Abstract
The Ebola virus (EBOV) VP35 protein blocks the virus-induced phosphorylation and activation of interferon regulatory factor 3 (IRF-3), a transcription factor critical for the induction of alpha/beta interferon (IFN-α/β) expression. However, the mechanism(s) by which this blockage occurs remains incompletely defined. We now provide evidence that VP35 possesses double-stranded RNA (dsRNA)-binding activity. Specifically, VP35 bound to poly(rI) · poly(rC)-coated Sepharose beads but not control beads. In contrast, two VP35 point mutants, R312A and K309A, were found to be greatly impaired in their dsRNA-binding activity. Competition assays showed that VP35 interacted specifically with poly(rI) · poly(rC), poly(rA) · poly(rU), or in vitro-transcribed dsRNAs derived from EBOV sequences, and not with single-stranded RNAs (ssRNAs) or double-stranded DNA. We then screened wild-type and mutant VP35s for their ability to target different components of the signaling pathways that activate IRF-3. These experiments indicate that VP35 blocks activation of IRF-3 induced by overexpression of RIG-I, a cellular helicase recently implicated in the activation of IRF-3 by either virus or dsRNA. Interestingly, the VP35 mutants impaired for dsRNA binding have a decreased but measurable IFN antagonist activity in these assays. Additionally, wild-type and dsRNA-binding-mutant VP35s were found to have equivalent abilities to inhibit activation of the IFN-β promoter induced by overexpression of IPS-1, a recently identified signaling molecule downstream of RIG-I, or by overexpression of the IRF-3 kinases IKKɛ and TBK-1. These data support the hypothesis that dsRNA binding may contribute to VP35 IFN antagonist function. However, additional mechanisms of inhibition, at a point proximal to the IRF-3 kinases, most likely also exist.
Activation of alpha/beta interferon (IFN-α/β) production is a key step in the innate immune response to viral infection. Double-stranded RNA (dsRNA) has long been used as an experimental inducer of IFN-α/β and is potentially synthesized during the replication of many viruses. Thus, viral dsRNA has been hypothesized to be a trigger of cellular antiviral responses (29). A number of cellular dsRNA recognition proteins have been implicated in the IFN-induced antiviral response to infection. These include the dsRNA-dependent protein kinase PKR, the 2′,5′-oligoadenylate synthase, and ADAR1 (20, 44, 59, 63). More recently, two IFN-induced, caspase recruiting domain (CARD)-containing, DExD/H family helicases, the retinoic acid-inducible gene I (RIG-I) protein and the melanoma differentiation-associated gene 5 (MDA-5) protein, have been implicated as key sensors of viral infection (1, 30, 56, 66, 67). These proteins are activated by viral infection, possibly through recognition of dsRNA or of ribonucleoprotein complexes produced during infection, and transduce downstream signaling to activate the IFN-α/β responses (34).
The transcription factor interferon regulatory factor 3 (IRF-3) plays a critical role in the activation of the IFN-α/β gene. A cytoplasmic protein in its inactive state, IRF-3 becomes hyperphosphorylated on serine and threonine residues, dimerizes, and accumulates in the nucleus, where it participates in initial IFN-α/β gene expression (37, 68, 69). RIG-I and MDA-5 activate IRF-3 in response to dsRNA or to viral infection upstream of the IRF-3 kinases TBK-1 and IKKɛ (18, 30, 54, 66). This signaling appears to involve the homotypic interaction of the CARDs of the helicases with another CARD-containing protein termed alternatively IPS-1, MAVS, VISA, or Cardif (31, 41, 53, 65).
Viruses have evolved a variety of mechanisms to avoid recognition or to block the antiviral responses mediated by IFNs (2, 19). Several such proteins also exhibit dsRNA-binding activity. Examples of dsRNA-binding proteins that counteract cellular antiviral responses include the NS1 proteins of influenza A (NS1A) and B (NS1B) viruses, the E3L protein of vaccinia virus, the σ3 protein of reovirus, and the pTRS1 protein of human cytomegalovirus (5, 11, 12, 14, 16, 17, 22, 27, 57). Although dsRNA binding appears to contribute to IFN antagonist function, NS1A, NS1B, and E3L possess additional dsRNA-binding-independent mechanisms to inhibit the cellular antiviral response.
The Ebola virus (EBOV) VP35 protein has been found to inhibit IFN-α/β production and the activation of IRF-3 (3, 4, 9, 23, 48). Recently, it was suggested that VP35 may contain a dsRNA-binding motif similar to that found in the NS1 protein of influenza A virus (23). It was further suggested that this dsRNA-binding activity may be required for the ability of VP35 to inhibit IFN production (23). Here, we provide evidence that VP35 has dsRNA-binding activity that may contribute to IFN antagonism. It is likely that VP35 also possesses a dsRNA-binding-independent mechanism(s) that targets a point at or downstream of the IRF-3 kinases TBK-1 and IKKɛ.
MATERIALS AND METHODS
Reagents.
CNBr-activated Sepharose 4B and polyribonucleotide poly(rI) · poly(rC) (pIC) were obtained from Amersham Biosciences, United Kingdom. Rabbit anti-hIRF-3 (sc-9082) was from Santa Cruz Biotechnology (California). Sheep polyclonal anti-human IFN-β was from PBL Biomedical Laboratories (New Jersey). Monoclonal anti-FLAG (M2), poly(rA) · poly(rU) (pAU), poly(rU), and poly(rA) were from Sigma (Missouri). Monoclonal anti-Ebola virus Zaire VP35 N-terminus (6C5) and C-terminus (17C6) antibodies were generated in collaboration with the Mount Sinai Hybridoma Center. Recombinant human IFN-β was from Calbiochem (San Diego, CA).
Cell lines and virus.
HEK293, 293T, and Vero cells were maintained in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml). All cell lines were cultured at 37°C and 5% CO2. Sendai virus Cantell (SeV) and a Newcastle disease virus expressing green fluorescent protein (NDV-GFP), described previously (46), were grown in 10-day-old embryonated chicken eggs for 2 days at 37°C.
Plasmids.
The Ebola virus Zaire VP35 open reading frame (ORF) was subcloned from a pcDNA3 expression vector, described elsewhere (4), into the mammalian expression vector pCAGGS (45). The VP35 point mutants R312A and K309A were generated by site-directed mutagenesis with the Quik-Change-XL kit (Stratagene). The RIG-I ORF was subcloned from a pEF-BOS vector (56) into pCAGGS, and an N-terminal FLAG tag sequence was added during cloning. IPS-1 (IFN-β promoter stimulator 1) was PCR amplified from a human 5751684 pCMV-SPORT6 clone obtained from the ATCC and cloned into pCAGGS with an amino-terminal hemagglutinin tag. Plasmids encoding human FLAG-tagged TBK-1 and IKKɛ were kindly provided by John Hiscott (McGill University) and have been described previously (54). The reporter vectors pHISG-54-CAT (CAT, chloramphenicol acetyltransferase) and pIFN-β-CAT have been described previously (8, 61). Firefly luciferase cloned into pCAGGS was used as a transfection control.
Reporter gene assays.
293T cells (1.6 × 106) were transfected with the indicated amount of expression plasmid DNA together with the ISG54-CAT or IFN-β-CAT reporter plasmids by the calcium phosphate precipitation method (51). Twenty-four h posttransfection, cells were lysed or infected with SeV (multiplicity of infection [MOI] of 8) for 1 h. Twelve h postinfection, cells were lysed with reporter lysis buffer (Promega) and CAT activities were determined (51). Firefly luciferase activity was assayed as recommended by the manufacturer (Promega) and was used to normalize CAT activity. Reporter gene activation is expressed as induction (fold) over an uninduced, empty-vector-transfected control.
Interferon bioassay.
