Abstract
Newcastle disease virus (NDV) is a negative-strand RNA virus with oncolytic activity against human tumors. Its effectiveness against tumors and safety in normal tissue have been demonstrated in several clinical studies. Here we show that the spread of NDV infection is drastically different in normal cell lines than in tumor cell lines and that the two cell types respond differently to beta interferon (IFN-β) treatment. NDV rapidly replicated and killed HT-1080 human fibrosarcoma cells but spread poorly in CCD-1122Sk human skin fibroblast cells. Pretreatment with endogenous or exogenous IFN-β completely inhibited NDV replication in normal cells but had little or no effect in tumor cells. Thus, the outcome of NDV infection appeared to depend on the response of uninfected cells to IFN-β. To investigate their differences in IFN responsiveness, we analyzed and compared the expression and activation of components of the IFN signal transduction pathway in these two types of cells. The levels of phosphorylated STAT1 and STAT2 and that of the ISGF3 complex were markedly reduced in IFN-β-treated tumor cells. Moreover, cDNA microarray analysis revealed significantly fewer IFN-regulated genes in the HT-1080 cells than in the CDD-1122Sk cells. This finding suggests that tumor cells demonstrate a less-than-optimum antiviral response because of a lesion in their IFN signal transduction pathway. The rapid spread of NDV in HT-1080 cells appears to be caused by their deficient expression of anti-NDV proteins upon exposure to IFN-β.
The interest in using Newcastle disease virus (NDV), an avian Paramyxovirus, as an antineoplastic agent has increased steadily in recent years (4, 10, 35, 36). Several preclinical and clinical studies, including phase I and phase II studies, have shown some success (9, 15, 42). Independent of its immune-stimulating property, NDV's ability to selectively lyse tumor cells is one rationale for using this virus as an antitumor agent. In clinical studies, some strains of NDV (natural or derived), such as 73-T (7), Italian (48), and PV701 (42), caused human tumors to regress without affecting the surrounding normal tissue (30, 42, 43, 47, 48). A previous study in which the virus yield of strain 73-T was measured in the supernatant of infected cell lines (tumor cells and normal cells) found that NDV efficiently and selectively replicates in and kills tumor cells but not normal cells (47). Also, mice with tumor xenografts showed complete regression of the tumors after they were injected with NDV; mice without tumors that were injected with the virus remained healthy (47).
The type I interferons (IFNs)—IFN-α and IFN-β—are the main cytokines for innate immune response against viral infections. This is exemplified by the fact that mice genetically lacking some of the components of the IFN signal transduction system are highly susceptible to viral infections (6, 14, 21, 32, 33). The general mechanism leading to the production of antiviral products has been well characterized. Type I IFN binding to cell-specific receptors causes clustering of receptor subunits and activation of receptor-associated Janus kinases JAK1 and TYK2, which in turn phosphorylate and activate the signal transducers and activators of transcription STAT1 and STAT2. The STAT proteins heterodimerize and form a complex with IRF9, a member of the IFN regulatory factor family of proteins. This trimeric complex, known as ISGF3, provides DNA recognition and transactivation by binding to IFN-stimulated response elements (ISRE) in the target genes. Production of IFN-stimulated gene (ISG) products leads to the creation of an antiviral state in the target cells that blocks viral replication. Thus, one of the consequences of IFN production by virus-infected cells is the protection of other, uninfected cells and the prevention of further virus replication and growth.
Several types of mammalian cells secrete a high level of IFN upon infection with NDV (23-25, 29, 31, 52, 56); however, the mechanism by which NDV kills human tumor cells and the role of IFN in the spread of this virus in human cells is not entirely known. Many types of tumor cells harbor defects in IFN response (1, 11, 38, 44, 50, 54, 55), and some viruses, such as the vesicular stomatitis virus, exploit this defect to cause virus-mediated oncolysis (2, 37, 51).
In this study, we investigated the activity of several different strains of NDV in normal human cell lines and human tumor cell lines to increase our understanding of the mechanisms that control the vastly different outcomes of NDV infection in these two types of cells. We also examined the response of uninfected normal cells and tumor cells to the antiviral factor IFN-β. Finally, we analyzed the expression of IFN-β-regulated genes upon IFN stimulation and compared the differences between the changes found in normal cells and those found in tumor cells.
MATERIALS AND METHODS
Viruses and cell lines.
The Beaudette C strain of NDV was used throughout this study. In addition, the following strains were used for growth and replication studies: Australian-Victoria, California, Kansas, La Sota, and 73-T. All of the viral strains were propagated in DF1 chicken fibroblast cells (ATCC, Manassas, VA) with Dulbecco's modified Eagle medium (DMEM; BioWhittaker, Inc., Walkersville, MD) containing 10% fetal bovine serum (FBS; Sigma, St. Louis, MO) or 5% chicken allantoic fluid (for NDV strain La Sota).
The following cell lines and corresponding culture media were used: normal human skin fibroblast cells (CCD-1122Sk; ATCC, Manassas, VA), Iscove's modified Dulbecco's medium with 10% FBS; normal human lung fibroblast cells (MRC5; BioWhittaker), Eagle's minimal essential medium with 10% FBS; normal human prostate epithelial cells (PrEC; Clonetics, San Diego, CA), the manufacturer's instructions were followed; normal human skeletal muscle cells (SKMC; Clonetics), the manufacturer's instructions were followed; normal human mammary epithelial cells (HMEC; Clonetics), the manufacturer's instructions were followed; human prostate epithelial cells transfected with a plasmid carrying one copy of human papillomavirus 18 and transformed with Ki-Ras by using the Kirsten murine sarcoma virus (RWPE-2; ATCC), ATCC complete growth medium; human fibrosarcoma (HT-1080; ATCC), Eagle's minimal essential medium with 10% FBS; human cervical carcinoma (C-33A; ATCC), MEM with 10% FBS; human colorectal adenocarcinoma (SW-620; ATCC), Leibovitz's L15 medium with 10% FBS; human lung adenosquamous carcinoma (NCI-H596; ATCC), RPMI with 10% FBS.
Virus growth and production in vitro.
NDV was propagated and titrated in DF1 cells. For growth studies, the cells were grown to 80% to 90% confluence in a six-well plate and infected with NDV in DMEM with 0.15% bovine serum albumin at a multiplicity of infection (MOI) of 0.001. At 1 h postinfection, the inoculum was removed and the cells were washed twice with phosphate-buffered saline (PBS) and then covered with 2 ml of the recommended medium. Cells were then incubated in a 37°C incubator. For nonvirulent NDV strain La Sota, the medium was supplemented with 5% chicken allantoic fluid as a source of proteases that cleave the F protein. An aliquot of the culture supernatant was collected every 24 h for 4 days for estimation of the virus titer. The procedure was repeated three times, and the standard deviation was calculated.
NDV assay of culture supernatants.
Plaque assays were performed to determine the level of NDV infection in the culture supernatant. DF1 cells were adsorbed with different dilutions of the culture supernatants. At 1 h postinfection, the inoculum was removed and the cells were overlaid with Opti-MEM medium supplemented with 2% FBS and 0.9% (wt/vol) methylcellulose (Sigma) in the presence of 2% FBS. For NDV strain La Sota, the above-mentioned overlay included 5% chicken allantoic fluid. The cells were incubated at 37°C for 4 days or until plaques appeared. The cells were then stained with neutral red, and the plaques was counted.
Immunoflourescence of infected cells.
Cells were grown in glass chamber slides (Nalge Nunc International, Rochester, NY). The cells were infected with NDV at different MOIs. At 12 or 24 h postinfection, the medium was aspirated and the cells were rinsed in PBS and fixed with 3.7% formaldehyde. The slides were transferred to a coplin jar and treated with cold acetone for 7 min. The slides were then rinsed three times with PBS for 2 min per rinse. The cells were incubated with a monoclonal antibody against NDV hemagglutinin-neuraminidase (HN) for 45 min at 37°C. After the slides were rinsed in PBS thrice, the cells were incubated with fluorescein isothiocyanate-coupled secondary anti-mouse antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) at a dilution of 1:3,000. The cells were then rinsed in PBS thrice and mounted with p-phenyldiamine (Sigma) mount. The cells were then examined under a confocal microscope.
