Abstract
The CD81 tetraspanin was first identified as a hepatitis C virus (HCV) receptor by its ability to bind the soluble ectodomain of envelope glycoprotein E2 (sE2). More recently, it has been suggested that CD81 is necessary but not sufficient for HCV entry into target cells. Here we present further evidence that putative human hepatocyte-specific factors act in concert with CD81 to mediate sE2 binding and HCV pseudoparticle (HCVpp) entry. Moreover, we show that CD81-mediated HCVpp entry entails E2 binding to residues in the large extracellular loop as well as molecular events mediated by the transmembrane and intracellular domains of CD81. The concept that CD81 receptor function progresses in stages is further supported by our finding that anti-CD81 monoclonal antibodies inhibit HCVpp entry by different mechanisms. The half-life of CD81-HCVpp binding was determined to be approximately 17 min, and we propose that binding is followed by CD81 oligomerization, partitioning into cholesterol-rich membrane domains, or other, lateral protein-protein interactions. This results in the formation of a receptor-virus complex that undergoes endocytosis and pH-dependent membrane fusion.
The infectious hepatitis C virus (HCV) particle is enveloped by a lipid membrane comprising heterodimers of E1 and E2 envelope glycoproteins (E1E2) (34). Hepatocytes are the major target cells of HCV (6, 23, 39), and recent evidence indicates that the tissue tropism of this virus is specifically restricted by E1E2 (24, 42). The exact functions of E1 and E2 remain to be determined, but there is now ample evidence demonstrating that E2 is the receptor-binding subunit of the HCV envelope. The CD81 tetraspanin was first identified as a putative HCV receptor by its ability to bind specifically and with a high affinity to the soluble ectodomain of E2 (sE2) (33). Even though this transmembrane domain (TM domain)-truncated protein is expressed in the absence of E1, it appears to accurately mimic certain structural and functional properties of native E2 and has been a useful tool for studying E2-CD81 interactions (15, 18, 36).
With the advent of HCV pseudoparticles (HCVpp) and replicating molecular clones, a significant body of data has been generated indicating that CD81 is necessary but not sufficient for viral entry into target cells (2, 13, 26, 43, 45, 47). Firstly, all nonhepatic, CD81-positive human cells tested to date are resistant to HCVpp (25). Moreover, most available human hepatoma cell lines are CD81 positive, but only three are susceptible to HCVpp entry (2, 3, 13, 22). One CD81-negative human hepatoma cell line, however, becomes susceptible to HCVpp when modified to express CD81. Murine and hamster cells of nonhepatic origin engineered to express human CD81 remain resistant to HCVpp, and transgenic mice expressing human CD81 do not become susceptible to HCV infection (2, 13, 27, 45). Finally, we previously showed that HCVpp entry is inhibited by an anti-CD81 monoclonal antibody (MAb) at a step following HCV attachment to target cells (13). Taken together, these observations led us to propose that CD81 functions as an entry coreceptor and that one or more human hepatocyte-specific molecules act as the primary attachment receptor that accounts for the restricted tropism of HCV. It is notable that CD81 internalization is extremely slow (32, 40) and therefore incompatible with the kinetics of viral entry (T. Dragic, unpublished results). HCV binding to CD81 as well as another receptor or CD81 interactions with other membrane proteins might therefore be necessary to shunt the HCV-receptor complex into a more efficient endocytic pathway.
The tetraspanin web, which is associated with CD81, is an amalgam of different tetraspanins, transmembrane proteins, and signaling enzymes that partition into detergent-resistant, cholesterol-rich regions of the plasma membrane (25). Different domains of CD81 appear to interact with different components of the tetraspanin web: the intracellular (ICL) domain of CD81 associates with signaling enzymes (5, 44, 46), palmitoylation of intracellular cysteine residues is essential for CD81 multimerization and partitioning into cholesterol-rich membrane domains (8-12), polar residues within the TM domain participate in inter- and intramolecular TM helix packing (38), and the large extracellular loop (LEL) plays a role in CD81 multimerization and the assembly of multiprotein complexes within the tetraspanin web (20, 38, 46). Moreover, the LEL is the binding site for HCV E2 (14, 16-18, 21, 28, 31-33, 35, 45). To date, the role of the CD81 transmembrane and intracellular domains in HCV entry has not been explored.
In this study, we investigated the determinants of CD81 receptor function. Our analyses indicate that sE2 binding correlates with CD81 expression only on cells that are not permissive to HCVpp entry and therefore do not express the putative entry receptor. Moreover, sE2 binding to these cells was almost completely inhibited by an anti-CD81 MAb, whereas complete inhibition of binding to permissive cells could not be achieved. Structural determinants of CD81 receptor function were also investigated, and we found that residues in LEL that are important for sE2 binding are also important for HCVpp entry. Surprisingly, mutagenesis of several transmembrane and intracellular residues impaired HCVpp entry while slightly increasing sE2 binding. Finally, we investigated two commercially available MAbs for their binding to CD81 and inhibition of HCVpp entry. Whereas the difference in half-maximal binding concentrations (EC50s) between the two MAbs was 70-fold, the difference in their half-maximal inhibitory concentrations (IC50s) was 260-fold. This suggests that anti-CD81 MAbs inhibit HCVpp entry through different mechanisms. Experiments to determine the time course of inhibition of entry showed that the CD81-HCVpp binding step of entry occurs with a half-life (t1/2) of ∼17 min. Together, our results lead us to postulate that E2 binding to the LEL is followed by recruitment and oligomerization of CD81 or other cell surface molecules, which lead to internalization of HCV particles and subsequent stages of viral entry.