Conditioned media (100 μl) from 293T-transfected, SeV-infected cells were UV treated to inactivate infectious SeV and overlaid onto Vero cells seeded in a 96-well black microtiter plate (Costar, Corning, NY). After 24 h of treatment, the Vero cells were infected with the IFN-sensitive virus NDV-GFP for 1 h at an MOI of 6. Twenty-four h postinfection, viral replication was assessed by fluorescence microscopy and GFP fluorescence was quantified in a FLUOstar OPTIMA plate reader (BMG Labtechnologies, North Carolina) set with excitation and emission wavelengths at 485 and 530 nm, respectively. To verify the presence of IFN-β, a neutralizing anti-hIFN-β was added (at a final concentration of ∼10 μg/ml) to the indicated conditioned medium 30 min before it was overlaid onto Vero cells. Recombinant hIFN-β was added directly to Vero cells in a series of twofold dilutions 24 h prior to NDV-GFP infection to induce antiviral effect.
Poly(rI) · poly(rC)-Sepharose coprecipitation and Western blotting.
293T cells were transfected by the calcium phosphate precipitation method with the indicated plasmids. Twenty-four h posttransfection cells were lysed in 500 μl of lysis buffer (50 mM Tris, pH 8.0, 280 mM NaCl, 1% Igepal, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol), supplemented with 1 mM orthovanadate, 1 mM dithiothreitol, and a cocktail of protease inhibitors (Roche). Clarified cell lysates were mixed with the indicated amount of pIC-Sepharose suspension, previously washed with phosphate-buffered saline. Beads and cell lysates were incubated for 2 h at 4°C with gentle agitation. Then beads were washed 10 times with lysis buffer, and precipitated proteins were separated by 12% polyacrylamide gel electrophoresis (PAGE) under denaturing and reducing conditions. After electrophoresis, proteins were transferred to a polyvinylidene difluoride membrane, blocked in 5% nonfat dry milk dissolved in Tris-buffered saline, and then probed with the indicated primary antibody. Secondary antibodies conjugated to horseradish peroxidase and a chemiluminescence detection system (Perkin-Elmer) were used to visualize the antigens. To assess the specificity of dsRNA binding, cell lysates were mock treated or incubated with soluble competitors including pIC, pAU, in vitro-generated dsRNA, poly(rU), poly(rA), and double-stranded DNA (dsDNA) before pIC-Sepharose coprecipitation. Precise molar concentrations of these synthetic RNAs are not available, because the manufacturers do not provide information regarding their length. In vitro-transcribed dsRNAs of 200, 400, 600, 800, and 1,000 bp were also used as competitors. These dsRNAs were generated with the T7 RiboMax Express RNAi system (Promega) using PCR fragments amplified from an EBOV VP35 cDNA. Forward PCR primers were designed with a 5′-T7 promoter sequence followed by the first 20 nucleotides of the VP35 ORF. Reverse primers annealed at different positions of the VP35 ORF to generate PCR products of the desired length.
Bacterially produced VP35.
The carboxy-terminal 171 amino acids of the Zaire Ebola virus VP35 were expressed with a carboxy-terminal six-histidine tag, from pET22b(+) in Escherichia coli Origami B host strains. Bacterially produced protein was purified from cell lysates with a Talon Cobalt metal affinity bead column. After loading, the column was washed with HEPES-buffered saline containing 17 mM imidazole to remove any contaminants and then eluted with HEPES-buffered saline containing 250 mM imidazole.
IRF-3 dimer formation.
To assess the activation of endogenous IRF-3, we performed a native PAGE that resolves IRF-3 monomers and dimers, as described previously (42). HEK293 cells were transfected with the indicated plasmids using Lipofectamine 2000 (Invitrogen). Twenty-four h posttransfection, cells were infected with SeV (MOI of 8) for 1 h. Eight h postinfection, cells were lysed in 50 μl of 50 mM Tris-HCl, pH 8.0, 1% Igepal, 150 mM NaCl, supplemented with 5 mM orthovanadate and a cocktail of protease inhibitors (Roche). Lysates were clarified by centrifugation and loaded onto a 7.5% native polyacrylamide gel that had been prerun for 30 min at 40 mA, with and without 0.2% sodium deoxycholate in the cathode and anode chambers, respectively. The samples were electrophoresed at 25 mA for 50 min. Western blot assays were performed as previously described. Rabbit anti-hIRF-3 antibody was used at a 1:500 dilution.
RESULTS
VP35 binds specifically to dsRNA.
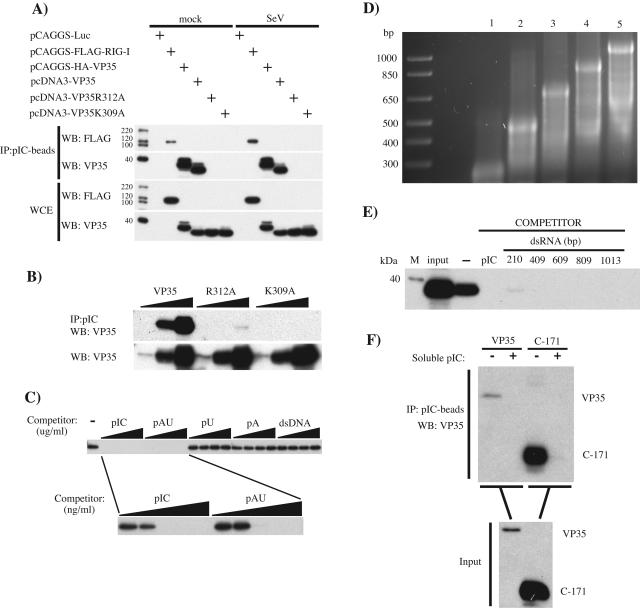
Wild-type VP35 was previously hypothesized to bind dsRNA, and two VP35 point mutants, R312A and K309A, were predicted to be impaired in dsRNA binding and to impair IFN antagonist function (23). We therefore assessed the ability of these VP35s to bind to dsRNA with an in vitro “pull-down” assay. 293T cells were transfected with hemagglutinin-tagged VP35, untagged wild-type VP35, or untagged VP35 mutant (R312A or K309A) expression plasmids. Also included as controls were plasmids encoding firefly luciferase or a previously described dsRNA-binding protein, RIG-I (67). Lysates from the transfected cells were then incubated with pIC-Sepharose beads with a lysate/beads ratio of 0.05. As shown in Fig. 1A, RIG-I and wild-type VP35 coprecipitated with pIC beads, whereas neither R312A nor K309A detectably bound to the pIC beads. To address the possibility that activation of the IFN-α/β response might modulate the function of VP35, we compared dsRNA-binding activities in mock-infected and SeV-infected cells but found no differences in binding by either wild-type or mutant VP35 (Fig. 1A). Note that no VP35 was coprecipitated with “empty” Sepharose beads (data not shown). Finally, we confirmed that all VP35 constructs were expressed at similar levels by Western blotting of the cell lysates used for the pull-down assays (Fig. 1A).
FIG. 1.