Assessment of the anti-NDV factor(s) in supernatant from infected cells.
The supernatants from infected cells in culture were collected at 72 h postinfection. NDV was removed from the supernatants via ultrafiltration with Centricon YM-100 filters (Millipore, Billerica, MA) according to the manufacturer's protocol. Fresh cells were treated with the filtered supernatant for 15 h. After removal of the supernatant, the cells were infected with NDV at an MOI of 0.001. The culture supernatants were assayed for NDV at 24, 48, 72, and 96 h postinfection.
To identify the anti-NDV factor(s) in the supernatant, we performed an experiment similar to the one described above, except that the supernatant was incubated with a combination of antibodies that neutralize IFN-α and IFN-β (Βiosource, Camarillo, CA) for 1 h at room temperature before it was overlaid on the cells. Two concentrations, 300 U and 1,000 U, of each antibody were examined.
IFN assay of the supernatant.
NDV infection of different cell lines was performed as described above. The supernatant from the infected cells in culture was collected at different times postinfection, and an IFN-β enzyme-linked immunosorbent assay (ELISA) kit (Biosource) was used to detect IFN-β in the media according to the manufacturer's protocol.
Immunoblot analysis of NDV proteins.
The cells were grown to 80% to 90% confluence in a six-well plate and either pretreated with 500 IU of IFN-β (Βiosource) for 15 h before infection or infected directly with NDV at an MOI of 1. The infection medium used was DMEM with 0.15% bovine serum albumin. At 1 h postinfection, the inoculum was removed, the cells were washed twice with PBS, covered with 2 ml of the recommended medium, and then incubated at 37°C. The cells were lysed 48 h later with RIPA buffer (50 mM Tris-HCl [pH 7.4], 10 mM EDTA, 150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 1% deoxycholate, 1% Triton X-100), and 5 μg of total protein was loaded onto a 10% SDS-polyacrylamide gel (precast gel from Invitrogen, Carlsbad, CA). The resolved proteins were transferred onto a polyvinylidene difluoride membrane and probed with polyclonal chicken antibodies specific for NDV and a horseradish peroxidase-conjugated anti-chicken secondary antibody (Jackson ImmunoResearch Laboratories). The membrane was exposed to a chemiluminescent substrate and detected by autoradiography.
Immunoblot analysis of IFN signaling intermediates.
The cells were treated with 500 IU/ml IFN-β for 15, 30, 60, 120, 240, or 360 min. The cells were harvested and lysed in RIPA buffer, and the protein concentration was determined by the Micro BCA method (Pierce, Rockford, IL). For analysis of STAT1, STAT2, and IRF9, we loaded equal amounts of protein onto a 10% SDS-polyacrylamide gel, transferred them onto a polyvinylidene difluoride membrane, and detected the protein with antibodies specific for STAT1 and STAT2 (rabbit polyclonal antibodies from Santa Cruz Biotechnologies, Santa Cruz, CA), phosphorylated forms of STAT1 (mouse monoclonal antibody to Tyr 701; Cell Signaling Technology, Danvers, MA), phosphorylated forms of STAT2 (mouse monoclonal antibody to Tyr 690; Cell Signaling Technology), and IRF9 (Santa Cruz Biotechnologies). For analysis of JAK1 and TYK2, the proteins were immunoprecipitated by antibodies specific to JAK1 kinase and TYK 2 kinase (mouse monoclonal antibody from BD Transduction Labs, Franklin Lakes, NJ). After resolution on the gel, the proteins were immunoblotted and probed with rabbit monoclonal antibodies against the JAK1 and TYK2 kinases or those against phosphorylated forms of the JAK1 kinase (Tyr 1022/1023) or the TYK2 kinase (Tyr 1054/1055) (Cell Signaling Technology). The blots were probed with the specific horseradish peroxidase-conjugated secondary antibody to determine the relative amounts of proteins.
Electrophoretic mobility shift assay (EMSA).
The biotinylated probe was prepared by end labeling the ISRE oligonucleotide sequences (AGGAAATAGAAACTG)2 with biotin via the Biotin 3′ End DNA Labeling Kit (Pierce). Cells were treated with IFN-β for 30 min, and then nuclear extracts were prepared with the NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit (Pierce). The LightShift Chemiluminescent EMSA Kit (Pierce) was used to perform the binding reaction between the extract and the labeled ISRE. The complexes were resolved on a precast native Tris-borate-EDTA polyacrylamide gel (Invitrogen) and transferred to a nylon membrane before cross-linking the DNA to the membrane. Biotinylated DNA was detected with streptavidin-horseradish peroxidase conjugate and chemiluminescent substrate. The membrane was visualized by autoradiography.
RNA isolation and preparation of cDNA for microarray analysis.
HT-1080 and CCD-1122Sk cells were grown in 10-cm dishes until 80% confluent and then treated with IFN-β (500 IU/ml) for 15 h. Control cells were left untreated. Total RNA was isolated by TRIzol reagent (Invitrogen) according to the manufacturer's protocol. To assess the integrity of the total RNA, we tested a small amount of the sample on a 2100 Bioanalyzer Lab-on-a-Chip system (Agilent Technologies, Palo Alto, CA). The RNA sample was confirmed to have two strong peaks representing the 18S and 28S rRNA fragments. The 28S/18S ratio was approximately 2, and there was very little low-molecular-weight degradation product. The RNA was reverse transcribed with dT primer, followed by synthesis of second-strand DNA. Double-stranded DNA was transcribed in vitro incorporating biotinylated ribonucleotides. The cRNA was fragmented to minimize steric hindrance during the hybridization step.
Microarray analysis.
The labeled target was hybridized to the human gene chip UG133A (Affymetrix, Santa Clara, CA), which represents 22,283 human genes, according to the manufacturer's protocol. In brief, the hybridization was performed for 18 h at 45°C. The arrays were washed and stained with streptavidin conjugated to phycoerythrin in the Affymetrix automated fluidics station. The arrays were then scanned, and an image file was produced. The image file was converted with Affymetrix Microarray Suite software (version 5.0). Expression signals were scaled to a target intensity of 500 and log transformed. Arrays were omitted if the scaling factor exceeded 3 standard deviations of the mean or if the ratio of 3′ to 5′ mRNA for β-actin or glyceraldehyde-3-phosphate dehydrogenase was greater than 3. The data was analyzed with Spotfire DecisionSite software (Spotfire, Somerville, MA). The cDNA microarrays were analyzed by a statistical algorithm that calculated a signal value and a detection P value for each gene. A P value of <0.4 was considered as present call for a gene, 0.4 to 0.6 as marginal, and >0.6 as absent.
RESULTS
Spread of NDV among normal cells differs from that among tumor cells.
To assess the ability of NDV to infect and yield virus in normal human cells and human tumor cells, we conducted multicycle growth kinetic studies of the Beaudette C strain of NDV. We infected the cells at a low multiplicity (MOI = 0.001) and then determined the virus titer in the culture supernatant at different times postinfection. Infection of human normal cells (i.e., CCD-1122Sk, MRC5, and PrEC cells) produced lower virus yields throughout the 5 days of infection than did parallel infections of several human tumor cell lines (data not shown).
At the peak of infection (72 h postinfection), HT-1080 cells, NCI-H596 cells, HPV-18 cells, and ras gene-transformed human prostate epithelial cells (RWPE-1) produced more than 1,000 times more virus particles than did normal CCD-1122Sk and MRC5 cells (Table 1). Moreover, in contrast to normal cells, all of the tumor cells were killed and obliterated at this stage of virus infection. These results confirm the inherent ability of NDV to grow rapidly and replicate in human tumor cells; however, in the C-33A cell line, NDV infection yielded 10 to 100 times fewer virus particles than the number obtained from the other infected tumor cell lines. Also, the titers of NDV were at least 10-fold higher in primary PrEC cells than they were in the other infected normal cells. This finding suggests that some tumor cells and normal cells may not show a typical response to NDV infection.
TABLE 1.