MATERIALS AND METHODS
Cells and antibodies.
Human NKNT3 hepatoma cells were provided by I. Fox (University of Nebraska Medical Center, Omaha), human Huh-7 hepatoma cells were provided by R. Chowdhurry (Albert Einstein College of Medicine, Bronx, NY), and murine Hepa hepatoma cells were provided by D. Neufeld (Albert Einstein College of Medicine, Bronx, NY). All other cell lines used in this study were obtained from the American Type Culture Collection. Cells were cultured under standard conditions. CD81-positive derivatives of HepG2 and 3T3 cells were generated and cultured as previously described (13). CD81-positive U87, U937, and Hepa cells were generated by transduction with the pQCXIP retroviral vector (Clontech) designed to express the human CD81 gene along with the puromycin selection marker. CD81-positive clones were selected in puromycin (2 μg/ml; Sigma), and the expression of CD81 was confirmed by flow cytometry after labeling with anti-CD81 MAbs. JS81 and fluorescein isothiocyanate-conjugated JS81 were purchased from Pharmingen, 1.3.3.22 was purchased from Chemicon, O.N.165 was purchased from U.S. Biologicals, and 1D6 was purchased from Serotec. The phycoerythrin (PE)-conjugated hamster anti-mouse CD81 MAb Eat2 was purchased from Becton Dickinson Pharmingen. The anti-E2 MAb H53 was kindly provided by J. Dubuisson (Institut Pasteur, Lille, France). A nonspecific, isotype-matched murine immunoglobulin G (IgG) from Pharmingen was used as a negative control. A PE-conjugated goat anti-mouse antibody (Caltag Laboratories) was used for flow cytometry analyses.
Binding of anti-CD81 MAbs to cell lines.
Cells were detached with 2 mM EDTA in phosphate-buffered saline (PBS), resuspended, and washed once in Dulbecco's medium-PBS supplemented with 1% bovine serum albumin and 0.05% sodium azide. Cells (2 × 105) were incubated for 1 h at 4°C with serially diluted anti-CD81 MAbs, washed twice, and labeled with PE-conjugated goat anti-mouse IgG. After fixation with 1% formaldehyde, MAb binding was quantified by flow cytometry (mean fluorescence intensity [MFI]), using a FACSCalibur instrument (Becton Dickinson).
sE2 binding and inhibition.
Cells (2 × 105) were incubated with 2 μg/ml of sE2 (Austral Biologicals) for 1 h at room temperature in Dulbecco's medium-PBS supplemented with 1% bovine serum albumin and 0.05% sodium azide and were then washed twice with PBS. Soluble E2 binding to cells was detected by flow cytometry after labeling with the anti-E2 MAb H53 (1:100) followed by a PE-conjugated goat anti-mouse IgG (1:100). For inhibition of sE2 binding, cells were incubated for 1 h at 4°C with a nonspecific murine IgG (5 μg/ml), JS81 (5 μg/ml), or 1D6 (100 μg/ml), followed by incubation with soluble E2 (2 μg/ml). Binding of soluble E2 was detected with H53-coated FluoSpheres (10 beads/cell) and measured by flow cytometry. The beads were generated by coupling 15 μl of NeutrAvidin-labeled FluoSpheres (505/515 nm, 1.0 μm; Molecular Probes) to 3 μg of biotin-SP-conjugated goat anti-mouse F(ab′)2 (Jackson Immunoresearch) for 2 h at 37°C. Complexes were washed twice and incubated overnight at 4°C with H53 (1:100).
Production of HCVpp and entry into target cells.
293T cells (5 × 106) were collected in Dulbecco's modified Eagle's medium without antibiotics and transfected with 36 μl of Lipofectamine 2000 (Invitrogen) mixed with 4 μl of NLluc+ Env− reporter vector and 8 μl of E1E2 expression vector according to the manufacturer's instructions. Cell culture supernatants were collected at 48 h posttransfection and clarified with 0.45-μm filters (Millipore). Target cells (4 × 104) were infected with 250 μl of supernatant containing HCVpp, and luciferase activity (in relative light units [RLU]) was measured in cell lysates 48 h after infection, using a luciferase assay system (Promega). For inhibition-of-entry studies, anti-CD81 MAbs JS81, 1.3.3.22, O.N.165, and 1D6 were serially diluted, starting with 2 μg/ml for JS81, 1.3.3.22, and O.N.165 and with 160 μg/ml for 1D6. MAbs were added to target cells immediately prior to infection with HCVpp. For experiments to determine the time course of inhibition, cold HCVpp were added to cells and prebound by spinning for 1 h at 2,000 rpm at 4°C. Cells were washed twice with cold PBS, and warm medium containing MAbs was added at different time points from 0 to 3 h. After the addition of MAbs, cells were kept at 37°C for the duration of the experiment.
CD81 mutagenesis.