Ebola virus VP35 binds to dsRNA. (A) 293T cells were transfected with 500 ng of the indicated protein expression plasmids (Luc, firefly luciferase). Twenty-four h posttransfection, cells were either mock infected (mock) or infected with SeV at an MOI of 8. Twelve h postinfection, cell lysates were prepared and incubated with pIC-Sepharose for 2 h at 4°C. Protein complexes were collected by centrifugation, washed 10 times, and separated by 12% sodium dodecyl sulfate-PAGE. Proteins were transferred to a polyvinylidene difluoride membrane and visualized by Western blotting using anti-Flag M2 (WB: FLAG) or anti-VP35 6C5 (WB: VP35) monoclonal antibodies. To determine levels of protein expression, a fraction (2%) of the whole-cell extracts (WCE) was separated for Western blotting. (B) To assess the degree of impairment in dsRNA binding by the VP35 mutants, increasing amounts of lysates from cells expressing wild-type VP35 (VP35) or a VP35 mutant (R312A or K309A) were incubated with a fixed amount of pIC-Sepharose to obtain cell lysate/bead ratios of 0.05, 0.5, and 5 (wedges), and binding was assessed as in panel A. Input levels of wild-type and mutant VP35 proteins were assessed by Western blotting (WB: VP35). (C) The specificity for dsRNA binding was determined by competition assay. Soluble pIC, pAU, poly(rU) (pU), poly(rA) (pA), or dsDNA (from salmon sperm) at 12.5, 25, 50, and 100 μg/ml was added to cell lysates prior to addition of pIC-Sepharose (wedges, upper panel). Soluble dsRNA molecules were further tested at concentrations of 1, 10, 100, 1,000, and 10,000 ng/ml before pIC-Sepharose was added (wedges, lower panel). (D) Viral RNA was synthesized in vitro by using a T7-driven promoter cloned in front of VP35 sequences such that either positive-sense or negative-sense transcripts of different lengths were generated. Complementary ssRNAs were annealed in vitro to obtain the corresponding dsRNAs. These dsRNAs were analyzed on an agarose gel. Lanes: 1, 210 bp; 2, 409 bp; 3, 609 bp; 4, 809 bp; 5, 1,013 bp. A DNA ladder is present in the leftmost lane and contains molecules of the indicated sizes in base pairs. (E) Cell lysates containing wild-type VP35 were incubated without competitor (−) or with the indicated in vitro-transcribed dsRNA competitor molecules (see panel D) before pIC-Sepharose was added. The concentration of each dsRNA was 30 nM for each binding reaction. As a control, soluble pIC was used at 20 μg/ml. VP35 was detected by Western blotting as described previously. (F) The carboxy-terminal 171 amino acids of wild-type VP35 (C-171) were produced in a bacterial expression system with a C-terminal His tag and purified using a Talon Cobalt metal affinity column. Purified protein was used at ∼80 ng/ml in pIC-Sepharose binding assays as described previously, without (−) or with (+) soluble pIC as competitor. As a positive control, lysates from wild-type VP35-transfected 293T cells were run side by side. Input represents 2% of the total protein used for the immunoprecipitation. VP35 was detected with the C-terminal 10C7 monoclonal antibody.
Next, we further assessed the extent to which the dsRNA-binding activity of the R312A and K309A mutants was impaired. Increasing amounts of cell lysates (5, 50, and 500 μl) containing equivalent amounts of wild-type or mutant VP35s were incubated with a fixed amount of pIC beads such that the ratios of cell lysates to pIC beads were 0.05, 0.5, and 5, respectively. By this protocol, we were able to coprecipitate wild-type VP35 when either 50 or 500 μl of cell lysates was used (Fig. 1B). The point mutants R312A and K309A were virtually undetectable in the Western blot, although R312A showed a very weak binding activity at the highest volume of cell lysate used (Fig. 1B). To address the binding specificity of VP35 for dsRNA, different soluble synthetic polynucleotides were added to VP35-containing lysates prior to addition of pIC beads. Only the dsRNA molecules pIC and pAU were able to compete with pIC-Sepharose for binding VP35 (Fig. 1C). The single-stranded RNAs (ssRNAs) poly(rU) and poly(rA) and dsDNA (salmon sperm DNA) were unable to compete with pIC-Sepharose at the tested concentrations. The specificity of VP35 for dsRNA-type molecules was underscored by the relatively low amount of soluble pIC and pAU needed for the competition assay. As little as 100 ng/ml of synthetic pIC or pAU was sufficient to abrogate binding of VP35 to the pIC beads, while 100 μg of the ssRNA or dsDNA molecules was insufficient to block VP35 binding (Fig. 1C).
Next, we wanted to assess the ability of VP35 to bind to dsRNA molecules with a more “natural” composition (i.e., molecules with a nucleotide composition that might actually be encountered by VP35 in infected cells). Towards this end, dsRNA molecules were generated from single-stranded RNAs in vitro transcribed from a VP35 cDNA in a size range of 200 to 1,000 bp (Fig. 1D). Each of these molecules, when used at approximately 30 nM, was an effective soluble competitor, inhibiting VP35 binding to pIC-Sepharose (Fig. 1E). Serving as a positive control for this experiment was soluble pIC at 20 μg/ml (Fig. 1E).
To investigate whether VP35 can bind directly to dsRNA, we attempted to produce the full-length protein in a bacterial expression system. However, soluble forms of the protein could be obtained only when the carboxy-terminal 171 amino acids (C-171) were expressed and purified. Because this region contains a putative dsRNA-binding motif (23), we tested this mutant protein in our binding assays. As shown in Fig. 1F, C-171 was coprecipitated with pIC beads and soluble pIC could efficiently compete for C-171 binding (Fig. 1F).
Cumulatively, these data suggest that VP35 directly binds to dsRNA through its carboxy terminus and that the mutations R312A and K309A each significantly impair this activity. Although additional biochemical studies will further elucidate the interaction between VP35 and dsRNA, we next asked whether this activity plays a critical role in the “IFN-antagonist” activity of VP35.
dsRNA-binding mutants of VP35 inhibit IFN-β gene expression induced by SeV infection.
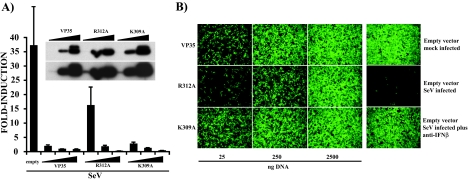
dsRNA-binding activity is a property shared by several viral proteins that inhibit host IFN responses, including the influenza A virus NS1 protein, the influenza B virus NS1 protein, and the vaccinia virus E3L protein (16, 17, 57, 64). It is also a property of several IFN-induced proteins implicated in the host antiviral response whose enzymatic activity is modulated by dsRNA, including PKR, OAS, and ADAR1 (52). To determine the extent to which dsRNA binding may influence the ability of VP35 to inhibit IRF-3 activation and IFN-α/β production, we compared the abilities of wild-type VP35, R312A, and K309A to inhibit the SeV-induced activation of an IFN-β promoter reporter plasmid that expresses the CAT enzyme. Although these mutants were previously reported to be impaired for IFN antagonist activity, we wished to perform a more quantitative comparison of these two mutants and to interpret these results in light of the dsRNA binding data presented above. Upon viral infection, reporter gene expression was strongly induced in empty-vector-transfected cells (Fig. 2A). Wild-type VP35 inhibited this activation to background levels at all plasmid DNA concentrations tested. Interestingly, transfection of VP35 and K309A (25 ng, 250 ng, or 2,500 ng of plasmid) resulted in comparable levels of inhibition (Fig. 2A). The R312A mutant also inhibited reporter gene activation when 250 and 2,500 ng of plasmid were transfected, but at 25 ng, the inhibitory activity substantially diminished (Fig. 2A). The differences in reporter gene activation were not due to differences in protein expression because all constructs showed similar protein levels (Fig. 2A, inset). These data demonstrate that the R312A mutant is a less effective inhibitor of SeV-induced IFN response than are wild-type VP35 and K309A, at least when these proteins are expressed at relatively low levels.
FIG. 2.