NDV yield at 72 h postinfection (MOI = 0.001) in normal and tumor cell lines
| Cell line | Origina | NDV titer (PFU/ml) |
|---|---|---|
| Normal cells | ||
| CCD 1122Sk | Skin fibroblast | 4 × 103 |
| MRC5 | Lung fibroblast | 8 × 103 |
| PrEC | Prostate epithelium | 7 × 104 |
| Tumor cells | ||
| HT-1080 | Fibrosarcoma | 5 × 106 |
| NCI-H596 | Lung adenosquamous carcinoma | 6 × 106 |
| C-33A | Cervical carcinoma | 3 × 105 |
| RWPE-2b | Prostate epithelium | 8 × 106 |
| SW-620 | Colorectal adenocarcinoma | 2 × 106 |
All cells were of human origin.
RWPE-2 cells were transfected with the HPV-18 genome and transformed with Ki-ras.
NDV infection of all of the normal cell lines at a high multiplicity (MOI, ≥2) produced a rapid virus yield and death of the cells (data not shown). The virus yield in normal cells infected with NDV at an MOI of 2 was in excess of 105 PFU at 72 h postinfection. This result demonstrates that the poor virus yield after low-MOI infection of normal cells is due to the limited spread of viral infection. An extremely high yield of NDV after low-MOI infection of tumor cells resulted in rapid and excessive cytopathology and death of those cells.
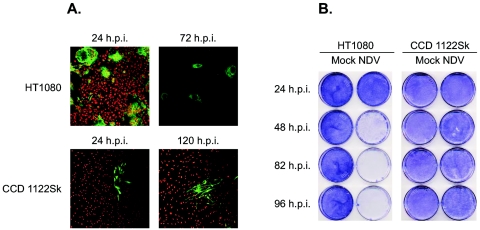
We observed the cells by immunofluorescence after staining the viral HN on the surface of the infected cells (Fig. 1A). Twenty-four hours after infection of HT-1080 cells with the Beaudette C strain (MOI = 0.001), we observed huge multinucleated cells. Three days later, almost all of the cells were dead and detached from the surface of the plate. Similar results were obtained with other tumor cell lines (data not shown). Thus, the important hallmarks of NDV infection of tumor cells were the rapid spread and killing of the cells. In contrast, infection of CCD-1122Sk cells resulted in 5% to 10% of the cells being infected at 24 h and only 15% to 20% at 120 h. The fate of both types of infected cells was also observed on crystal violet staining (Fig. 1B). Thus, in normal cells infected at a low MOI, the spread and progress of NDV infection were limited.
FIG. 1.
The spread of NDV infection differs in normal and tumor cell lines. Cells seeded on chamber slides (A) or 100-mm cell culture dishes (B) were infected with the Beaudette C strain of NDV at an MOI of 0.001. (A) The cells were fluorescently stained with antibodies against NDV HN (green) and then observed with a confocal microscope. Human fibrosarcoma cells (HT-1080) showed a severe cytopathic effect 24 h postinfection (h.p.i.). At 72 h postinfection, almost all of the cells were detached from the surface. The normal skin fibroblast cells (CCD-1122Sk) showed a limited cytopathic effect at 24 h postinfection and progressed very little, even after 120 h. (B) The cells were stained with crystal violet at various times postinfection; untreated (Mock) cells were also observed. In contrast to the normal cells, the tumor cells were dead and detached from the dish at 72 h postinfection.
Different NDV strains show similar growth phenotypes.
A few selected strains of NDV have been utilized for clinical therapy of tumors that have been effectively lysed by these strains. The relative nontoxicity of NDV toward normal human cells has also been demonstrated. One such widely used strain is the mouse tumor-adapted 73-T strain, which was derived from Lederle's NDV by 73 passages in vitro and 13 passages in vivo in murine Ehrlich ascites tumor cells (7). Growth of NDV strain 73-T in different human tumor and normal cell lines has shown that this strain efficiently and selectively replicates in and kills tumor cells but not normal cells (47). The growth of the other NDV strains has not been studied under controlled laboratory conditions; therefore, their selectivity for tumor cells has not been confirmed.
As mentioned above, the Beaudette C strain showed selectivity of rapid growth and replication in tumor cells and poor replication and slow spread in normal cells. We then examined four other strains of NDV (Kansas, California, La Sota, and Australian-Victoria) to see how they replicate and grow in the HT-1080 and CCD-1122Sk cell lines (Table 2). The strains were infected at a low multiplicity (MOI = 0.001), and the characteristics of their infections were compared with those of the 73-T strain. We observed that the growth of these strains in normal and tumor cells was comparable to that of the 73-T strain. Thus, different strains of NDV are inherently tumor selective and may be equally useful as antitumor therapeutic agents.
TABLE 2.
Virus yield at 72 h postinfection of normal and tumor cell lines infected with different strains of NDV
| Cell line | Titer (PFU/ml) of NDV strain:
|
||||
|---|---|---|---|---|---|
| Australian- Victoria | Kansas | California | La Sota | 73-T | |
| Normal cells | |||||
| CCD 1122Sk | 3 × 103 | 3 × 103 | 1 × 104 | 2 × 103 | 7 × 102 |
| MRC5 | 6 × 103 | 2 × 104 | 4 × 104 | 6 × 103 | 1 × 103 |
| PrEC | 1 × 104 | 9 × 104 | 3 × 104 | 9 × 103 | 8 × 102 |
| Tumor cells | |||||
| HT-1080 | 4 × 106 | 1 × 106 | 2 × 106 | 9 × 105 | 4 × 105 |
| NCI-H596 | 5 × 106 | 1 × 106 | 2 × 106 | 1 × 106 | 1 × 106 |
| C-33A | 4 × 105 | 2 × 105 | 2 × 105 | 5 × 105 | 1 × 105 |
| SW-620 | 7 × 105 | 3 × 105 | 1 × 105 | 4 × 105 | 1 × 105 |
Infected cells secrete an anti-NDV factor(s) that protects uninfected normal cells but not tumor cells.
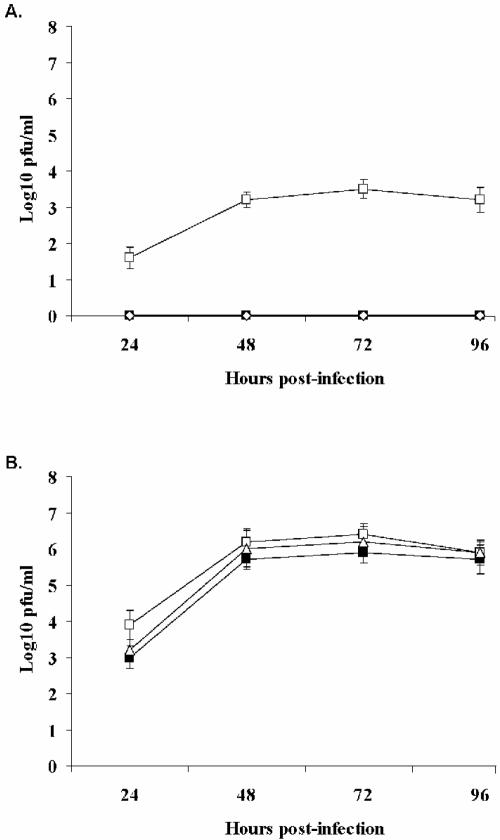
To investigate the poor spread of NDV in normal cells in culture, we determined whether culture supernatant from NDV-infected cells had the ability to prevent infection of other cells. CCD-1122Sk cells and HT-1080 cells were infected with NDV strain Beaudette C (MOI = 0.001), and supernatants were collected at 60 h postinfection. The supernatants were clarified by ultrafiltration to remove all virus particles and then applied to fresh CCD-1122Sk cells, which were incubated for 15 h. The cells were then challenged with NDV strain Beaudette C (MOI = 0.001), and the viral titer of the supernatants was measured at different times after infection (Fig. 2).
FIG. 2.