The human and murine CD81 proteins were subcloned into the pcDNA3.1 vector (Invitrogen) using BamHI and NotI restriction enzyme sites. Human CD81-encoding DNA was a kind gift of S. Levy (Stanford University, Calif.). Murine CD81 was amplified from NIH 3T3 mRNA extracts using the primers 5′GTTCGGTTGCTGGATCCATGGGGGTGGAGGGCTGCACCAAATGC3′ and 5′TTCTGAGCATGGTGCTGTGCTGTGGCATCCGGAACAGCTCCGTGTACACCCCTACGACGTGCCCGACTACGCCTGAG CGGCCGCAGCAACCGAAC3′. Mutagenesis of LEL, transmembrane, and intracellular human CD81 residues was performed with a QuickChange kit from Stratagene according to the manufacturer's instructions. The primers used to delete the N terminus were 5′-GGGCTGGATCCATGGGCAGCGCCGGCAGCGCCATCAAGTACCTGCTCTTCGTCTTCAATTTCGTC3′ and 5′GTACTATGACGTACCTGACTATGCTTCACTGTGAGCGGCCGCAGCCC3′, and those used to delete the C terminus were 5′GGGCTGGATCCATGGGAGTGGAGGGCTGCACCAAGTGCATCAAG3′ and 5′GAGATGATCCTGAGCATGTGCTGTGCTGTGGCGGCAGCGCCGGCAGCGCCTGAGCGGCCGC AGCCC3′. Nucleotide sequencing was performed to ascertain the presence of the appropriate mutations in the CD81 coding sequence.
HCVpp entry mediated by CD81 mutants.
CD81-negative HepG2 hepatoma cells (5 × 104) were transfected with the pcDNA3.1 vector expressing wild-type or mutant CD81, using Lipofectamine 2000 according to the manufacturer's instructions for this cell line. After 48 h, transfected HepG2 cells were infected with HCVpp and incubated for another 48 h before measurements of luciferase activity. CD81 mutant expression levels were quantified by flow cytometry analyses (results are reported as percent wild-type CD81 expression) after labeling of cells with JS81-fluorescein isothiocyanate (1:100) or 1D6 (10 μg/ml) and PE-conjugated goat anti-mouse IgG (1:100). Mouse CD81 was detected after labeling of cells with PE-conjugated Eat2. To calculate HCVpp entry mediated by CD81 mutants, the following formula was used: (RLUmutant/MFImutant)/(RLUwild type/MFIwild type) × 100.
RESULTS
CD81 expression levels do not correlate with sE2 binding in HCV-permissive cell lines.
We and others have previously shown that there is no correlation between CD81 expression and HCVpp entry (3, 13, 22). This is true even for human hepatoma cell lines. For example, Huh-7 and Hep3B cells, which are permissive for HCVpp entry, express less CD81 than entry-resistant H1H and NKNT3 cells (Fig. 1A). CD81-negative HepG2 hepatoma cells, however, become permissive for HCVpp when engineered to express this receptor, but the level of entry is an order of magnitude lower than that observed in Huh-7 or Hep3B cells (2, 13). Moreover, all nonhepatic CD81-positive human cell lines tested to date are resistant to HCVpp entry (2, 13, 22). We have now identified two CD81-negative, nonhepatic human cell lines. U87 astroglioma cells and U937 promonocytic cells remain resistant to HCVpp entry when modified to express CD81 (Fig. 1A). Similarly, murine 3T3 fibroblasts and Hepa hepatoma cells remain resistant to HCVpp entry when engineered to express human CD81 (Fig. 1A). These results reinforce previous conclusions that a human hepatocyte-specific factor is required for HCVpp entry into target cells.
FIG. 1.
Expression of CD81 on the cell surface and sE2 binding. (A) CD81 expression was determined with different cell lines, including human hepatoma cells permissive to HCVpp (checkered bars), human cells nonpermissive to HCVpp (solid bars), and murine cells (hatched bars), as indicated along the x axis. CD81 was quantified by flow cytometry (MFI) after labeling with an anti-CD81 MAb (JS81). Results are means of three independent experiments ± standard deviations (SD). Permissivity (+, 104 to 105 RLU; ++, 105 to 106 RLU) or resistance (−) of cells to HCVpp is indicated under the bar graph. (B) sE2 binding to cells from panel A, as indicated along the x axis, was quantified by flow cytometry (MFI) after labeling of cells with the anti-E2 MAb H53 followed by a PE-conjugated anti-mouse IgG antibody. Results are means of three independent experiments ± SD. (C) Anti-CD81 MAbs JS81 (black bars) and 1D6 (hatched bars) were used to inhibit sE2 binding to a subset of cells from panel A. Residual sE2 on cells was quantified by flow cytometry (MFI) after labeling with H53-coupled fluorescent beads. The percentage of inhibition of sE2 binding is indicated for each cell line and was calculated relative to sE2 binding in the presence of a nonspecific isotype-matched murine IgG. Values are means of three independent experiments ± standard deviations.
We also sought to determine whether there is a correlation between CD81 expression levels and sE2 binding to cells. These two parameters were quantified by flow cytometry with human and nonhuman cell lines of different origins. No sE2 binding was observed with CD81-negative, HCVpp entry-resistant cells, including human U87 and U937 cells as well as murine 3T3 and Hepa cells (Fig. 1B). However, sE2 binding was readily detected with the CD81-positive derivatives of these cell lines as well as the naturally CD81-positive human HeLa, 293T, H1H, and NKNT3 cells (Fig. 1B). Overall, there was a good correlation between CD81 expression levels and sE2 binding to these nonpermissive cells. This, however, did not hold true for HCVpp-permissive cells: sE2 binding to Huh-7 cells was threefold higher than binding to Hep3B cells, even though the two cell types express similar levels of CD81 (Fig. 1B). Moreover, CD81-negative HepG2 cells bound sE2, and this was significantly increased by CD81 expression.