Wild-type and dsRNA-binding mutant VP35s inhibit IFN-β gene expression induced by SeV infection. (A) Increasing concentrations (25, 250, or 2,500 ng, indicated by wedges) of plasmids expressing VP35 and the K309A and R312A mutants were transfected into 293T cells together with 300 ng each of an IFN-β-CAT reporter and a constitutive pCAGGS-firefly luciferase reporter. Twenty-four h posttransfection, cells were infected with Sendai virus (MOI of 8), and 12 h postinfection, CAT and luciferase activities were determined. Values are expressed as induction (fold) over an empty-plasmid mock-infected control (not shown). Virus-induced CAT activity was normalized to firefly luciferase activity. Error bars indicate standard deviations of at least three independent experiments. Expression levels of the different VP35 constructs were determined by Western blotting (inset). Blots were probed with a monoclonal antibody to VP35 (6C5). Two exposures of the same blot are shown; expression levels in the 25-ng samples were detected only when the film was overexposed (inset, lower panel). (B) After UV irradiation, a series of twofold dilutions of conditioned media from the experiment described in panel A was overlaid onto Vero cells in a 96-well plate. Twenty-four h after treatment, cells were infected with NDV-GFP (MOI of 6), and 24 h postinfection, virus replication was examined by fluorescence microscopy. Shown are the results obtained when the conditioned media from 293T cells transfected with 25, 250, or 2,500 ng of VP35 and R312A and K309A mutant plasmids were used. Right column: in the empty-vector mock-infected panel, NDV-GFP replication is readily detected by the presence of green fluorescence. The empty-vector SeV-infected panel lacks GFP expression, demonstrating the antiviral state created by IFN. IFN is present in the conditioned media due to the Sendai virus infection of the transfected 293T cells. This is demonstrated in the empty-vector SeV-infected plus anti-IFN-β panel, where neutralizing anti-IFN-β antibody rescues NDV-GFP replication. All panels are from cells treated with a 16-fold dilution of the conditioned medium.
Next, we assessed the abilities of wild-type VP35 and dsRNA-binding mutants to inhibit the SeV-induced production of endogenous IFN-β by employing an IFN bioassay. In these experiments, 293T cells were transfected with empty expression plasmid or with plasmids that expressed wild-type VP35, R312A, or K309A. One day posttransfection, the cells were mock infected or infected with SeV (MOI of 8) to induce IFN production. One day later, the media from these cells were harvested and irradiated with UV light to inactivate any infectious SeV. The inactivated medium was then subjected to twofold serial dilutions and transferred to Vero cells. IFN present in this medium (induced by the SeV) would be expected to induce an antiviral state in the Vero cells. After an overnight incubation, the medium was removed, and the Vero cells were then infected with NDV-GFP (MOI of 6). Expression of GFP was then used as an indicator of virus replication. SeV infection of empty-vector-transfected 293T cells resulted in the production of IFN which blocked NDV-GFP replication (Fig. 2B, compare empty-vector, mock-infected to empty-vector, SeV-infected panels). That IFN-β largely mediates the antiviral effect in this assay was demonstrated by the ability of an anti-IFN-β antibody to neutralize the antiviral activity (Fig. 2B, empty vector, infected plus anti-IFN-β). This last test also argues against any effect, on NDV-GFP replication in Vero cells, of live SeV that resisted the UV treatment. Transfection of even low amounts of plasmids expressing wild-type VP35 and K309A rescued NDV-GFP replication in Vero cells, demonstrating the capacity of these two proteins to antagonize the production of IFN-β (Fig. 2B). The R312A mutant was also able to rescue NDV-GFP infection when 250 or 2,500 ng of expression plasmid was transfected. This last mutant was clearly less effective than the other constructs in blocking IFN production (Fig. 2B, middle row). Thus, the IFN bioassay results mirror the reporter gene assay, where the R312A mutant is impaired relative to wild-type VP35 and the K309A mutant. However, both dsRNA-binding mutants retain some ability to inhibit SeV-induced IFN-β production.
Wild-type VP35 and dsRNA-binding mutants can inhibit IFN-β gene activation mediated by RIG-I.
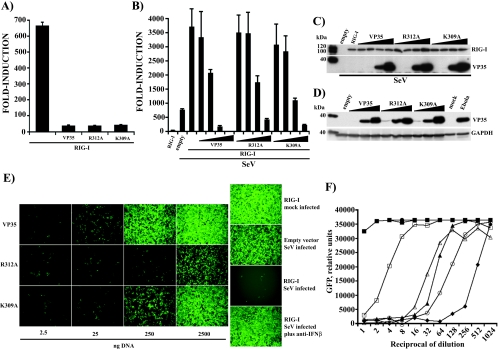
The RIG-I protein reportedly plays a key role in sensing viral infection and activating IFN-α/β production (30, 56, 67). RIG-I recognizes dsRNA, possibly through its DExD/H box helicase domain, and activates downstream signaling through its N-terminal CARD, resulting in the activation of IRF-3 (30, 56, 67). Given VP35's ability to inhibit the activation of IRF-3 (3), we investigated whether wild-type VP35 and the dsRNA-binding mutants could block the RIG-I-activated signaling pathway. First, we tested the ability of the VP35 constructs to block the RIG-I-induced activation of the IRF-3-responsive ISG54 promoter. We found that in the absence of viral infection, overexpression of RIG-I in 293T cells was sufficient to activate this promoter. As shown in Fig. 3A, transfection of 250 ng of RIG-I plasmid resulted in a greater-than-600-fold induction of the promoter relative to empty plasmid transfection control. When 2,500 ng of wild-type VP35 or dsRNA-binding mutants was cotransfected with RIG-I, a strong inhibition of reporter gene activation was seen, indicating that wild-type and dsRNA-binding mutant forms of VP35 could inhibit virus-independent signaling by RIG-I (Fig. 3A). However, RIG-I functions in the virus-induced activation of IFN responses, and expression of RIG-I followed by virus infection results in a synergistic activation of the IFN-β gene (56, 67). We therefore also tested the abilities of the VP35 constructs to inhibit reporter gene activation in RIG-I-transfected, SeV-infected cells. Towards this end, we transfected small amounts (25 ng) of RIG-I plasmid DNA and increasing concentrations of wild-type or mutant VP35 plasmid (2.5, 25, 250, and 2,500 ng), together with an IFN-β-CAT reporter gene. Under these conditions, overexpression of RIG-I alone resulted in a 30-fold induction of reporter gene. Upon viral infection, reporter gene induction was greater than 700-fold in empty-plasmid-transfected cells. When 25 ng of RIG-I expression plasmid was transfected in cells that were subsequently infected with SeV, reporter gene induction increased to nearly 4,000-fold (Fig. 3B). Remarkably, wild-type VP35 efficiently blocked this synergistic activation following transfection of 2,500 and 250 ng of expression plasmid. Decreasing the amounts of VP35 plasmid to 25 and 2.5 ng resulted in a loss of inhibitory activity (Fig. 3B). The R312A and K309A mutants were also able to inhibit reporter gene activation but only when higher amounts of expression plasmid were transfected. IFN antagonist activity was lost rapidly (compared to wild-type VP35) as the mutant DNA was diluted. To verify that the differences in reporter gene activation were not due to differences in protein expression, extracts from transfected cells were analyzed by Western blot assay. Comparable amounts of RIG-I were detected in all transfections (Fig. 3C, top panel). The different VP35s were readily detected where the two highest concentrations of plasmid DNA were transfected but could not be detected at the two lowest concentrations, correlating with the diminished IFN-antagonist function seen in these transfections (Fig. 3C, lower panel). Based on these data, the dsRNA-binding mutants retain an ability to inhibit reporter gene activation induced by the combination of SeV infection and RIG-I overexpression, but the mutants are less potent than wild-type VP35. To determine whether our ectopic expression of VP35 results in protein levels comparable to what are seen in virus-infected cells, we performed Western blot assays for VP35 on transfected-cell extracts and EBOV-infected Vero cell extracts (MOI of 1, 48 h postinfection). VP35 was expressed in EBOV-infected cells at levels comparable to that seen in cells transfected with 250 or 2,500 ng of expression plasmid (Fig. 3D). We therefore conclude that we are studying biologically relevant levels of VP35 in our transfection experiments.
FIG. 3.
