible that Egypt bee virus, isolated in Egypt in 1977 from phenotypically normal, diseased adults (10), can also be added to this list since it is serologically related to DWV (8, 9, 13) and shares its particle structure, protein profile, slow accumulation during pupal propagation, and instability during purification. The minor differences between DWV, KV, and VDV-1 are heavily concentrated in the L protein region. It would be interesting to see whether the DWV-associated symptoms, apparently absent in KV and VDV-1, map to this region, as predicted from the involvement of the L protein in disease pathogenesis in other viruses (31, 40). Whether these differences are enough to warrant separate nomenclature for these viruses remains to be decided. Both the nucleotide and amino acid differences between DWV, KV, and VDV-1 are very much in line with the variability found among strains of other bee viruses (1, 5, 11, 24, 36). Furthermore, in light of what has been known about DWV for several decades, neither KV nor VDV-1 is particularly unique. Most bee viruses thus far discovered, including DWV, are maintained as unapparent chronic infections among the bee population, with the occasional outbreak leading to clinical symptoms (3, 8, 9, 13). The absence of gross symptoms for KV and VDV-1 may simply be a reflection of this state. Even in the presence of mites, it can take months or years before honeybee populations accumulate a high enough virus titer to show symptoms. Several bee viruses are concentrated in the hypopharyngeal glands in bee brains (7, 13, 26, 27), from which they are transmitted to the larvae as part of the glandular secretions (royal jelly) used to feed young larvae. The discovery of KV in the bee brains is therefore not surprising and may be equally due to the mode of virus transmission and to the appearance of aggressive behavior in bees. Finally, it is entirely possible that these viruses have multiple effects on their hosts depending on the virus titer and the tissues affected. In the absence of the major ecological, biological, and molecular differences required for species designation (70), it is our contention that KV and possibly VDV-1 are biological and geographic variants of a virus that historically has been known as DWV.
Acknowledgments
G.L. and J.R.M. contributed equally to this work. Many thanks to Brenda Ball, IARC-Rothamsted, for virus strains and for preliminary testing of our symptomatic bees; to Anne Stanley at Hershey Medical School and Michael Ellis at Colorado State for the N-terminal sequence determinations; to Norberto Milani, University of Udine, for providing samples; to Giuliana Botti for technical help and critical suggestions; and to A. Dal-Piaz for mass spectrometry.
This work was supported by grants from the Pennsylvania Department of Agriculture (ME441833), the Binational Agriculture Research and Development Foundation (US-3205-01R), the Italian Ministry of Health, and the Italian Agency for Agriculture (A.M.A. Project).
REFERENCES
- 1.Allen, M. F., and B. V. Ball. 1995. Characterisation and serological relationships of strains of Kashmir bee virus. Ann. Appl. Biol. 126:471-484. [Google Scholar]
- 2.Allen, M. F., and B. V. Ball. 1996. The incidence and world distribution of honeybee viruses. Bee World 77:141-162. [Google Scholar]
- 3.Anderson, D. L., and A. J. Gibbs. 1988. Inapparent virus infections and their interactions in pupae of the honey bee (Apis mellifera Linnaeus) in Australia. J. Gen. Virol. 69:1617-1625. [Google Scholar]
- 4.Anderson, D. L., and J. W. H. Trueman. 2002. Varroa jacobsoni (Acari: Varroidae) is more than one species. Exp. Appl. Acarol. 24:165-189. [DOI] [PubMed] [Google Scholar]
- 5.Antunez, K., B. D'Alessandro, E. Corbella, and P. Zunino. 2005. Detection of chronic bee paralysis virus and acute bee paralysis virus in Uruguayan honeybees. J. Invertebr. Pathol. 90:69-72. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 7.Bailey, L. 1969. The multiplication and spread of sacbrood virus of bees. Ann. Appl. Biol. 63:483-491. [DOI] [PubMed] [Google Scholar]
- 8.Bailey, L., and B. V. Ball. 1991. Honey bee pathology, 2nd ed. Academic Press, London, England.