To further confirm that sE2 binding to permissive cells is mediated by CD81 as well as other factors, we investigated the inhibition of sE2 binding by anti-CD81 MAbs JS81 and 1D6, which inhibit HCVpp entry (13). sE2 binding to CD81-positive, non-HCVpp-permissive cells, such as U87-CD81 or 293T cells, was inhibited approximately 80% by JS81 (Fig. 1C). In contrast, binding of sE2 to permissive cells, such as Huh-7 and HepG2-CD81 cells, was inhibited approximately 50% (Fig. 1C). Finally, binding to HepG2 cells was not inhibited by JS81 (<10%). The differences in binding inhibition between the three sets of cells were statistically significant (P ≤ 0.02; P values depended on the cell lines compared in a two-tailed t test), but those between cells belonging to the same group were not (P ≥ 0.4). Similar results were observed with the MAb 1D6, using Huh-7 and 293T cells (Fig. 1C). We therefore concluded that E2 binding to nonpermissive cells is mediated solely by CD81, whereas binding to permissive cells is mediated by CD81 and another molecule.
Residues in LEL are required for HCVpp entry and sE2 binding to target cells.
The species specificity of viral infection is often determined by envelope glycoprotein interactions with cellular receptors. Since CD81 proteins of several uninfectable species nonetheless bind sE2, it is unlikely that CD81 is the determinant of HCV species specificity. Even so, sequence comparisons of different CD81 proteins have provided insights into the structural requirements for HCV receptor function. Murine CD81 was previously shown not to bind sE2 (1), and here we demonstrate that it also does not mediate HCVpp entry when transfected into HepG2 cells (see Fig. 3A). The murine and human CD81 proteins are homologous except for 3 residues in the small extracellular loop and 17 residues in the LEL (Fig. 2). Since the LEL is the sE2 binding component of CD81, we generated mutants with substitutions in human CD81 at all 17 positions that differ in murine CD81. We also generated mutants with substitutions at two positions that differ in rat CD81, which also does not mediate sE2 binding (Fig. 2) (16). Mutant CD81 proteins were characterized for expression as well as the ability to mediate HCVpp entry and sE2 binding. Notably, all of the mutants were expressed at levels similar to that of the wild-type protein (Fig. 3A).
FIG. 3.
Role of specific CD81 LEL residues in HCVpp entry and sE2 binding. (A) HCVpp entry was tested in HepG2 cells transfected with pcDNA3.1, human wild-type CD81, murine wild-type CD81 (all in white bars), and human CD81 mutants, as indicated along the x axis. Human residues were replaced by alanine (black bars) or by corresponding murine (shaded bars) or rat (hatched bars) CD81 residues. HCVpp entry was calculated as a percentage of the entry level into cells expressing human wild-type CD81. The expression of CD81 mutants was quantified by flow cytometry after labeling of cells with JS81 (or 1D6 for the A164T mutant) and was expressed as a percentage of human wild-type CD81 expression. The percentage of HCVpp entry was further normalized for CD81 expression. Values are means of three independent experiments ± SD. (B) Binding of sE2 to CD81 mutants with significantly decreased HCVpp entry was quantified by flow cytometry. HepG2 cells were transfected with pcDNA3.1, human wild-type CD81, murine CD81, and CD81 mutants. Transfected cells were incubated with sE2, followed by labeling with H53 and a PE-conjugated anti-mouse IgG. Binding was analyzed by flow cytometry and expressed as a percentage of sE2 binding to HepG2 cells expressing human wild-type CD81. The percentage of sE2 binding was further normalized for mutant expression levels. Values are averages of three independent experiments ± SD.
FIG. 2.
CD81 sequence and structure. (A) Comparison of human, mouse, and rat CD81 LEL sequences. Residues differing between human and mouse CD81 proteins are shaded in gray. Most of these residues are also different in rat CD81 and are therefore not indicated. The two additional residues that differ in rat CD81 are shaded in black. (B) Predicted two-dimensional structure of CD81, including the two extracellular loops, four transmembrane helices, and the intracellular loop along with the intracellular N-terminal and C-terminal tails. LEL residues that were mutagenized in this study are shown using the same color code as that in panel A. Polar transmembrane residues and intracellular cysteines that were mutagenized are indicated in white circles and squares, respectively.
We determined that alanine substitutions at residues K171, I182, and F186 decreased HCVpp entry >50% compared to the wild type (P ≤ 0.0006; the P value depended on the mutant tested and was determined by a two-tailed t test), with the greatest effect being exerted by the F-to-A substitution at position 186 (Fig. 3A). The I181A mutation decreased entry by approximately 30% (P = 0.002). Substitutions of these four residues also significantly decreased sE2 binding to CD81 compared to the wild-type receptor (P ≤ 0.0006) (Fig. 3B). Surprisingly, the K171A mutant bound sE2 as poorly as the F186A mutant, even though it was more efficient at mediating HCVpp entry. We also substituted human residues for corresponding murine or rat CD81 residues at these four critical positions. The K171R, I181S, I182L, and F186L mutants were all more efficient than the alanine mutants at mediating HCVpp entry and sE2 binding (Fig. 3A and B), indicating that conservation of the side chain charge and bulk at these positions is important for receptor activity. Finally, alanine substitutions and human-to-mouse and human-to-rat substitutions at all other positions, including positions 184, 188, and 196, which were reported by others to affect CD81-E2 binding, had no major effect on entry. Based on our observations, we concluded that at least four LEL residues are required for the E2-CD81 interaction and that effects on E2 binding always translate into effects on HCVpp entry.
CD81 transmembrane and intracellular residues are required for HCVpp entry but not sE2 binding.