Wild-type VP35 and dsRNA-binding mutants inhibit IFN-β gene activation mediated by RIG-I. (A) 293T cells were transfected with 250 ng of RIG-I expression plasmid with and without 2,500 ng of VP35 or R312A or K309A mutant plasmid. An ISG54-CAT reporter gene and a constitutively expressed firefly luciferase control reporter were also transfected at 300 ng. CAT and luciferase activities were measured 24 h posttransfection. Values are expressed as induction (fold) over an empty-vector-transfected control. Error bars represent standard deviations of at least three independent experiments. (B) 293T cells were transfected with 25 ng of RIG-I expression plasmid alone or with 2.5, 25, 250, and 2,500 ng of the indicated VP35 constructs (increasing amounts indicated by the wedges). Twenty-four h posttransfection, cells were mock infected or infected with Sendai virus (MOI of 8). The IFN-β-CAT reporter gene and the firefly luciferase transfection control plasmids were used at 300 ng. CAT and luciferase activities were measured 12 h postinfection. Values are expressed as before. Error bars represent standard deviations of at least three independent experiments. (C) Expression of RIG-I, wild-type VP35, and VP35 mutants from the experiment described in panel B was assessed by Western blotting using monoclonal anti-FLAG and anti-VP35 antibodies. (D) 293T cells were transfected with 25, 250, and 2,500 ng (increasing amounts indicated by the wedges) of each VP35 construct (wild-type VP35, R312A, or K309A) as described above, and cell extracts were prepared 24 h posttransfection. Lysates from Vero cells that were mock infected or infected with Zaire EBOV (MOI of 1) were prepared 48 h postinfection and gamma irradiated to eliminate infectious virus. VP35 expression was detected by Western blot assay as before. A monoclonal anti-human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was used as a loading control. (E) An IFN bioassay was performed as described before (see Fig. 2B) with UV-irradiated, twofold dilutions of conditioned media from the experiment described in panel B before. The main panels represent supernatants from 293T cells transfected with 2.5, 25, 250, and 2,500 ng of VP35 plasmid. Shown are data from cells treated with 64-fold-diluted conditioned media. (F) The IFN bioassay was analyzed with a fluorescence plate reader. Data for all twofold dilutions of conditioned media are presented. ▪, empty vector mock infected; •, RIG-I, mock infected; ▴, empty vector, SeV infected; ⧫, RIG-I, SeV infected; □, VP35 plus RIG-I, SeV infected; ▵, K309A plus RIG-I, SeV infected; ○, R312A plus RIG-I, SeV infected. Only the data from transfections using 2.5 μg of plasmid DNA are shown. y-axis values are relative GFP fluorescence units. x-axis values are the reciprocal of the dilutions of the conditioned media. This is a representative result of an experiment replicated three times.
To support the results obtained from the IFN-β reporter assay, we performed an IFN bioassay with the media from these transfected cells. The limited amounts (25 ng) of RIG-I plasmid transfected in this experiment did not result in a significant production of IFN-β, as assessed by the bioassay (Fig. 3E, right side, top panel). When cells containing RIG-I were infected with SeV, IFN-β secretion was greatly enhanced compared with the empty-vector, SeV-infected control (Fig. 3E, right panel, compare empty-vector SeV-infected to RIG-I, SeV-infected panels). It should be noted that in Fig. 3E, the cell supernatants were diluted 64-fold, whereas in the previous experiment (Fig. 2B), the supernatants were diluted 16-fold. This is why in Fig. 3E the medium from empty-vector, SeV-infected sample does not greatly reduce NDV-GFP replication. Again, addition of a neutralizing anti-IFN-β antibody restored NDV-GFP infection (Fig. 3E, RIG-I, SeV-infected plus anti-IFN-β). Transfection of 250 or 2,500 ng of VP35 plasmid inhibited the IFN-β secretion induced by RIG-I plus SeV (Fig. 3E, VP35 panels). In this experiment, each of the dsRNA-binding mutants displayed reduced IFN-inhibitory activity relative to wild-type VP35. The activity of the R312A mutant was clearly reduced, even when 2,500 ng of plasmid was transfected, although some ability to inhibit IFN-β production was still evident (Fig. 3E, R312A panels). The K309A mutant displayed greater IFN-β-antagonist activity, but it remained less effective than wild-type VP35 (Fig. 3E, K309A panels). Results of the IFN bioassay were also analyzed in a fluorescence plate reader (Fig. 3F). The media from empty-vector or RIG-I transfections and mock infection did not produce levels of IFN-β sufficient to induce an antiviral state in Vero cells; therefore, in cells treated with these supernatants, NDV-GFP showed the highest levels of replication as assessed by GFP fluorescence (Fig. 3F, filled circles and squares, respectively). The medium from SeV-infected, RIG-I-transfected cells had to be diluted approximately 1,000-fold before detectable IFN activity was lost (Fig. 3F, filled diamonds). In contrast, a 128-fold dilution was required to eliminate IFN activity from empty-vector-transfected, SeV-infected cells, confirming the synergistic effect observed in the IFN-β reporter assay (Fig. 3F, filled triangles). When VP35 was transfected at 2,500 ng in SeV-infected cells, only an 8- to 16-fold dilution was required to eliminate IFN activity from the medium (Fig. 3F, empty squares). Cells transfected with 2,500 ng of the dsRNA-binding mutants displayed intermediate levels of IFN-β production after SeV infection, and the R312A mutant (Fig. 3F, empty circles) was again more impaired than the K309A mutant (Fig. 3F, empty triangles). Both mutants, however, retained some ability to suppress IFN-β production. These results indicate that, under conditions of enhanced IFN induction, the two dsRNA-binding mutants are less effective IFN antagonists than wild-type VP35; however, the mutants retain some activity.
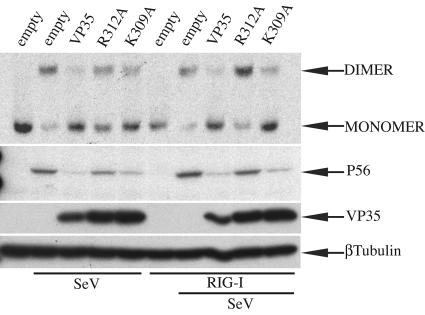
Effect of VP35 and dsRNA-binding mutants on virus- and RIG-I-mediated activation of IRF-3.
IRF-3 plays a key role in the dsRNA- and virus-induced activation of IFN-α/β- and IFN-stimulated gene expression (37, 62, 69). Upon viral infection, IRF-3 is phosphorylated at critical serine/threonine residues at its C terminus, triggering its homodimerization and association with cotransactivators in the nucleus (28, 37, 42, 68). IRF-3 dimerization has frequently been used as a measure of its activation and can be monitored by a native PAGE assay (28, 42). Here, we found that wild-type VP35 inhibited, compared to empty-plasmid-infected control, endogenous IRF-3 dimer formation induced by SeV (Fig. 4, top panel). Only weak IRF-3 dimer formation was detected in K309A-expressing cells. In contrast, IRF-3 dimerization was readily detected in cells transfected with the R312A mutant, although dimer levels remained lower than that seen in the empty-vector, SeV-infected control (Fig. 4, top panel). When RIG-I was overexpressed in cells infected with SeV, endogenous IRF-3 dimer formation was again blocked in cells expressing wild-type VP35 and K309A. In contrast, IRF-3 dimerization was enhanced in cells expressing the R312A mutant (Fig. 4, top panel). Next, we examined whether the observed patterns of IRF-3 activation correlated with the transcription of an endogenous, IRF-3-responsive gene, the human 561 gene. The product of this gene, the P56 protein, is readily induced during viral infection in an IRF-3-dependent manner (21, 47). The same cell extracts used for IRF-3 dimer formation were analyzed by Western blotting with anti-P56 antibody. Empty-vector-transfected, mock-infected cell lysates did not contain detectable levels of P56, but by 8 h post-SeV infection P56 was clearly induced (Fig. 4, middle panel). Wild-type VP35 and K309A were strong inhibitors of P56 expression induced either by virus alone or by RIG-I plus virus. The R312A mutant was not able to completely inhibit P56 expression induced by virus infection, and the impairment of the R312A mutant was even more evident in the cells with RIG-I plus virus. Thus, the R312A mutant was less potent than either wild-type VP35 or the K309A mutant in preventing IRF-3 activation and subsequent gene activation.
FIG. 4.