- 9.Bailey, L., and B. V. Ball. 1994. Honey bee viruses, p. 654-660. In R. G. Webster and A. Granoff (ed.), Encyclopedia of virology. Academic Press, San Diego, Calif.
- 10.Bailey, L., M. Carpenter, and R. D. Woods. 1979. Egypt bee virus and Australian isolates of Kashmir bee virus. J. Gen. Virol. 43:641-647. [Google Scholar]
- 11.Bakonyi, T., E. Grabensteiner, J. Kolodziejek, M. Rusvai, G. Topolska, W. Ritter, and N. Nowotny. 2002. Phylogenetic analysis of acute bee paralysis virus strains. Appl. Environ. Microbiol. 68:6446-6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ball, B. V., and M. F. Allen. 1988. The prevalence of pathogens in honey bee (Apis mellifera) colonies infested with the parasitic mite Varroa jacobsoni. Ann. Appl. Biol. 113:237-244. [Google Scholar]
- 13.Ball, B. V., and L. Bailey. 1997. Viruses, p. 11-31. In R. A. Morse and K. Flottum (ed.), Honey bee pests, predators and diseases, 3rd ed. A. I. Root, Medina, Ohio.
- 14.Basavappa, R., R. Syed, O. Flore, J. P. Icenogle, D. J. Filman, and J. M. Hogle. 1994. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9Å resolution. Protein Sci. 3:1651-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bazan, J. F., and R. J. Fletterick. 1988. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc. Natl. Acad. Sci. USA 85:7872-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blair, W. S., and B. L. Semler. 1991. Role for the P4 amino acid residue in substrate utilization by the poliovirus 3CD proteinase. J. Virol. 65:6111-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowen-Walker, P. L., S. J. Martin, and A. Gunn. 1999. The transmission of Deformed Wing Virus between honeybees (Apis mellifera L.) by ectoparasitic mite Varroa jacobsoni Oud. J. Invertebr. Pathol. 73:101-106. [DOI] [PubMed] [Google Scholar]
- 18.Calderon, R. A., J. van Veen, H. G. Arce, and M. E. Esquivel. 2003. Presence of deformed wing virus and Kashmir bee virus in Africanized honey bee colonies in Costa Rica infested with Varroa destructor. Bee World 84:112-116. [Google Scholar]
- 19.Chen, Y. P., I. B. Smith, A. M. Collins, J. S. Pettis, and M. F. Feldlaufer. 2004. Detection of deformed wing virus infection in honey bees, Apis mellifera L., in the United States. Am. Bee J. 144:557-559. [Google Scholar]
- 20.Chen, Y. P., J. A. Higgins, and M. F. Feldlaufer. 2005. Quantitative real-time reverse transcription-PCR analysis of deformed wing virus infection in the honeybee (Apis mellifera L.). Appl. Environ. Microbiol. 71:436-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi, H., T. Sasaki, T. Tomita, M. Kobayashi, and S. Kawase. 1992. Processing of structural polypeptides of infectious flacherie virus of silkworm, Bombyx mori: VP1 and VP4 are derived from VP0. J. Invertebr. Pathol. 60:113-116. [DOI] [PubMed] [Google Scholar]
- 22.Curry, S., E. Fry, W. Blakemore, R. Abu-Ghazaleh, T. Jackson, A. King, S. Lea, J. Newman, and D. Stuart. 1997. Dissecting the roles of Vp0 cleavage and RNA packaging in picornavirus capsid stabilization: the structure of empty capsids of foot and mouth disease virus. J. Virol. 71:9743-9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daly, H. V., D. DeJong, and N. D. Stone. 1988. Effect of parasitism by Varroa jacobsoni on morphometrics of Africanized worker honey-bees. J. Apic. Res. 27:126-130. [Google Scholar]
- 24.de Miranda, J. R., M. Shen, and S. M. Camazine. 2002. Molecular characterization of deformed wing virus, p. 265-270. In E. H. Erickson, R. E. Page, and A. A. Hanna (ed.), Proceedings of the 2nd International Conference on Africanized Honey Bees and Bee Mites. A. I. Root, Medina, Ohio.