CD81 homo- and heterooligomerization as well as its association with cholesterol contribute to the assembly of multiprotein complexes in detergent-resistant membrane domains. Different components of CD81 are involved in these interactions (Fig. 2B). Palmitoylation of intracellular cysteine residues is essential for multimerization as well as for partitioning into cholesterol-rich domains (8, 10, 12). Highly conserved polar residues within the TM domain participate in inter- and intramolecular TM helix packing (38). The intracellular domain of CD81 associates with the signaling enzymes protein kinase C (PKC) and phosphatidylinositol 4-kinase (5, 44, 46). We therefore mutated residues within these domains in order to test their roles in HCVpp entry and sE2 binding (Fig. 2B). Notably, all of the mutants were expressed at levels similar to that of the wild-type protein (Fig. 4A).
FIG. 4.
Role of specific CD81 transmembrane and intracytoplasmic residues in HCVpp entry and sE2 binding. (A) HCVpp entry was tested in HepG2 cells transfected with pcDNA3.1 (white bars), human wild-type CD81 (white bars), or CD81 mutants (black bars), as indicated along the x axis. HCVpp entry was calculated as a percentage of the entry level into cells expressing human wild-type CD81. The expression of CD81 mutants was quantified by flow cytometry after labeling of cells with JS81 and was expressed as a percentage of human wild-type CD81 expression. The percentage of HCVpp entry was further normalized for CD81 expression. Values are means of three independent experiments ± SD. (B) Binding of sE2 to CD81 mutants with significantly decreased HCVpp entry was quantified by flow cytometry. HepG2 cells were transfected with pcDNA3.1, human wild-type CD81, and CD81 mutants. Transfected cells were incubated with sE2, followed by labeling with H53 and PE-conjugated anti-mouse IgG. Binding was analyzed by flow cytometry and expressed as a percentage of E2 binding to HepG2 cells expressing human wild-type CD81. The percentage of sE2 binding was further normalized for mutant expression levels. Values are means of three independent experiments ± SD.
Deleting the N terminus of CD81 had no impact on viral entry, whereas deletion of the C terminus decreased entry 30% compared to that of the wild-type receptor (P = 0.01, as determined by a two-tailed t test) (Fig. 4A). A similar decrease in HCVpp entry was observed when all five cysteine residues in the intracellular domains of CD81 were mutated to alanine in order to abrogate receptor palmitoylation (P = 0.0004) (Fig. 4A). Within the transmembrane domain, the C80A and N18A/E219A substitutions decreased entry by approximately 40% (P ≤ 0.0001) (Fig. 4A). Alanine substitutions for C97, C104, N18, E105, and E219, however, had no significant effects on viral entry. Surprisingly, all of the TM and ICL domain mutants with decreased HCVpp entry exhibited increased sE2 binding compared to wild-type CD81 (P ≤ 0.0007; the P value depended on the mutant examined) (Fig. 4B). We therefore propose that the observed decrease in HCVpp entry is due to indirect mechanisms, such as the disruption of CD81 oligomerization or interactions with other proteins, and not to a direct disruption of CD81-E2 binding.
Anti-CD81 MAbs inhibit HCVpp entry by different mechanisms.
Four commercially available anti-CD81 MAbs were compared for the abilities to bind cell surface-associated CD81 and to inhibit HCVpp entry. Three of the four MAbs, JS81, O.N.165, and 1.3.3.22, exhibited very similar binding profiles for Huh-7 cells, with half-maximal binding concentrations (EC50s) around 0.21 μg/ml (Fig. 5A and C; data not shown). Moreover, these MAbs exhibited similar binding patterns for several other human cell lines, suggesting the absence of cell type-specific CD81 isoforms and conformations (data not shown). MAb 1D6, however, exhibited significantly less binding to Huh-7 cells (Fig. 5A and C), with an EC50 of 13.9 μg/ml.
FIG. 5.
Binding to cells and inhibition of HCVpp entry by anti-CD81 MAbs. (A) Different concentrations of MAbs JS81 (squares) and 1D6 (triangles) ranging over 2 orders of magnitude were added to Huh-7 cells, and binding was quantified by flow cytometry (MFI) after cell labeling with a PE-conjugated anti-mouse IgG. Binding is expressed as a percentage of the MFI at the highest, saturating concentration of MAb, beyond which no further effect was observed. Values are means of three independent experiments ± SD. (B) Different concentrations of MAbs JS81 (squares) and 1D6 (triangles) were added to Huh-7 cells, which were then infected with HCVpp. Luciferase activity was measured in cell lysates 48 h after infection. The percentage of HCVpp entry was calculated relative to entry in the absence of anti-CD81 MAbs. A 100% inhibition of HCVpp entry was set at the MAb concentration beyond which no further effect was observed. Values are means of three independent experiments ± SD. (C) Calculated EC50 and IC50 values for JS81 and 1D6 after nonlinear curve fitting using GraphPad Prism 4 software.
When we tested the ability of the different anti-CD81 MAbs to inhibit HCVpp entry, we found that the EC50 and the half-maximal inhibitory concentration (IC50) were nearly identical for MAb 1D6 (Fig. 5C). Surprisingly, for JS81 and similar MAbs, the EC50 was fourfold higher than the IC50 (0.048 μg/ml). This means that JS81 need only occupy one of every four CD81 molecules in order to completely block HCVpp entry. In contrast, 1D6 must occupy all of the available receptor sites in order to completely inhibit viral entry. Our observations suggest that there is more than one mechanism of inhibiting CD81-mediated HCVpp entry. We postulate that 1D6 inhibits E2-CD81 binding, whereas JS81 may inhibit postbinding steps such as receptor oligomerization (as well as inhibiting E2-CD81 binding, as shown in Fig. 1C).