Pretreatment of cells with supernatant from NDV-infected CCD-1122Sk or HT-1080 cells in culture inhibited the growth of NDV in normal cells but not in tumor cells. At 60 h postinfection, supernatants were collected, clarified, and then applied to uninfected cells for 15 h. The cells were then infected with NDV (MOI = 0.001), and viral titers were determined. (A) Viral titers in CCD-1122Sk cell cultures without pretreatment (open squares) and after pretreatment with supernatant from HT-1080 cells (filled square) or CCD-1122Sk cells (diamonds). (B) Viral titers in HT-1080 cell cultures without pretreatment (open squares) and after pretreatment with supernatant from HT-1080 cells (filled squares) or CCD-1122Sk cells (triangles).
NDV production was completely inhibited in the normal cells after treatment. This finding suggests that infected CCD-1122Sk cells and HT-1080 cells secrete a soluble anti-NDV factor(s) that protects normal cells from NDV infection. When HT-1080 cells were similarly pretreated before infection, the NDV production did not differ significantly from that of the control infection (Fig. 2B). Although both infected normal and infected tumor cells secreted an anti-NDV factor(s), only normal cells responded to the protective effect.
IFN is the main anti-NDV factor secreted by CCD-1122Sk and HT-1080 cells.
IFN is the most common antiviral protein secreted by virus-infected cells; therefore, we investigated whether the anti-NDV factor was IFN. The supernatant was incubated with antibodies against IFN-α and -β before it was used to pretreat CCD-1122Sk cells. After pretreatment, cells were infected with NDV at a low multiplicity (MOI = 0.001). NDV infection after the pretreatment of CCD-1122Sk cultures with IFN antibody-treated supernatant from the infected HT-1080 cells restored viral growth to levels similar to that of the control cells (Fig. 3B). Similarly, pretreatment of CCD-1122Sk cells with the IFN antibody-treated supernatant from infected CCD-1122Sk cells significantly neutralized the effect of the supernatant (Fig. 3A). IFN is thus the primary anti-NDV factor that is released by both HT-1080 and CCD-1122Sk cells.
FIG. 3.
Pretreatment of CCD-1122Sk cells with IFN antibody-treated supernatant has little effect on NDV growth. NDV-infected culture supernatants from either normal cells (CCD-1122Sk) (A) or tumor cells (HT-1080) (B) were treated with neutralizing antibodies against IFN-α and IFN-β before pretreatment of fresh CCD-1122Sk cells. The cells were then infected with NDV at an MOI of 0.001. Viral titers were determined in the culture medium at various times postinfection. The viral titers of control infections (filled squares) differed markedly from those of cultures that were pretreated with just the supernatant (triangles) but not from those of cultures that were pretreated with antibody-treated supernatant (open squares).
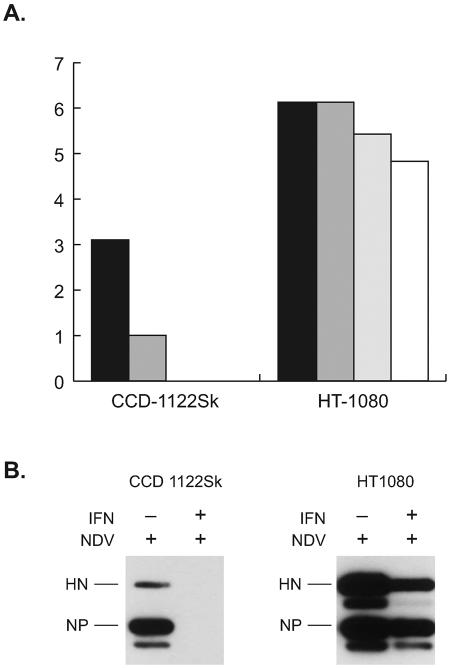
NDV-infected cells in culture secrete IFN-β.
We monitored the amount of IFN secreted by NDV-infected cells. The level of biologically active IFN was calculated by the sandwich ELISA method. Both normal and tumor cells secreted moderate to high levels of IFN-β after infection with NDV (Fig. 4); however, no IFN-α was detected in either cell line. Low levels of IFN-α were detected in tumor cells infected with NDV, but only at a very high multiplicity (MOI = 5) (data not shown). These results indicate that IFN-β is the principal antiviral factor secreted by NDV-infected normal cells and tumor cells.
FIG. 4.
Both HT-1080 and CCD-1122Sk secrete IFN-β. Culture medium was collected at various time points postinfection, and the levels of biologically active human IFN-β were determined by the human IFN-β ELISA kit. The levels of IFN-β secreted by normal CCD-1122Sk cells (black bars) and HT-1080 tumor cells (white bars) are shown.
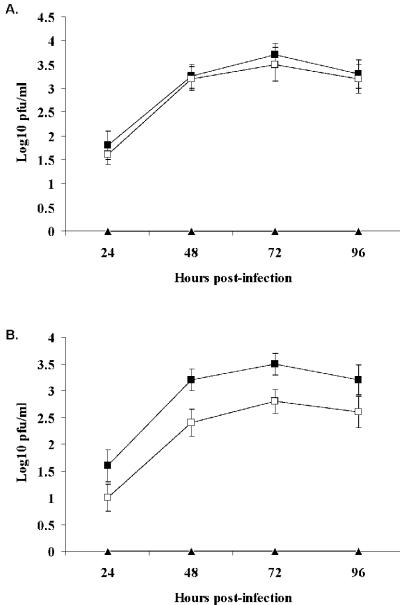
NDV replication in CCD-1122Sk cells, but not in HT-1080 cells, is severely inhibited by exogenous IFN-β treatment.
Pretreatment of CCD-1122Sk cells with 100 or 200 IU/ml IFN-β for 15 h before the cells were infected with NDV at a low multiplicity (MOI = 0.001) inhibited NDV replication (Fig. 5A). The same pretreatment of HT-1080 cells did not significantly reduce the virus yield or replication (Fig. 5A), but pretreatment with 1,000 IU/ml IFN-β did nominally reduce NDV production. In contrast, IFN-α pretreatment altered NDV infection only at very high doses in either of the cell types (data not shown). When the cells were assayed for NDV protein production, we observed a complete shutdown of NDV proteins NP and HN in pretreated (500 IU/ml IFN-β) CCD-1122Sk cells but not in pretreated HT-1080 cells (Fig. 5B). These results indicate that normal cells are extremely sensitive to the antiviral effect of naturally secreted or exogenous IFN-β, whereas tumor cells are relatively resistant.
FIG. 5.
NDV growth and replication are suppressed by pretreatment with exogenous IFN-β in CCD-1122Sk normal cells but not in HT-1080 tumor cells. (Α) NDV titers at 72 h postinfection in cells that were either left untreated (black bars) or pretreated with different concentrations of IFN-β (dark gray bars, 100 IU; gray bars, 200 IU; open bars, 1,000 IU) for 15 h. (B) NDV protein synthesis in infected cells with (+) or without (−) pretreatment with 500 IU of IFN-β for 15 h. After infection, the cells were lysed and immunoprobed with NDV polyclonal antibody. Pretreatment with IFN-β inhibited the HN and NP proteins in normal cells only.
IFN-resistant HT-1080 tumor cells have reduced levels of ISGF3 complexes.
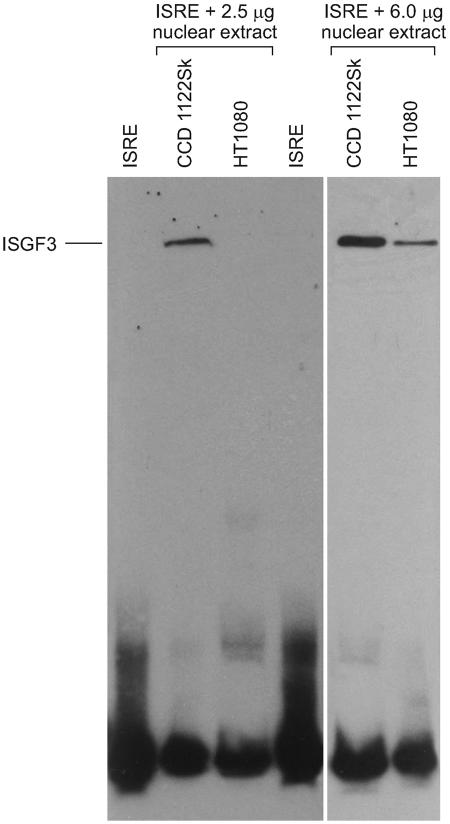
The rapid spread of NDV in tumor cells, even those pretreated with IFN-β, showed that anti-NDV responses are compromised in tumor cells. Therefore, we investigated whether IFN treatment in uninfected tumor cells induced the same level of ISGF3 complexes as it does in normal cells. EMSAs were performed on nuclear extracts prepared from IFN-treated HT-1080 cells and CCD-1122Sk cells (Fig. 6). An ISRE-binding complex was induced in CCD-1122Sk cells. A 200-fold molar excess of unlabeled ISRE prevented the signal shift. This finding confirmed that the shift results from a specific protein-DNA interaction (not shown). The ISGF3 complex was not detected in HT-1080 cells but was present at a relatively reduced level when the quantity of the extract used in the binding reaction was increased to 6 μg. Thus, tumor cells are poor responders to IFN treatment because the absence of ISGF3 complexes in these cells prevents the creation of an antiviral state that would block viral replication.