Effect of VP35 and dsRNA-binding mutants on virus- and RIG-I-mediated activation of IRF-3. HEK293 cells were transfected with 4 μg of plasmids that express wild-type VP35 (VP35), R312A, and K309A, with or without 40 ng of RIG-I plasmid as indicated. Twenty-four h posttransfection, cells were either mock SeV infected or infected with Sendai virus (MOI of 8), as indicated. Eight h postinfection, cells were lysed and proteins were separated in a continuous 7.5% native gel that was prerun with and without 0.2% sodium deoxycholate in the cathode and anode chambers, respectively. Endogenous IRF-3 was detected by Western blotting with a primary rabbit anti-hIRF-3 (1:500) antibody and a secondary goat anti-rabbit immunoglobulin G-horseradish peroxidase (1:5,000) antibody (top panel). Expression of P56 and the different forms of VP35 in the same cell lysates was analyzed by Western blotting following separation of proteins by 12.5% sodium dodecyl sulfate-PAGE. A mouse anti-human β-tubulin antibody was used as a loading control.
Wild-type VP35 and dsRNA-binding mutants can impair IFN-β reporter gene activation induced by IPS-1, TBK1, and IKKɛ.
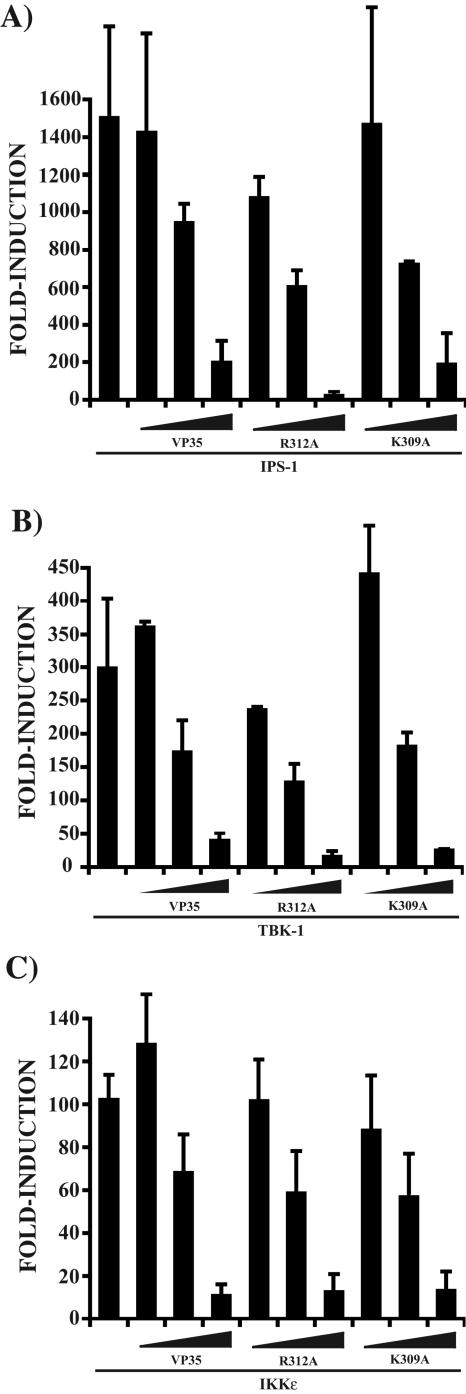
Because the dsRNA-binding mutants of VP35 retained the ability to antagonize IFN-β production induced by viral infection and RIG-I, we sought to investigate the abilities of these mutants to impair signaling components downstream of RIG-I, components which are not known to either sense or interact directly with dsRNA. The recently described CARD-containing protein IPS-1, also known as MAVS, VISA, or Cardif, has been shown to function downstream of RIG-I and serve as an adaptor molecule for RIG-I signaling (31, 41, 53, 65). When we overexpressed IPS-1 in 293T cells, the IFN-β promoter was strongly induced, as previously reported. Notably, wild-type VP35, K309A, and R312A all showed a similar concentration-dependent inhibition of reporter gene activation by IPS-1 (Fig. 5A). Next, to further understand where VP35 may affect RIG-I/IRF-3 signaling, we investigated the effect of the VP35s on the activation of the IFN-β promoter by the IRF-3 kinases TBK1 and IKKɛ. These kinases are responsible for the phosphorylation of IRF-3 (18, 54) and function downstream of IPS-1 (31, 53). Overexpression of TBK1 and IKKɛ was able to strongly induce reporter gene activation, as previously reported. Surprisingly, as was observed with IPS-1, wild-type VP35 and dsRNA-binding mutants showed a similar concentration-dependent inhibition of reporter gene activation by the kinases (Fig. 5B and C).
FIG. 5.
Wild-type VP35 and dsRNA-binding mutants block IPS-1-, TBK-1-, and IKKɛ-induced IFN-β gene activation. (A) 293T cells were transfected with 25, 250, and 2,500 ng (wedges) of the indicated VP35 constructs together with 25 ng of IPS-1 expression plasmid. Additionally, all transfections contained 300 ng each of IFN-β-CAT reporter and pCAGGS-firefly luciferase plasmids. Error bars represent standard deviations of at least three independent experiments. (B) The experiment was performed as in panel A, but with 50 ng of TBK-1 expression plasmid as the activator of gene expression. (C) The experiment was performed as in panel A, but with 50 ng of IKKɛ expression plasmid as the activator of gene expression.
Cumulatively, these data indicate that the dsRNA-binding activity of VP35 may play a role in its ability to inhibit signaling to IRF-3 by virus infection. However, because wild-type VP35 and the dsRNA-binding mutants can equally block downstream signaling components of the virus-RIG-I pathway, mechanisms independent of dsRNA-binding activity most likely contribute to VP35's IFN antagonist activity.
DISCUSSION
Production of dsRNA species during viral replication has long been considered a potential trigger of innate antiviral responses. Recently, the DExD/H box helicases RIG-I and MDA-5 were shown to function as viral dsRNA sensors leading to the production of IFN-α/β (1, 56, 66, 67). Viruses have evolved mechanisms to evade this recognition, and some encode proteins such as the E3L protein of vaccinia virus, the NS1 proteins of influenza A and B viruses, and the σ3 protein of reovirus that reportedly antagonize the host antiviral response, at least in part, by binding to dsRNA (5, 16, 17, 64).
Recently, it was suggested that the EBOV VP35 protein may have dsRNA-binding activity and that this function might be required for its IFN antagonist properties (23). This report provides the first experimental demonstration of a dsRNA-binding activity associated with VP35. We used an established “pull-down” assay in which pIC covalently bound to Sepharose beads is used to coprecipitate a possible dsRNA-binding protein. Such assays have recently been used to demonstrate dsRNA-binding activity for the cellular protein RIG-I and the human cytomegalovirus protein pTRS1 (22, 67). We found that VP35, present in whole-cell extracts of mock-infected or SeV-infected 293T cells, can be coprecipitated with pIC-Sepharose beads. Several lines of evidence argue that the interaction of VP35 with the pIC beads is specific for dsRNA. First, VP35 did not detectably coprecipitate with Sepharose beads to which no nucleic acid had been cross-linked (data not shown). Second, neither synthetic homo-oligomeric ssRNA molecules [poly(rA) and poly(rU)] nor dsDNA could, at any concentration tested, compete with the pIC beads for binding to VP35. In contrast, three types of dsRNA molecules did effectively compete with the pIC beads for binding to VP35. Soluble pIC and pAU were able to compete with pIC-Sepharose for VP35 binding, even when the soluble dsRNAs were present in amounts 1,000-fold lower than the amounts of single-stranded or dsDNA molecules tested. In addition, dsRNA molecules of a more “natural” structure and complexity, ranging in size from approximately 200 to 1,000 bp, which were in vitro transcribed from an EBOV cDNA, also effectively competed for binding to VP35.