Time course of CD81-mediated stages of entry.
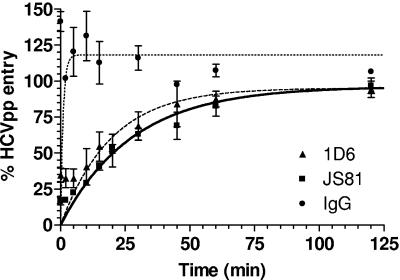
In order to determine the duration of CD81-mediated HCVpp entry, we performed experiments to examine the time course of inhibition. After prebinding of HCVpp to cells at 4°C, synchronized viral entry was allowed to proceed at 37°C. Anti-CD81 MAbs were added at different time points postattachment at concentrations of >EC90. Note that under these assay conditions, wherein 100% of CD81 was occupied by the MAbs, we were only measuring the time course of E2 binding inhibition. The duration of subsequent CD81-mediated stages of HCVpp entry cannot be measured by this assay. The half-maximal times (t1/2) required for 1D6 and JS81 to exert their inhibitory effects were found to be 14 and 19 min, respectively (Fig. 6). The differences between the two time courses were not statistically significant (P ≤ 0.1; the P value depended on the time point and was determined by a two-tailed t test), and we therefore concluded that the t1/2 of CD81-mediated HCVpp binding is approximately 17 min.
FIG. 6.
Time course of inhibition of HCVpp entry. Anti-CD81 MAbs were added to cells that were prebound to viral particles by centrifugation at 4°C. Luciferase activities were measured at 48 h postinfection and expressed relative to entry in the absence of antibody. Values are means of three independent experiments ± SD.
DISCUSSION
In this study, several aspects of CD81 receptor function were investigated. We initially sought to determine whether there is a correlation between sE2 binding and CD81 cell surface expression levels. sE2 binding was directly proportional to CD81 expression only for cell lines that are not permissive to HCVpp entry. Moreover, sE2 binding to nonpermissive cells could be inhibited by anti-CD81 MAbs, indicating that binding was entirely dependent on CD81. In contrast, cell lines that are permissive to HCVpp entry did not bind sE2 in proportion to their CD81 expression levels, and sE2 binding to these cells could not be inhibited >50% with anti-CD81 MAbs. Notably, the level of HCVpp entry into permissive cells was also not proportional to CD81 expression. Our observations further confirm the existence of accessory cell surface molecules that bind sE2 and participate in HCV internalization and entry. These are presumably the molecules that mediate sE2 binding to HepG2 cells in the absence of CD81 and determine the absolute levels of HCVpp entry. Interestingly, several reports have shown that HCVpp, virions, and virus-like particles do not bind to CD81, suggesting that there are differences between membrane-associated and soluble E2 (30, 41). Native, heterodimer-associated E2 might require an interaction with the primary receptor to enable its binding to CD81, whereas soluble E2 may adopt a conformation already primed for CD81 binding.
Before the advent of experimental models of HCV entry and replication, numerous studies relied on sE2 to probe viral interactions with CD81. CD81 proteins of other species variably bind HCV sE2. For example, tamarin and chimpanzee CD81 proteins bind sE2 as well as human CD81 does, but rat and mouse CD81 proteins do not bind HCV sE2 (1, 16, 29). These comparative studies identified the LEL as the major E2 binding site on CD81. In particular, it was shown that residues L162, I182, N184, and F186 are required for sE2 binding to CD81 (14). More recent studies demonstrated that CD81 F186L, E188K, and D196E mutants are impaired for sE2 binding but, nonetheless, mediate wild-type levels of HCVpp entry (28, 45). Our mutagenesis data, based on sequence differences between human and mouse (as well as rat) CD81 proteins, confirm the importance of K171, I181, I182, and F186 in sE2 binding as well as HCVpp entry. However, in our experimental system, mutants with substitutions of residues N184, E188, and D196 all exhibited wild-type receptor phenotypes. We did not test the role of residue L162. Moreover, for LEL mutants, impairment of HCVpp entry always translated into impairment of sE2 binding. We therefore concluded that the LEL is the major E2 binding site and does not appear to play additional roles in HCV entry and CD81 receptor function.
We also investigated the possible role in HCV entry of CD81 transmembrane and intracellular residues that are involved in multimerization, intra- and intermolecular packing, and partitioning into cholesterol-rich membrane domains. Several of these mutants, including a C-terminal deletion mutant, a palmitoylation mutant (with all five intracellular C residues replaced with A residues), and polar transmembrane residue mutants, were impaired for entry but exhibited increased sE2 binding. The mechanism of entry impairment still remains to be determined, but we propose that CD81 multimerization and partitioning into cholesterol-rich membrane domains participate in HCV entry, probably by influencing receptor recruitment to the site of virus binding as well as internalization of the receptor-virus complex. Note that McKeating et al. (28) demonstrated that a CD9 chimera comprising only the LEL of CD81 mediates HCVpp entry, implying that whatever accessory functions may be required for entry are not specific for the transmembrane and intracellular domains of CD81. However, CD81 and CD9 are among the most closely related tetraspanins and are often found in association with the same proteins (4, 7, 19, 20, 37, 46). CD81 chimeras with other tetraspanins may therefore yield different results.