- 25.de Miranda, J. R., M. Drebot, S. Tyler, M. Shen, C. E. Cameron, D. B. Stoltz, and S. M. Camazine. 2004. Complete nucleotide sequence of Kashmir bee virus and comparison with acute bee paralysis virus. J. Gen. Virol. 85:2263-2270. [DOI] [PubMed] [Google Scholar]
- 26.Denholm, C. H. 1999. Inducible honey bee viruses associated with Varroa jacobsoni. Ph.D. thesis. Keele University, Staffordshire, England.
- 27.Du, Z. L., and Z. B. Zhang. 1985. Ultrastructural change in the hypopharyngeal glands of worker honey bees (Apis cerana) infected with sacbrood virus. Zool. Res. 6:155-162. [Google Scholar]
- 28.Fujiyuki, T., H. Takeuchi, M. Ono, S. Ohka, T. Sasaki, A. Nomoto, and T. Kubo. 2004. Novel insect picorna-like virus identified in the brains of aggressive worker honeybees. J. Virol. 78:1093-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Genersch, E., C. Yue, I. Fries, and J. R. de Miranda. 2006. Detection of deformed wing virus (DWV), a honey bee viral pathogen, in bumble bees (Bombus terrestris and Bombus pascuorum) with wing deformities. J. Invertebr. Pathol. 91:61-63. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh, R. C., B. V. Ball, M. M. Willcocks, and M. J. Carter. 1999. The nucleotide sequence of sacbrood virus of honey bee: an insect picorna-like virus. J. Gen. Virol. 80:1541-1549. [DOI] [PubMed] [Google Scholar]
- 31.Glaser, W., R. Cencic, and T. Skern. 2001. Foot-and-mouth disease virus leader proteinase: involvement of C-terminal residues in self-processing and cleavage of eIF4GI. J. Biol. Chem. 276:35473-35481. [DOI] [PubMed] [Google Scholar]
- 32.Gorbalenya, A. E., A. P. Donchenko, V. M. Blinov, and E. V. Koonin. 1989. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases: a distinct protein superfamily with a common structural fold. FEBS Lett. 243:103-114. [DOI] [PubMed] [Google Scholar]
- 33.Gorbalenya, A. E., and E. V. Koonin. 1989. Virus proteins containing the purine nucleotide-binding proteins. Nucleic Acids Res. 17:8413-8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Govan, V. A., N. Leat, M. Allsopp, and S. Davison. 2000. Analysis of the complete genome sequence of acute bee paralysis virus shows that it belongs to novel group of insect-infecting RNA viruses. Virology 277:457-463. [DOI] [PubMed] [Google Scholar]
- 36.Grabensteiner, E., W. Ritter, M. J. Carter, S. Davison, H. Pechhacker, J. Kolodziejek, O. Boecking, I. Derakhshifar, R. Moosbeckhofer, E. Licek, and N. Nowotny. 2001. Sacbrood virus of the honeybee (Apis mellifera): rapid identification and phylogenetic analysis using reverse transcription-PCR. Clin. Diagn. Lab. Immunol. 8:93-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gromeier, M., E. Wimmer, and A. E. Gorbalenya. 1999. Genetics, pathogenesis and evolution of picornaviruses, p. 287-343. In E. Domingo, R. G. Webster, and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, London, England.