To address whether VP35 may directly interact with dsRNA, we sought to use purified, bacterially expressed VP35. Although full-length VP35 could not be produced in a soluble form, we were finally able to produce limited amounts of a soluble, truncated VP35 (amino acids 171 to 340). The mutant protein was coprecipitated with pIC-Sepharose beads, and soluble pIC could efficiently compete for binding, suggesting that VP35 can directly interact with dsRNA. An important goal will be to produce dsRNA-binding, mutant versions of C-171 and perform more-extensive biochemical comparisons of these proteins' dsRNA-binding affinities.
Given the ability of VP35 to inhibit IRF-3 activation and IFN-β production and given its dsRNA-binding activity, we examined the abilities of wild-type and mutant VP35s to inhibit the virus-induced IFN response. As was previously reported, wild-type VP35 inhibits SeV-induced activation of IRF-3 and production of IFN-β (3, 23). The two dsRNA-binding mutants were previously found to be largely inactive (R312A) and of intermediate activity (K309A) when tested for their abilities to inhibit SeV-induced activation of the IRF-3-responsive ISG56 promoter (23). In the present study, by titrating the amounts of each VP35 construct, we find a similar pattern, where the R312A mutant is more impaired than the K309A mutant, even though neither mutant is able to detectably bind dsRNA. Most significantly, however, we find that both mutants retain some ability to block IFN-β promoter activation, endogenous IFN-β protein production, and the activation of IRF-3. Thus, as noted above when SeV alone is used as an activator, the dsRNA-binding activity may not be absolutely essential for the IFN antagonist activity of VP35.
The current model to explain how virus infection activates IFN-α/β gene expression places RIG-I early in the pathway that leads to activation of IRF-3, at least in somatic and conventional dendritic cells (30, 35). It should be noted, however, that another cellular RNA helicase, MDA-5, has also been identified as a sensor of virus infection that leads to IRF-3 activation and IFN production (1, 66). Activation of these cellular helicases leads to a signaling pathway that requires IPS-1, followed by the phosphorylation of IRF-3 by TBK-1 and IKKɛ (7, 25, 31, 65).
Given that both RIG-I and MDA-5 are thought to be dsRNA-activated sensors of virus infection, we asked whether wild-type and mutant VP35s would inhibit signaling through RIG-I. VP35 and its mutants were able to inhibit IRF-3-responsive ISG54 promoter activation when RIG-I was overexpressed, suggesting that VP35 acts at the level of RIG-I or downstream of RIG-I in the signaling pathways leading to IRF-3 activation. In further support of this conclusion, when relatively small amounts of RIG-I were expressed in cells that were subsequently infected with SeV, a potent synergistic activation of IFN-β expression was detected, and VP35 retained its ability to block this activation. As was seen when virus alone was used as an activator, the dsRNA-binding mutants were both impaired relative to wild-type VP35; however, the K309A mutant was again less impaired than was R312A. To connect the gene expression data with activation of IRF-3, we directly examined activation of endogenous IRF-3, in the absence or presence of wild-type and mutant VP35s, by a dimerization assay. Consistent with our other findings, the R312A mutant was less effective at inhibiting IRF-3 dimerization than was wild-type VP35 or K309A. This was true whether SeV or SeV plus RIG-I was used as an activator. Examination of P56, the product of an IRF-3-responsive endogenous gene, provided further confirmation of this conclusion. It is important to note that there is a general correlation between our reporter gene data and our data examining IRF-3 dimerization and the activation of P56 gene expression. In all cases, wild-type VP35 is a more potent inhibitor of IRF-3 than is K309A, and K309A is a more potent inhibitor than R312A. However, this assay does not readily distinguish between different samples where IRF-3 is strongly activated in both. Thus, although SeV infection alone induced less total reporter gene expression than did expression of RIG-I plus virus infection, we cannot see differences in IRF-3 dimer levels between cells activated by SeV alone and cells that received both RIG-I and Sendai virus. This apparent discrepancy likely reflects several facts. The activation of the reporter gene is assessed by the accumulation over time of the CAT enzyme, which is a relatively stable protein with a half-life of about 50 h (58). On the other hand, endogenous proteins like IRF-3 go through a dynamic process of phosphorylation-dependent activation and phosphorylation-dependent degradation, especially during viral infection, when the reactions occur with faster kinetics (37). Therefore, our Western blots represent a “snapshot” of IRF-3 activation. Further, during our SeV infection, it is likely that 100% of cells are infected and therefore the observed patterns of endogenous gene activation are representative of the whole cell population. By contrast, the VP35s will affect endogenous gene activation only in transfected cells. Although transfection efficiency in 293T cells is usually high, it is unlikely to reach 100%. Thus, while all transfected cells receiving the reporter gene will harbor the different VP35 constructs and have a direct effect on the reporter gene induction, the patterns of endogenous gene activation will reflect the average effect of those cells successfully transfected with the VP35s. What is clear is that both dsRNA mutants retained a strong IFN antagonist function during viral infection but were impaired when tested under strong stimulatory conditions (i.e., RIG-I plus viral infection). However, the K309A mutant appears to be a more efficient IFN antagonist than the R312A mutant.
Finally, we assessed the abilities of wild-type and mutant VP35s to target those components of the IRF-3-activating signaling pathways that lie downstream of RIG-I. Our results indicate that wild-type and dsRNA-binding mutant VP35s had equal abilities to inhibit activation of the IFN-β promoter induced by overexpression of IPS-1, a recently identified signaling molecule downstream of RIG-I, and by overexpression of the IRF-3 kinases TBK-1 and IKKɛ. These data support the hypothesis that, while dsRNA-binding activity may contribute to the VP35 IFN antagonist function, it is likely that VP35 has a dsRNA-independent function by impairing signaling at or downstream of TBK-1 and IKKɛ. Evidence also exists for dsRNA-binding-independent targeting of IFN production by the influenza A virus NS1 protein and the influenza B virus NS1 protein (16, 17).
It should also be kept in mind that dsRNA-binding activity may also affect other functions of VP35. The ability of VP35 to inhibit other aspects of the host innate antiviral response has not yet been examined in detail. In addition to suppressing IFN production, the influenza virus NS1 protein and the vaccinia virus E3L protein also inhibit other antiviral functions including dsRNA-activated proteins with antiviral activities, such as PKR, 2′,5′-oligoadenylate synthetases, and the RNA-specific adenosine deaminase ADAR1 (6, 13, 15, 24, 33, 38, 39, 49, 50, 55, 60). E3L also inhibits apoptosis (32, 40), while the influenza B virus NS1 protein inhibits ISG15-ylation (70-72). There is also evidence that NS1 and E3L can inhibit RNA silencing, although the role of RNA silencing in the antiviral response in mammalian cells remains unclear (10, 36). The ability of VP35 to antagonize these functions and the possible role of its dsRNA-binding activity in any inhibition may be of interest. In addition to interactions with host cell pathways, VP35 also plays an essential role in viral RNA synthesis and formation of viral nucleocapsid structures. dsRNA-binding activity could conceivably influence these functions as well (26, 43). The availability of the R312A and K309A mutants is a first step toward addressing these other potential functions of VP35 dsRNA-binding activity.
Acknowledgments
This work was supported by NIH grants to C.F.B., including AI053571 and AI059536. C.F.B. is a recipient of an Ellison Medical Foundation New Scholar Award in Global Infectious Disease. W.B.C. is supported by a fellowship awarded by Northeast Biodefense Center-Lipkin, PI (AI057158). M.G. and Y.-M.L. were supported by NIH grant AI060389, a Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Diseases award, and the Ellison Medical Foundation New Scholars in Global Infectious Disease Research program (ID-NS-0032). M.G. is the Nancy C. and Jeffery A. Marcus Scholar in Medical Research in Honor of Dr. Bill S. Vowell. E.O.S. is supported by AI053423 from the NIH and a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
We thank Ganes C. Sen (Cleveland Clinic) for providing anti-P56 antibody and John Hiscott (McGill University), David E. Levy (New York University), and Adolfo García-Sastre (Mount Sinai School of Medicine) for providing plasmids. We thank Ben K. Chen (Mount Sinai School of Medicine) for providing access to and training on the fluorescence plate reader. We thank Lawrence W. Leung for assistance with evaluating levels of VP35 in transfected versus infected cells and for critical reading of the manuscript and Mauricio Sanchez for excellent technical assistance.