FIG. 6.
IFN-β treatment stimulates the formation ISGF3 complexes in CCD-1122Sk normal cells but not in HT-1080 tumor cells. Cells were treated with IFN-β for 30 min prior to harvest. Nuclear extracts were prepared, and 2.5 or 6 μg of the extract was bound with biotinylated ISRE sequence probes. The complexes were resolved on a Tris-borate-EDTA polyacrylamide gel and analyzed by EMSA with the use of biotin-labeled ISRE probes.
HT-1080 cells and CCD-1122Sk cells differ in their relative amounts of phosphorylated STAT1 and STAT2 proteins.
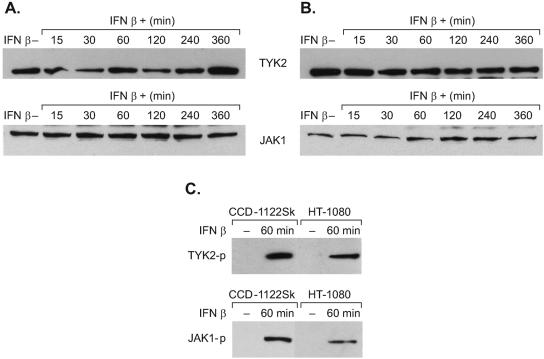
Because HT-1080 cells had reduced levels of ISGF3 complexes, it was of interest to determine what effect, if any, IFN has on the expression of IFN-signaling components in HT-1080 and CCD-1122Sk cells. The cells were treated with IFN-β and then harvested at different time points and probed by immunoblot analysis with a panel of antibodies specific for JAK1, TYK2, STAT1, STAT2, and IRF9. In addition, they were probed for the presence of phosphotyrosine forms of the JAK1 (Y1022/1023) and TYK2 (Y1054/1055) kinases and the phosphotyrosine forms of the STAT1 (Y701) and STAT2 (Y690) proteins. The levels of the tyrosine kinases JAK1 and TYK2 were similar in the untreated cells and throughout the period of IFN-β treatment in both cell types (Fig. 7). In addition, the IFN-β treatment induced phosphorylation of the JAK1 and TYK2 kinases in both cell types without significant differences in the levels of the proteins (Fig. 7C). IFN-β-induced phosphorylation of JAK kinases in HT-1080 cells indicated that neither binding of the IFN to the receptor nor the subsequent oligomerization of the receptor subunit is likely inhibited in those cells. Also, the levels of the STAT1, STAT2, and IRF9 proteins did not change significantly in either untreated or IFN-treated cells (Fig. 8). However, the phosphotyrosine forms of STAT1 and STAT2 progressively decreased throughout the IFN-β treatment (Fig. 8). The IFN-β-resistant HT-1080 cell line contained markedly less phosphorylated STAT1 and STAT2 proteins than did the IFN-β-sensitive CCD-1122Sk cell line. Because STAT proteins are indispensable for dimerization, nuclear translocation, and DNA binding, reduced levels of phosphorylated STAT1 and STAT2 could result in the observed deficiency of ISGF3 complexes.
FIG. 7.
Detection of JAK1 and TYK2 in CCD-1122Sk (A) and HT-1080 (B) cells either left untreated (−) or treated (+) with IFN-β for various times. Total untreated cell lysates were prepared after 360 min in culture, and those of treated cells were prepared 15 to 360 min after IFN-β treatment. The proteins were then immunoblotted and probed with antibodies specific for the unphosphorylated proteins. (C) The levels of phosphorylated TYK2 and JAK1 (TYK2-p and JAK1-p) were measured in both cell types 60 min after IFN-β treatment.
FIG. 8.
Relative amounts of STAT1, STAT2, their phosphorylated forms (STAT1-p and STAT2-p), and IRF9 were measured in CCD-1122Sk (A) and HT-1080 cells (B). The cells were harvested at different times after treatment; untreated cells were harvested after 360 min, and IFN-β-treated cells were harvested from 15 to 360 min after treatment. Proteins were immunoblotted and detected with polyclonal anti-IRF9, anti-STAT1, anti-STAT2, anti-phosphotyrosine STAT1, or anti-phosphotyrosine STAT2 antibodies.
Microarray analysis of tumor cells and normal cells showed a significant difference in the number of IFN-induced genes and the intensity of their expression.
The trimeric ISGF3 complex provides DNA recognition and transactivation of IFN-inducible genes, thereby linking IFN signaling at the cell surface with the production of antiviral proteins and generation of an effective antiviral response. Therefore, the low level of ISGF3 complexes probably hinders this response in tumor cells. We performed gene expression analysis of IFN-stimulated cells to examine the consequences of low levels of ISGF3 for the production of IFN-stimulated gene products. The percentages of genes detected in the two cell types were similar, regardless of treatment: 50.3% for untreated HT-1080 cells, 47.2% for IFN-treated HT-1080 cells, 46.5% for untreated CCD-1122Sk cells, and 46.9% for IFN-treated CCD-1122Sk cells. Most of the transcripts were either unchanged or changed less than twofold after IFN stimulation. There were 399 gene transcripts that were differentially regulated with a twofold or greater change in expression. The number of genes that were differentially expressed was 356 in CCD-1122Sk cells and 91 in HT-1080 cells, with a 48-gene overlap (Table 3).
TABLE 3.
Genes differentially regulated in normal cells and tumor cells treated with IFN-β
| Cell line | No. of genes
|
No. of IFN-β-regulated genesa
|
|||
|---|---|---|---|---|---|
| Unique | Overlapping | Total | Downregulated | Upregulated | |
| CCD-1122Sk | 308 | 48 | 356 | 40 | 316 |
| HT-1080 | 43 | 48 | 91 | 10 | 81 |
A twofold or greater difference in the signal intensity of a gene after IFN-β treatment was required for the gene's expression to be considered changed.
When we compared the signal intensities of differentially regulated genes by using hierarchical clustering (data not shown), we found that several clusters were upregulated in normal cells but not in tumor cells. The 48 overlapping genes showed a 3- to 200-fold upregulation in CCD-1122Sk cells and only a 2- to 10-fold upregulation in HT-1080 cells. To better compare each gene in this group, we calculated the ratios of fold change in signal intensity of each gene in normal cells with that in tumor cells. The ratios ranged from 2 to 62; thus, the number and intensity of the differentially expressed genes were significantly higher in normal cells than in tumor cells. A partial list of these 48 gene transcripts and their magnitude of upregulation are presented in Table 4. Almost all of the gene transcripts were known ISG products that have antiviral, growth-inhibitory, apoptosis-inducing, and immune response functions.
TABLE 4.