- 38.Guarné, A., J. Tormo, R. Kirchweger, D. Pfistermueller, I. Fita, and T. Skern. 1998. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 17:7469-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harlow, E., and D. P. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Hinton, T. M., N. Ross-Smith, S. Warner, G. J. Belsham, and B. S. Crabb. 2002. Conservation of L and 3C protease activities across distantly related aphthoviruses. J. Gen. Virol. 83:3111-3121. [DOI] [PubMed] [Google Scholar]
- 41.Isawa, H., S. Asano, K. Sahara, T. Iizuka, and H. Bando. 1998. Analysis of genetic information of an insect picorna-like virus, infectious flacherie virus of silkworm: evidence for evolutionary relationships among insect, mammalian and plant picorna(-like) viruses. Arch. Virol. 143:127-143. [DOI] [PubMed] [Google Scholar]
- 42.Koch, W., and W. Ritter. 1991. Experimental examinations concerning the problem of deformed emerging bees after infestation with Varroa jacobsoni. J. Vet. Med. 38:337-344. [DOI] [PubMed] [Google Scholar]
- 43.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand viruses: implication of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 44.Kovac, H., and K. Crailsheim. 1988. Lifespan of Apis mellifera Carnica Pollm. infested by Varroa jacobsoni in relation to season and extent of infestation. J. Apic. Res. 27:230-238. [Google Scholar]
- 45.Lang, A. S., A. I. Culley, and C. A. Suttle. 2004. Genome sequence and characterization of a virus (HaRNAV), which is related to picorna-like viruses that infect the marine toxic bloom-forming alga, Heterosigma akashiwo. Virology 320:206-217. [DOI] [PubMed] [Google Scholar]
- 46.Leat, N., B. Ball, V. Govan, and S. Davison. 2000. Analysis of the complete genome sequence of black queen cell virus, a picorna-like virus of honey bees. J. Gen. Virol. 81:2111-2119. [DOI] [PubMed] [Google Scholar]
- 47.LeGendre, N., M. Mansfield, A. Weiss, and P. Matsudaira. 1993. Purification of proteins and peptides by SDS-PAGE, p. 71-101. In P. Matsudaira (ed.), A practical guide to protein and peptide purification for microsequencing, 2nd ed. Academic Press, San Diego, Calif.
- 48.Liljas, L., J. Tate, T. Lin, P. Christian, and J. E. Johnson. 2002. Evolutionary and taxonomic implications of conserved structural motifs between picornaviruses and insect picorna-like viruses. Arch. Virol. 147:59-84. [DOI] [PubMed] [Google Scholar]
- 49.Marcangeli, J., L. Monetti, and N. Fernandez. 1992. Malformations produced by Varroa jacobsoni on Apis mellifera in the province of Buenos Aires, Argentina. Apidologie 23:399-402. [Google Scholar]
- 50.Martin, S. J. 2001. The role of Varroa and viral pathogens in the collapse of honeybee colonies: a modeling approach. J. Appl. Ecol. 38:1082-1093. [Google Scholar]
- 51.Mayo, M. A. 2002. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 147:1655-1663. [DOI] [PubMed] [Google Scholar]
- 52.Moore, N. F., and S. M. Eley. 1991. Picornaviridae: picornaviruses of invertebrates, p. 371-386. In J. R. Adams and J. R. Bonami (ed.), Atlas of invertebrate viruses. CRC Press, Boca Raton, Fla.
- 53.Moore, N. F., B. Reavy, and L. A. King. 1985. General characteristics, gene organization and expression of small RNA viruses of insects. J. Gen. Virol. 66:647-659. [Google Scholar]
- 54.Nordström, S. 2000. Virus infections and varroa mite infestations in honey bee colonies. Ph.D. thesis. Swedish University of Agricultural Sciences, Uppsala, Sweden.