In attempting to further elucidate the receptor function of CD81, we also studied the binding and inhibitory properties of several commercially available anti-CD81 MAbs. We thus identified two types of antibodies with very different properties. For MAb 1D6, the EC50 for CD81 binding equals the IC50 for HCVpp entry inhibition. In other words, there is a one-to-one molar ratio between receptor occupancy by the MAb and inhibition of entry. In contrast, the EC50 of JS81 (as well as those of O.N.165 and 1.3.3.22) is fourfold higher than the IC50, translating into a one-to-four molar ratio between receptor occupancy and inhibition of entry. This means that for 100% of viral entry to be inhibited, only 25% of CD81 molecules need to be occupied by JS81. Put differently, there is a 70-fold difference in half-maximal binding concentrations between the two MAbs but a 260-fold difference in half-maximal inhibitory concentrations. These observations imply that, unlike 1D6, JS81 inhibits more than just E2-CD81 binding. We propose that this antibody additionally inhibits CD81 recruitment and oligomerization into the HCV-receptor entry complex, the very same functions that may be mediated by residues in the TM and intracellular domains of CD81.
The half-maximal time for HCVpp to undergo CD81-mediated entry was determined to be approximately 17 min. Because of our assay conditions, this is in fact the time required for HCVpp binding mediated by E2-CD81 interactions. We predict that this is preceded by receptor-induced E1E2 conformational changes and followed by assembly of CD81 into a virus-receptor complex. This entry complex then undergoes endocytosis and pH-dependent fusion (3, 22). It is notable that the internalization rate of CD81 is exceptionally slow (>12 h) (32, 40) and therefore incompatible with the rapid kinetics of HCV internalization and fusion (T. Dragic, unpublished results). This suggests that other molecules associated with the HCV entry complex must divert CD81 into a more efficient endocytic pathway. Alternatively, the interaction with CD81 may be transient, simply priming the virus for downstream entry events.
The CD81 tetraspanin was first identified as an HCV envelope glycoprotein E2-binding receptor and was more recently shown to be required for HCV entry into target cells. E2 proteins derived from a variety of isolates bind differently to CD81. Nonetheless, all isolates tested to date require CD81 for entry into target cells. This is true for both HCVpp and the newly discovered HCV replicating molecular clones. CD81 does not determine the species specificity of HCV, since CD81 proteins from some, but not all, uninfectable species can bind sE2. Moreover, CD81 cannot be the determinant of HCV liver tropism because it is expressed on a wide variety of human tissues. Our data further support the notion that human hepatocyte-specific molecules act together with CD81 to mediate viral entry into target cells. The effect of these molecules may be exerted before or after E2-CD81 binding (though we favor the latter) and may be necessary for HCV internalization and routing to fusion-permissive intracellular compartments.
Acknowledgments
We thank Jean Dubuisson for his continuing generosity in providing us with the anti-E2 MAb H53.
This work was supported by NIH grant AI060390 and the Burroughs Wellcome Fund for Investigators in Pathogenesis of Infectious Diseases. This work was also supported in part by the NIAID Centers for AIDS Research grant AI051519 to the Albert Einstein College of Medicine.
REFERENCES
- 1.Allander, T., X. Forns, S. U. Emerson, R. H. Purcell, and J. Bukh. 2000. Hepatitis C virus envelope protein E2 binds to CD81 of tamarins. Virology 277:358-367. [DOI] [PubMed] [Google Scholar]
- 2.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 4.Berditchevski, F. 2001. Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114:4143-4151. [DOI] [PubMed] [Google Scholar]
- 5.Berditchevski, F., K. F. Tolias, K. Wong, C. L. Carpenter, and M. E. Hemler. 1997. A novel link between integrins, transmembrane-4 superfamily proteins (CD63 and CD81), and phosphatidylinositol 4-kinase. J. Biol. Chem. 272:2595-2598. [DOI] [PubMed] [Google Scholar]
- 6.Boisvert, J., X. S. He, R. Cheung, E. B. Keeffe, T. Wright, and H. B. Greenberg. 2001. Quantitative analysis of hepatitis C virus in peripheral blood and liver: replication detected only in liver. J. Infect. Dis. 184:827-835. [DOI] [PubMed] [Google Scholar]
- 7.Charrin, S., F. Le Naour, V. Labas, M. Billard, J. P. Le Caer, J. F. Emile, M. A. Petit, C. Boucheix, and E. Rubinstein. 2003. EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 373:409-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charrin, S., S. Manie, M. Oualid, M. Billard, C. Boucheix, and E. Rubinstein. 2002. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett. 516:139-144. [DOI] [PubMed] [Google Scholar]
- 9.Charrin, S., S. Manie, C. Thiele, M. Billard, D. Gerlier, C. Boucheix, and E. Rubinstein. 2003. A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol. 33:2479-2489. [DOI] [PubMed] [Google Scholar]
- 10.Cherukuri, A., R. H. Carter, S. Brooks, W. Bornmann, R. Finn, C. S. Dowd, and S. K. Pierce. 2004. B cell signaling is regulated by induced palmitoylation of CD81. J. Biol. Chem. 279:31973-31982. [DOI] [PubMed] [Google Scholar]
- 11.Cherukuri, A., T. Shoham, H. W. Sohn, S. Levy, S. Brooks, R. Carter, and S. K. Pierce. 2004. The tetraspanin CD81 is necessary for partitioning of coligated CD19/CD21-B cell antigen receptor complexes into signaling-active lipid rafts. J. Immunol. 172:370-380. [DOI] [PubMed] [Google Scholar]
- 12.Clark, K. L., A. Oelke, M. E. Johnson, K. D. Eilert, P. C. Simpson, and S. C. Todd. 2004. CD81 associates with 14-3-3 in a redox-regulated palmitoylation-dependent manner. J. Biol. Chem. 279:19401-19406. [DOI] [PubMed] [Google Scholar]