Partial list of genes that are differentially regulated in both CCD-1122Sk and HT-1080 cells after treatment with IFN-βa
| Gene | Description or product | Ratio of fold changesb | Function(s) | Unigene cluster identification no. | GenBank accession no. |
|---|---|---|---|---|---|
| ISG 20 | IFN-stimulated gene, 20 kDa | 62.4 | Antiproliferative | NM_002201.2 | Hs.105434 |
| OAS1 | 2′,5′-Oligoadenylate synthetase 1 | 47.8 | Antiviral | NM_016816.1 | Hs.105434 |
| Cig5 | Viperin | 47.3 | Antiviral | AI337069 | Hs.17518 |
| C1orf29 | Chromosome 1 open reading frame 29 | 42.7 | Unknown | NM_006820.1 | Hs.389724 |
| GIP2 | IFN-α-inducible protein | 17.4 | Non-MHCc-restricted cytotoxicity | NM_005101.1 | Hs.458485 |
| Mx1 | Myxovirus resistance 1 | 17.0 | Antiviral | NM_002462.1 | Hs.436836 |
| IFIT1 | IFN-induced protein with tetratricopeptide repeats 1 | 16.1 | Unknown | NM_001548.1 | Hs.20315 |
| TNFSF10 | Tumor necrosis factor superfamily 10 | 12.9 | Apoptosis | NM_003810.1 | Hs.387871 |
| OAS2 | 2′-5′-Oligoadenylate synthetase 2, 69/71 kDa | 12.8 | Antiviral | NM_016817.1 | Hs.414332 |
| IFI27 | IFN-α-inducible protein 27 | 11.8 | Immune response and IFN signaling | NM_005532.1 | Hs.278613 |
| IFIT4 | IFN-induced protein with tetratricopeptide repeats 4 | 11.2 | Antiproliferative | NM_006187.1 | Hs.181874 |
| OAS3 | 2′-5′-Oligoadenylate synthetase 3, 100 kDa | 10.1 | Antiviral | NM_006187.1 | Hs.56009 |
| RIG-I | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide | 10.0 | Innate immune response | NM_014314.1 | Hs.438386 |
| MDA5 | Melanoma differentiation-associated protein 5 | 9.2 | Growth inhibition/apoptosis | NM_022168.1 | Hs.389539 |
| CEB1 | Cyclin E binding protein 1 | 8.4 | Regulator of CDK activity | NM_016323.1 | Hs.26663 |
| TLR3 | Toll-like receptor 3 | 7.5 | Innate antiviral response | NM_003265.1 | Hs.29499 |
| OASL | 2′-5′-Oligoadenylate synthetase like | 7.3 | Antiviral | AF_063612.1 | Hs.118633 |
| USP18 | Ubiquitin-specific protease 18 | 7.1 | Ubiquitin-specific protease activity | NM_017414.1 | Hs.38260 |
| GIP3 | IFN-α-inducible protein | 7.0 | Unknown | NM_022873.1 | Hs.287721 |
| MX2 | Myxovirus resistance 2 (mouse) | 6.8 | Antiviral | NM_002463.1 | Hs.926 |
This is a partial list of 48 overlapping genes that showed upregulation in the two cell types.
This ratio is a calculation of the fold change in the CCD 1122Sk transcript to that seen in the HT-1080 transcript.
MHC, major histocompatibility complex.
DISCUSSION
The effectiveness of NDV against tumors and its safety in normal tissue have been clearly proven in several clinical studies (8, 42, 49). Here we demonstrated for the first time that the spread of NDV is the primary differentiating factor that controls the final outcome of infection in normal cells and tumor cells. This finding has raised two key questions. First, do NDV-infected normal cells secrete a factor(s) that protects uninfected cells from further spread of the virus? Second, do NDV-infected tumor cells secrete the same factor(s), and if so, are they refractory to its effect? The spread of NDV was indeed inhibited in primary human skin fibroblast (CCD-1122Sk) cells as a result of secreted factors. By performing antibody neutralization, we showed that this secreted protective factor is predominantly IFN-β.
The antiviral effect of IFN was discovered in vitro in 1957 (22). Evidence that endogenous IFN has an antiviral effect was shown in mice treated with antibodies against IFN (18). Also, exogenous IFN-α/β protects animals and humans against several viruses (18). Previous studies have shown that large amounts of IFN are produced by various mammalian cell types when infected with NDV (23-25, 29, 31, 52, 56). Τhe secretion of IFN by the infected CCD-1122Sk cells prepared other uninfected cells for the viral attack, probably by stimulating the formation of anti-NDV proteins in those uninfected cells. We also showed that IFN was present in the supernatant of infected HT-1080 cells in culture but was ineffective in preventing the infection and spread of NDV. The HT-1080 cells were resistant to both secreted IFN and exogenous IFN-β treatment; therefore, NDV susceptibility in these cells probably results from impairment of the direct antiviral effects of IFN. In contrast, the uninfected CCD-1122Sk cells exposed to secreted IFN were immune to any incoming virus.
We then examined the responsiveness of uninfected HT-1080 cells and CCD-1122Sk cells to IFN by analyzing the expression and activation of components in the IFN signal transduction pathway, including the type I receptor-associated tyrosine kinases JAK-1 and TYK-2, components of the ISGF3 complex, and the level of the ISGF3 complex itself in the nucleus. Our data revealed no significant differences in the activation of the JAK1 and TYK2 kinases between the two cell lines. The activities of these kinases are modulated by the relative level of phosphorylation (5). Expression and activation of the kinases in the HT-1080 cells indicated that neither binding of IFN to the receptor nor subsequent oligomerization of the receptor subunit is probably inhibited.
Examination of the expression and activation of IFN signaling components revealed a marked reduction in the levels of phosphorylated STAT1 and STAT2 in IFN-β-resistant HT-1080 cells. Phosphorylation of Tyr701 in the STAT1 protein is mandatory for dimerization, nuclear translocation, and DNA binding (19). Also, the STAT2 transactivation domain containing phosphorylated Tyr690 provides the ISGF3 transactivation function (27, 46). As a result of these defects, HT-1080 cells also had deficiencies in their ISGF3 complexes. Previous studies have shown that mice deficient in STAT1 or STAT2 are susceptible to infection by a broad range of RNA and DNA viruses, including vesicular stomatitis virus, encephalomyocarditis virus, and herpes simplex virus (14, 21, 32, 33). In another study, a lethal viral disease in two infants was found to be a direct consequence of STAT1 deficiency (13). In cells lines derived from the two infants, the STAT1-containing ISGF3 complex was not activated and IFN did not inhibit virus replication. Thus, impairment of the STAT1-dependent response to IFN-α/β results in susceptibility to viral disease.
Given the marked reduction of these signal transduction proteins in IFN-resistant cells, it follows that in our study the transcription-mediated response to IFN would be severely affected. The number and intensity of expression of ISG transcripts, as analyzed by microarray, were significantly lower in HT-1080 cells than in CCD-1122Sk cells. The cellular outcome of viral infection of cells is ultimately dictated by ISG induction via activation of transcription at the ISRE. The IFN resistance of HT-1080 cells and the inability of these cells to prevent the spread of NDV may be attributed to the cellular lesions in their IFN signal transduction pathway and their deficient production of anti-NDV proteins.
The molecular basis for the reduction in the cellular levels of phosphorylated STAT1 and STAT2 in HT-1080 cells remains to be determined. Because the levels of STAT proteins were comparable in the two cell types but that of the phosphorylated forms was not, several possibilities must be investigated. One is a loss-of-function mutation in the alpha or beta subunit of the IFN receptor or in the JAK kinases of the tumor cells. Another possibility is a gain-of-function mutation in one of the suppressor of cytokine signaling proteins (26).
Microarray data revealed that HT-1080 cells produced fewer ISG transcripts than did CCD-1122Sk cells. Several of the ISG transcripts that were upregulated in normal cells are important for antiviral and innate immune responses (Table 4). The ISG product(s) that functions as an antiviral protein(s) against NDV is not known; hence, we cannot confirm any direct effect of the antiviral proteins on the prevention of NDV infection and spread. Additionally, IFNs activate other pathways that are STAT independent, including the insulin receptor substrate-phosphatidylinositol 3′-kinase cascade, the CBL-CrkL pathway, and the mitogen-activated protein kinase pathway (28). Some of the effectors in these pathways, such as the p38 mitogen-activated protein kinase, may play a crucial role in the induction of antiviral effects.
Defects in IFN response occur in tumor cells (1, 11, 38, 44, 50, 53-55). These lesions usually reside in the components of the IFN response pathway such as STAT1. Several melanoma and lymphoma cell lines contain reduced levels of STAT1 (38, 50). In our study, deficiency in STAT1 and STAT2 phosphorylation was observed in two of the three tumor cell lines examined (data not shown). These findings indicate that the IFN pathway is defective in most transformed cell lines and that this defect contributes to growth dysregulation in cancers and a concomitant loss of antiviral host defense. This brings up the question of whether other viruses can exploit these defects to facilitate oncolysis.