- 55.Nordström, S. 2003. Distribution of deformed wing virus within honey bee (Apis mellifera) brood cells infested with the ectoparasitic mite Varroa destructor. Exp. Appl. Acarol. 29:293-302. [DOI] [PubMed] [Google Scholar]
- 56.Nordström, S., I. Fries, A. Aarhus, H. Hansen, and S. Korpela. 1999. Virus infection in Nordic honeybee colonies with no, low or severe Varroa jacobsoni infestations. Apidologie 30:475-484. [Google Scholar]
- 57.Ongus, J. R., D. Peters, J.-M. Bonmatin, E. Bengsch, J. M. Vlak, and M. M. van Oers. 2004. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J. Gen. Virol. 85:3747-3755. [DOI] [PubMed] [Google Scholar]
- 58.Palmenberg, A. C. 1990. Proteolytic processing of picornaviral polyprotein. Annu. Rev. Microbiol. 44:603-623. [DOI] [PubMed] [Google Scholar]
- 59.Palmenberg, A. C., E. M. Kirby, M. R. Tanda, N. L. Drake, G. M. Duke, K. F. Potratz, and M. S. Collett. 1984. The nucleotide and deduced amino acid sequence of the encephalomyocarditis viral polyprotein coding region. Nucleic Acids Res. 12:2969-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roossinck, M. J. 1997. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 35:191-209. [DOI] [PubMed] [Google Scholar]
- 61.Ryan, M. D., and M. Flint. 1997. Virus-encoded proteases of the picornavirus super-group. J. Gen. Virol. 78:699-723. [DOI] [PubMed] [Google Scholar]
- 62.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 63.Shimanuki, H., N. W. Calderone, and D. A. Knox. 1994. Parasitic mite syndrome: the symptoms. Am. Bee J. 134:827-828. [Google Scholar]
- 64.Sumpter, D. J. T., and S. J. Martin. 2004. The dynamics of virus epidemics in varroa-infested honey bee colonies. J. Anim. Ecol. 73:51-63. [Google Scholar]
- 65.Swofford, D. L. 1998. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), version 4.0. Sinauer Associates, Sunderland, Mass.
- 66.Tentcheva, D., L. Gauthier, N. Zappulla, B. Dainat, F. Cousserans, M. E. Colin, and M. Bergoin. 2004. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl. Environ. Microbiol. 70:7185-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tentcheva, D., L. Gauthier, S. Jouve, L. Canabady-Rochelle, B. Dainat, F. Cousserans, M. E. Colin, B. V. Ball, and M. Bergoin. 2004. Polymerase chain reaction detection of deformed wing virus (DWV) in Apis mellifera and Varroa destructor. Apidologie 35:431-440. [Google Scholar]
- 68.Tentcheva, D., L. Gauthier, L. Bagny, J. Fievet, B. Dainat, F. Cousserans, M. E. Colin, and M. Bergoin. 2006. Comparative analysis of deformed wing virus (DWV) RNA in Apis mellifera L. and Varroa destructor. Apidologie 37:1-7. [Google Scholar]
- 69.Thomson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weigh matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, et al. 2000. Virus taxonomy: the classification and nomenclature of viruses. The seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 71.Wang, X., J. Zhang, J. Lu, F. Yi, C. Liu, and Y. Hu. 2004. Sequence analysis and genomic organization of a new insect picorna-like virus, Ectropis obliqua picorna-like virus, isolated from Ectopis obliqua. J. Gen. Virol. 85:1145-1151. [DOI] [PubMed] [Google Scholar]
- 72.Weinberg, K. P., and G. Madel. 1985. The influence of the mite Varroa jacobsoni Oud. on the protein concentration and the haemolymph volume of the blood of the worker bees and drones of the honey bee Apis mellifera. Apidologie 16:421-436. [Google Scholar]
- 73.Weitz, M., B. M. Baroudy, W. L. Maloy, J. R. Ticehurst, and R. H. Purcell. 1986. Detection of a genome-linked protein (VPg) of hepatitis A virus and its comparison with other picornaviral VPgs. J. Virol. 60:124-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson, J. E., M. J. Powell, S. E. Hoover, and P. Sarnow. 2000. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 20:4990-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu, C. Y., C. F. Lo, C. J. Huang, H. T. Yu, and C. H. Wang. 2002. The complete genome sequence of Perina nuda picorna-like virus, an insect-infecting RNA virus with a genome organization similar to that of mammalian picornavirus. Virology 294:312-323. [DOI] [PubMed] [Google Scholar]
- 76.Yang, X., and D. L. Cox-Foster. 2005. Impact of an ectoparasite on the immunity and pathology of an invertebrate: evidence for host immunosuppression and viral amplification. Proc. Natl. Acad. Sci. USA 102:7470-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yue, C., and E. Genersch. 2005. RT-PCR analysis of deformed wing virus (DWV) in honey bees (Apis mellifera) and mites (Varroa destructor). J. Gen. Virol. 86:3419-3425. [DOI] [PubMed] [Google Scholar]