- 13.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 101:7270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummer, H. E., K. A. Wilson, and P. Poumbourios. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143-11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flint, M., J. Dubuisson, C. Maidens, R. Harrop, G. R. Guile, P. Borrow, and J. A. McKeating. 2000. Functional characterization of intracellular and secreted forms of a truncated hepatitis C virus E2 glycoprotein. J. Virol. 74:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flint, M., J. M. Thomas, C. M. Maidens, C. Shotton, S. Levy, W. S. Barclay, and J. A. McKeating. 1999. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J. Virol. 73:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadlock, K. G., R. E. Lanford, S. Perkins, J. Rowe, Q. Yang, S. Levy, P. Pileri, S. Abrignani, and S. K. Foung. 2000. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J. Virol. 74:10407-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemler, M. E. 2001. Specific tetraspanin functions. J. Cell Biol. 155:1103-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hemler, M. E. 2003. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19:397-422. [DOI] [PubMed] [Google Scholar]
- 21.Higginbottom, A., E. R. Quinn, C. C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, N., T. Nakazawa, T. Mizutani, and K. Shimotohno. 1995. Susceptibility of human T-lymphotropic virus type I infected cell line MT-2 to hepatitis C virus infection. Biochem. Biophys. Res. Commun. 206:863-869. [DOI] [PubMed] [Google Scholar]
- 24.Lavillette, D., A. W. Tarr, C. Voisset, P. Donot, B. Bartosch, C. Bain, A. H. Patel, J. Dubuisson, J. K. Ball, and F. L. Cosset. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265-274. [DOI] [PubMed] [Google Scholar]
- 25.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89-109. [DOI] [PubMed] [Google Scholar]
- 26.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 27.Masciopinto, F., G. Freer, V. L. Burgio, S. Levy, L. Galli-Stampino, M. Bendinelli, M. Houghton, S. Abrignani, and Y. Uematsu. 2002. Expression of human CD81 in transgenic mice does not confer susceptibility to hepatitis C virus infection. Virology 304:187-196. [DOI] [PubMed] [Google Scholar]
- 28.McKeating, J. A., L. Q. Zhang, C. Logvinoff, M. Flint, J. Zhang, J. Yu, D. Butera, D. D. Ho, L. B. Dustin, C. M. Rice, and P. Balfe. 2004. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J. Virol. 78:8496-8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meola, A., A. Sbardellati, B. Bruni Ercole, M. Cerretani, M. Pezzanera, A. Ceccacci, A. Vitelli, S. Levy, A. Nicosia, C. Traboni, J. McKeating, and E. Scarselli. 2000. Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J. Virol. 74:5933-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owsianka, A., R. F. Clayton, L. D. Loomis-Price, J. A. McKeating, and A. H. Patel. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877-1883. [DOI] [PubMed] [Google Scholar]
- 31.Patel, A. H., J. Wood, F. Penin, J. Dubuisson, and J. A. McKeating. 2000. Construction and characterization of chimeric hepatitis C virus E2 glycoproteins: analysis of regions critical for glycoprotein aggregation and CD81 binding. J. Gen. Virol. 81:2873-2883. [DOI] [PubMed] [Google Scholar]
- 32.Petracca, R., F. Falugi, G. Galli, N. Norais, D. Rosa, S. Campagnoli, V. Burgio, E. Di Stasio, B. Giardina, M. Houghton, S. Abrignani, and G. Grandi. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 74:4824-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 34.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-1034. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 35.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 77:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosa, D., S. Campagnoli, C. Moretto, E. Guenzi, L. Cousens, M. Chin, C. Dong, A. J. Weiner, J. Y. Lau, Q. L. Choo, D. Chien, P. Pileri, M. Houghton, and S. Abrignani. 1996. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc. Natl. Acad. Sci. USA 93:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stipp, C. S., T. V. Kolesnikova, and M. E. Hemler. 2001. EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 276:40545-40554. [DOI] [PubMed] [Google Scholar]
- 38.Stipp, C. S., T. V. Kolesnikova, and M. E. Hemler. 2003. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 28:106-112. [DOI] [PubMed] [Google Scholar]
- 39.Sung, V. M., S. Shimodaira, A. L. Doughty, G. R. Picchio, H. Can, T. S. Yen, K. L. Lindsay, A. M. Levine, and M. M. Lai. 2003. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J. Virol. 77:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan, Y. J., S. P. Lim, P. Ng, P. Y. Goh, S. G. Lim, Y. H. Tan, and W. Hong. 2003. CD81 engineered with endocytotic signals mediates HCV cell entry: implications for receptor usage by HCV in vivo. Virology 308:250-269. [DOI] [PubMed] [Google Scholar]
- 41.Triyatni, M., J. Vergalla, A. R. Davis, K. G. Hadlock, S. K. Foung, and T. J. Liang. 2002. Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding. Virology 298:124-132. [DOI] [PubMed] [Google Scholar]
- 42.Voisset, C., and J. Dubuisson. 2004. Functional hepatitis C virus envelope glycoproteins. Biol. Cell 96:413-420. [DOI] [PubMed] [Google Scholar]
- 43.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yauch, R. L., F. Berditchevski, M. B. Harler, J. Reichner, and M. E. Hemler. 1998. Highly stoichiometric, stable, and specific association of integrin alpha3beta1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol. Biol. Cell 9:2751-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, X. A., A. L. Bontrager, and M. E. Hemler. 2001. Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific beta(1) integrins. J. Biol. Chem. 276:25005-25013. [DOI] [PubMed] [Google Scholar]
- 47.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]