Vesicular stomatitis virus causes oncolysis that is possibly mediated by loss of the antiviral response (2, 37, 51). Many viruses have evolved strategies to evade IFN-induced antiviral responses (17, 45). However, some paramyxoviruses and influenza viruses inhibit the IFN response only in a species-specific manner (3, 12, 39). The V protein in NDV provides IFN antagonist activity, but only in avian hosts (20, 40, 41). This finding suggests that the V protein cannot counteract the IFN response in human cells; thus, NDV fails to replicate in IFN-sensitive cell lines. However, host restriction is of limited consequence in tumor cells that are IFN resistant. Among influenza virus infections in which the NS1 protein counteracts the IFN-mediated antiviral response (16), only modified viruses like the NS1-deleted mutants cause oncolysis and fail to replicate in normal cells (34). The advantage of using NDV is that unmodified viruses can perform both of these functions because the host restriction of V protein prevents replication and spread in normal cells, and the IFN resistance in tumor cells allows NDV growth and oncolysis. Here we have shown that all of the NDV strains tested in this study selectively replicate and grow in tumor cells.
In summary, our comparison of ISG transcript production in an IFN-resistant, NDV-susceptible tumor cell line and an IFN-sensitive, NDV-resistant normal cell line revealed that tumor cells have reduced levels of phosphorylated STAT proteins and ISGF3 complexes. In addition, the less-than-optimum response of ISGs in the HT-1080 cells might explain their high susceptibility to NDV infection. These results indicate the need for further investigation into the oncolytic nature of NDV and the anti-NDV response in cells.
Acknowledgments
We acknowledge Geoffrey Neale and the Hartwell Center at St. Jude Children's Research Hospital for conducting microarray experiment and analyzing the data and Peter Murray for helpful suggestions.
This work was supported by Public Health Service grants AI38956 and AI055940 (to T.T.) from the National Institute of Allergy and Infectious Diseases and by American Lebanese Syrian Associated Charities.
REFERENCES
- 1.Abril, E., R. E. Mendez, A. Garcia, A. Serrano, T. Cabrera, F. Garrido, and F. Ruiz-Cabello. 1996. Characterization of a gastric tumor cell line defective in MHC class I inducibility by both alpha- and gamma-interferon. Tissue Antigens 47:391-398. [DOI] [PubMed] [Google Scholar]
- 2.Barber, G. N. 2005. VSV-tumor selective replication and protein translation. Oncogene 24:7710-7719. [DOI] [PubMed] [Google Scholar]
- 3.Basler, C., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus non-structural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. USA 98:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell, J. C., K. A. Garson, B. D. Lichty, and D. F. Stojdl. 2002. Oncolytic viruses: programmable tumor hunters. Curr. Gene Ther. 2:243-254. [DOI] [PubMed] [Google Scholar]
- 5.Briscoe, J., N. C. Rogers, B. A. Witthuhn, D. Watling, A. G. Harpur, A. F. Wilks, G. R. Stark, J. N. Ihle, and I. M. Kerr. 1996. Kinase negative mutants of JAK1 can sustain interferon-gamma-inducible gene expression but not an antiviral state. EMBO J. 15:799-809. [PMC free article] [PubMed] [Google Scholar]
- 6.Bray, M. 2001. The role of the type I interferon response in the resistance of mice to filovirus infection. J. Gen. Virol. 82:1365-1373. [DOI] [PubMed] [Google Scholar]
- 7.Cassel, W. A., and R. E. Garrett. 1965. Newcastle disease virus as an antineoplastic agent. Cancer 18:863-868. [DOI] [PubMed] [Google Scholar]
- 8.Cassel, W. A., and D. R. Murray. 1992. A ten-year follow-up on stage II malignant melanoma patients treated postsurgically with Newcastle disease virus oncolysate. Med. Oncol. Tumor Pharmacother. 9:169-171. [DOI] [PubMed] [Google Scholar]
- 9.Cassel, W. A., D. R. Murray, and H. S. Phillips. 1983. A phase II study on the postsurgical management of stage II malignant melanoma with a Newcastle disease virus oncolysate. Cancer 52:856-860. [DOI] [PubMed] [Google Scholar]
- 10.Chiocca, E. A. 2002. Oncolytic viruses. Nat. Rev. Cancer 2:938-950. [DOI] [PubMed] [Google Scholar]
- 11.Diaz, M. O., S. Ziemin, M. M. Le Beau, P. Pitha, S. D. Smith, R. R. Chilcote, and J. D. Rowley. 1988. Homozygous deletion of the α1- and β1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. USA 85:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupuis, S., E. Jouanguy, S. Al-Hajjar, C. Fieschi, I. Z. Al-Mohsen, S. Al-Jumaah, K. Yang, A. Chapgier, C. Eidenschenk, P. Eid, A. Al Ghonaium, H. Tufenkeji, H. Frayha, S. Gazlan, H. Al-Rayes, R. D. Schreiber, I. Gresser, and J. L. Casanova. 2003. Impaired response to interferon-α/β and lethal viral disease in human STAT 1 deficiency. Nat. Genet. 33:388-391. [DOI] [PubMed] [Google Scholar]
- 14.Durbin, J. E., R. Hackenmiller, M. C. Simon, and D. E. Levy. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443-450. [DOI] [PubMed] [Google Scholar]
- 15.Freeman, A. I., Z. Zakay-Rones, J. M. Gomori, E. Linetsky, L. Rasooly, E. Greenbaum, S. Rozenman-Yair, A. Panet, E. Libson, C. S. Irving, E. Galun, and T. Siegal. 2006. Phase I/II trial of intravenous NDV-HUJ oncolytic virus in recurrent glioblastoma multiforme. Mol. Ther. 13:221-228. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 17.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2002. Paramyxovirus strategies for evading the interferon response. Rev. Med. Virol. 12:337-357. [DOI] [PubMed] [Google Scholar]
- 18.Gresser, I. 1984. Role of interferon resistance to viral infection in vivo, p. 221-247. In J. Vilcek and E. De Maeyer (ed.), Interferons and the immune system. Elsevier Science Publishers B.V., Amsterdam, The Netherlands.
- 19.Horvath, C. M., and J. E. Darnell, Jr. 1996. The antiviral state induced by alpha interferon and gamma interferon requires transcriptionally active Stat1 proteins. J. Virol. 70:647-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, Z., S. Krishnamurthy, A. Panda, and S. K. Samal. 2003. Newcastle disease virus V protein is associated with viral pathogenesis and functions as an alpha interferon antagonist. J. Virol. 77:8676-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Improta, T., and R. Pine. 1997. Susceptibility to virus infection is determined by a STAT-mediated response to the autocrine effect of virus-induced type I interferon. Cytokine 9:383-393. [DOI] [PubMed] [Google Scholar]
- 22.Isaacs, A., and Lindenmann. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147:258-267. [PubMed] [Google Scholar]
- 23.Ito, Y., Y. Nagai, and K. Maeno. 1982. Interferon production in mouse spleen cells and mouse fibroblasts (L cells) stimulated by various strains of Newcastle disease virus. J. Gen. Virol. 62:349-352. [DOI] [PubMed] [Google Scholar]
- 24.Jameson, P., and S. E. Grossberg. 1979. Production of interferon by human tumor cells. Arch. Virol. 62:209-219. [DOI] [PubMed] [Google Scholar]
- 25.Kohase, M., and J. Vilcek. 1979. Interferon induction with Newcastle disease virus in FS4-cells: effect of priming with interferon and of virus inactivating treatments. Jpn. J. Med. Sci. Biol. 32:281-294. [DOI] [PubMed] [Google Scholar]
- 26.Krebs, D. L., and D. J. Hilton. 2001. SOCS proteins: negative regulators of cytokine signaling. Stem Cells 19:378-387. [DOI] [PubMed] [Google Scholar]
- 27.Leung, S., S. A. Qureshi, I. M. Kerr, J. E. Darnell, Jr., and G. R. Stark. 1995. Role of STAT2 in the alpha interferon signaling pathway. Mol. Cell. Biol. 15:1312-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, Y., K. K. Srivastava, and L. C. Platanias. 2004. Mechanisms of type I interferon signaling in normal and malignant cells. Arch. Immunol. Ther. Exp. 52:156-163. [PubMed] [Google Scholar]
- 29.Lomniczi, B. 1973. Studies on interferon production and interferon sensitivity of different strains of Newcastle disease virus. J. Gen. Virol. 21:305-313. [DOI] [PubMed] [Google Scholar]
- 30.Lorence, R. M., P. A. Rood, and K. W. Kelley. 1988. Newcastle disease virus as an antineoplastic agent: induction of tumor necrosis factor-α and augmentation of its cytotoxicity. J. Natl. Cancer Inst. 80:1305-1312. [DOI] [PubMed] [Google Scholar]
- 31.Marcus, P. I., C. Svitlik, and M. J. Sekellick. 1983. Interferon induction by viruses. X. A model for interferon induction by Newcastle disease virus. J. Gen. Virol. 64:2419-2431. [DOI] [PubMed] [Google Scholar]
- 32.Meraz, M. A., J. M. White, K. C. Sheehan, E. A. Bach, S. J. Rodig, A. S. Dighe, D. H. Kaplan, J. K. Riley, A. C. Greenlund, D. Campbell, K. Carver-Moore, R. N. DuBois, R. Clark, M. Aguet, and R. D. Schreiber. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84:431-442. [DOI] [PubMed] [Google Scholar]
- 33.Mrkic, B., J. Pavlovic, T. Rulicke, P. Volpe, C. J. Buchholz, D. Hourcade, J. P. Atkinson, A. Aguzzi, and R. Cattaneo. 1998. Measles virus spread and pathogenesis in genetically modified mice. J. Virol. 72:7420-7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muster, T., J. Rajtarova, M. Sachet, H. Unger, R. Fleischhacker, I. Romirer, A. Grassauer, A. Url, A. Garcia-Sastre, K. Wolff, H. Pehamberger, and M. Bergmann. 2004. Interferon resistance promotes oncolysis by influenza virus NS-1 deletion mutants. Int. J. Cancer 110:15-21. [DOI] [PubMed] [Google Scholar]
- 35.Nelson, N. J. 1999. Scientific interest in Newcastle disease virus is reviving. J. Natl. Cancer Inst. 91:1708-1710. [DOI] [PubMed] [Google Scholar]
- 36.Nemunaitis, J. 2002. Live viruses in cancer treatment. Oncology 16:1483-1492. [PubMed] [Google Scholar]
- 37.Obuchi, M., M. Fernandez, and G. N. Barber. 2003. Development of recombinant vesicular stomatitis viruses that exploit defects in host defense to augment specific oncolytic activity. J. Virol. 77:8843-8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pansky, A., P. Hildeband, E. Fasler-Kan, L. Baselgia, S. Ketterer, C. Beglinger, and M. H. Heim. 2000. Defective Jak-STAT signal transduction pathway in melanoma cells resistant to growth inhibition by interferon-α. Int. J. Cancer 85:720-725. [DOI] [PubMed] [Google Scholar]
- 39.Parisien, J. P., J. F. Lau, and C. M. Horvath. 2002. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J. Virol. 76:6435-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, M., A. Garcia-Sastre, J. F. Cros, C. F. Basler, and P. Palese. 2003. Newcastle disease virus V protein is a determinant of host range restriction. J. Virol. 77:9522-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pecora, A. L., N. Rizvi, G. I. Cohen, N. L. Meropol, D. Sterman, J. L. Marshall, S. Goldberg, P. Gross, J. D. O'Neil, W. S. Groene, M. S. Roberts, H. Rabin, M. K. Barmat, and R. M. Lorence. 2002. Phase I trial of intravenous administration of PV701, an oncolytic virus, in patients with advanced solid cancers. J. Clin. Oncol. 20:2251-2266. [DOI] [PubMed] [Google Scholar]
- 43.Phuangsab, A., R. M. Lorence, K. W. Reichard, M. E. Peeples, and R. J. Walter. 2001. Newcastle disease virus therapy of human tumor xenografts: antitumor effects of local or systemic administration. Cancer Lett. 172:27-36. [DOI] [PubMed] [Google Scholar]
- 44.Pitha, P. M. 2000. Introduction: interferon's connection to cancer. Semin. Cancer Biol. 10:69-72. [DOI] [PubMed] [Google Scholar]
- 45.Ploegh, H. L. 1998. Viral strategies of immune evasion. Science 280:248-253. [DOI] [PubMed] [Google Scholar]
- 46.Qureshi, S. A., S. Leung, I. M. Kerr, G. R. Stark, and J. E. Darnell, Jr. 1996. Function of Stat2 protein in transcriptional activation by alpha interferon. Mol. Cell. Biol. 16:288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reichard, K. W., R. M. Lorence, C. J. Cascino, M. E. Peeples, R. J. Walter, M. B. Fernando, H. M. Reyes, and J. A. Greager. 1992. Newcastle disease virus selectively kills human tumor cells. J. Surg. Res. 52:448-453. [DOI] [PubMed] [Google Scholar]
- 48.Schirrmacher, V., A. Griesbach, and T. Ahlert. 2001. Antitumor effects of Newcastle disease virus in vivo: local versus systemic effects. Int. J. Oncol. 18:945-952. [DOI] [PubMed] [Google Scholar]
- 49.Schirrmacher, V., and R. Heicappell. 1987. Prevention of metastatic spread by postoperative immunotherapy with virally modified autologous tumor cells. II. Establishment of specific systemic anti-tumor immunity. Clin. Exp. Metastasis 5:147-156. [DOI] [PubMed] [Google Scholar]
- 50.Sun, W. H., C. Pabon, Y. Alsayed, P. P. Huang, S. Jandeska, S. Uddin, L. C. Platanias, and S. T. Rosen. 1998. Interferon-α resistance in a cutaneous T-cell lymphoma cell line is associated with lack of STAT1 expression. Blood 91:570-576. [PubMed] [Google Scholar]
- 51.Stojdl, D. F., B. Lichty, S. Knowles, R. Marius, H. Atkins, N. Sonenberg, and J. C. Bell. 2000. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 6:821-825. [DOI] [PubMed] [Google Scholar]
- 52.Washburn, B., and V. Schirrmacher. 2002. Human tumor cell infection by Newcastle disease virus leads to upregulation of HLA and cell adhesion molecules and to induction of interferons, chemokines and finally apoptosis. Int. J. Oncol. 21:85-93. [DOI] [PubMed] [Google Scholar]
- 53.Wellbrock, C., C. Weisser, J. C. Hassel, P. Fischer, J. Becker, C. S. Vetter, I. Behrmann, M. Kortylewski, P. C. Heinrich, and M. Schartl. 2005. STAT5 contributes to interferon resistance of melanoma cells. Curr. Biol. 15:1629-1639. [DOI] [PubMed] [Google Scholar]
- 54.Wong, L. H., K. G. Krauer, I. Hatzinisiriou, M. J. Estcourt, P. Hersey, N. D. Tam, S. Edmondson, R. J. Devenish, and S. J. Ralph. 1997. Interferon-resistant human melanoma cells are deficient in ISGF3 components, Stat1, Stat2, and p48-ISGF3γ. J. Biol. Chem. 272:28779-28785. [DOI] [PubMed] [Google Scholar]
- 55.Xu, B., D. Grander, O. Sangfelt, and S. Einhorn. 1994. Primary leukemia cells resistant to α-interferon in vitro are defective in the activation of the DNA-binding factor interferon-stimulated gene factor 3. Blood 84:1942-1949. [PubMed] [Google Scholar]
- 56.Yeow, W.-S., M. W. Beilharz, and C. May Lai. 1997. The In vitro expression patterns of individual type I interferon genes in Newcastle disease virus infected murine splenocytes and fibroblasts. Int. J. Biochem. Cell. Biol. 29:513-520. [DOI] [PubMed] [Google Scholar]