REFERENCES
- 1.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basler, C. F., and A. Garcia-Sastre. 2002. Viruses and the type I interferon antiviral system: induction and evasion. Int. Rev. Immunol. 21:305-337. [DOI] [PubMed] [Google Scholar]
- 3.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beattie, E., K. L. Denzler, J. Tartaglia, M. E. Perkus, E. Paoletti, and B. L. Jacobs. 1995. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J. Virol. 69:499-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmann, M., A. Garcia-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bin, L. H., L. G. Xu, and H. B. Shu. 2003. TIRP, a novel Toll/interleukin-1 receptor (TIR) domain-containing adapter protein involved in TIR signaling. J. Biol. Chem. 278:24526-24532. [DOI] [PubMed] [Google Scholar]
- 8.Bluyssen, H. A., R. J. Vlietstra, A. van der Made, and J. Trapman. 1994. The interferon-stimulated gene 54 K promoter contains two adjacent functional interferon-stimulated response elements of different strength, which act synergistically for maximal interferon-alpha inducibility. Eur. J. Biochem. 220:395-402. [DOI] [PubMed] [Google Scholar]
- 9.Bosio, C. M., M. J. Aman, C. Grogan, R. Hogan, G. Ruthel, D. Negley, M. Mohamadzadeh, S. Bavari, and A. Schmaljohn. 2003. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J. Infect. Dis. 188:1630-1638. [DOI] [PubMed] [Google Scholar]
- 10.Bucher, E., H. Hemmes, P. de Haan, R. Goldbach, and M. Prins. 2004. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol. 85:983-991. [DOI] [PubMed] [Google Scholar]
- 11.Chang, H. W., and B. L. Jacobs. 1993. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology 194:537-547. [DOI] [PubMed] [Google Scholar]
- 12.Chang, H. W., L. H. Uribe, and B. L. Jacobs. 1995. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J. Virol. 69:6605-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, H. W., J. C. Watson, and B. L. Jacobs. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 89:4825-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dauber, B., G. Heins, and T. Wolff. 2004. The influenza B virus nonstructural NS1 protein is essential for efficient viral growth and antagonizes beta interferon induction. J. Virol. 78:1865-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies, M. V., H. W. Chang, B. L. Jacobs, and R. J. Kaufman. 1993. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J. Virol. 67:1688-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donelan, N. R., C. F. Basler, and A. Garcia-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 77:13257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donelan, N. R., B. Dauber, X. Wang, C. F. Basler, T. Wolff, and A. Garcia-Sastre. 2004. The N- and C-terminal domains of the NS1 protein of influenza B virus can independently inhibit IRF-3 and beta interferon promoter activation. J. Virol. 78:11574-11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Sastre, A. 2004. Identification and characterization of viral antagonists of type I interferon in negative-stranded RNA viruses. Curr. Top. Microbiol. Immunol. 283:249-280. [DOI] [PubMed] [Google Scholar]
- 20.Gribaudo, G., D. Lembo, G. Cavallo, S. Landolfo, and P. Lengyel. 1991. Interferon action: binding of viral RNA to the 40-kilodalton 2′-5′-oligoadenylate synthetase in interferon-treated HeLa cells infected with encephalomyocarditis virus. J. Virol. 65:1748-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo, J., K. L. Peters, and G. C. Sen. 2000. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 267:209-219. [DOI] [PubMed] [Google Scholar]
- 22.Hakki, M., and A. P. Geballe. 2005. Double-stranded RNA binding by human cytomegalovirus pTRS1. J. Virol. 79:7311-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartman, A. L., J. S. Towner, and S. T. Nichol. 2004. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology 328:177-184. [DOI] [PubMed] [Google Scholar]
- 24.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740-745. [DOI] [PubMed] [Google Scholar]
- 26.Huang, Y., L. Xu, Y. Sun, and G. J. Nabel. 2002. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell 10:307-316. [DOI] [PubMed] [Google Scholar]
- 27.Imani, F., and B. L. Jacobs. 1988. Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 sigma 3 protein. Proc. Natl. Acad. Sci. USA 85:7887-7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6:375-388. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs, B. L., and J. O. Langland. 1996. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 219:339-349. [DOI] [PubMed] [Google Scholar]
- 30.Kato, H., S. Sato, M. Yoneyama, M. Yamamoto, S. Uematsu, K. Matsui, T. Tsujimura, K. Takeda, T. Fujita, O. Takeuchi, and S. Akira. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19-28. [DOI] [PubMed] [Google Scholar]
- 31.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 32.Kibler, K. V., T. Shors, K. B. Perkins, C. C. Zeman, M. P. Banaszak, J. Biesterfeldt, J. O. Langland, and B. L. Jacobs. 1997. Double-stranded RNA is a trigger for apoptosis in vaccinia virus-infected cells. J. Virol. 71:1992-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langland, J. O., and B. L. Jacobs. 2004. Inhibition of PKR by vaccinia virus: role of the N- and C-terminal domains of E3L. Virology 324:419-429. [DOI] [PubMed] [Google Scholar]
- 34.Levy, D. E., I. Marie, and A. Prakash. 2003. Ringing the interferon alarm: differential regulation of gene expression at the interface between innate and adaptive immunity. Curr. Opin. Immunol. 15:52-58. [DOI] [PubMed] [Google Scholar]
- 35.Levy, D. E., and I. J. Marie. 2004. RIGging an antiviral defense—it's in the CARDs. Nat. Immunol. 5:699-701. [DOI] [PubMed] [Google Scholar]
- 36.Li, W. X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. Brydon, K. L. Johnson, A. Garcia-Sastre, L. A. Ball, P. Palese, and S. W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, Y., K. C. Wolff, B. L. Jacobs, and C. E. Samuel. 2001. Vaccinia virus E3L interferon resistance protein inhibits the interferon-induced adenosine deaminase A-to-I editing activity. Virology 289:378-387. [DOI] [PubMed] [Google Scholar]
- 39.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 40.Marques, J. T., D. Rebouillat, C. V. Ramana, J. Murakami, J. E. Hill, A. Gudkov, R. H. Silverman, G. R. Stark, and B. R. Williams. 2005. Down-regulation of p53 by double-stranded RNA modulates the antiviral response. J. Virol. 79:11105-11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 42.Mori, M., M. Yoneyama, T. Ito, K. Takahashi, F. Inagaki, and T. Fujita. 2004. Identification of Ser-386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J. Biol. Chem. 279:9698-9702. [DOI] [PubMed] [Google Scholar]
- 43.Muhlberger, E., M. Weik, V. E. Volchkov, H. D. Klenk, and S. Becker. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J. Virol. 73:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nanduri, S., B. W. Carpick, Y. Yang, B. R. Williams, and J. Qin. 1998. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 17:5458-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 46.Park, M.-S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters, K. L., H. L. Smith, G. R. Stark, and G. C. Sen. 2002. IRF-3-dependent, NFkappa B- and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. USA 99:6322-6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reid, S. P., W. B. Cardenas, and C. F. Basler. 2005. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology 341:179-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rivas, C., J. Gil, Z. Melkova, M. Esteban, and M. Diaz-Guerra. 1998. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2-5A synthetase enzyme. Virology 243:406-414. [DOI] [PubMed] [Google Scholar]
- 50.Romano, P. R., F. Zhang, S. L. Tan, M. T. Garcia-Barrio, M. G. Katze, T. E. Dever, and A. G. Hinnebusch. 1998. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol. Cell. Biol. 18:7304-7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 54.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 55.Sharp, T. V., F. Moonan, A. Romashko, B. Joshi, G. N. Barber, and R. Jagus. 1998. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology 250:302-315. [DOI] [PubMed] [Google Scholar]
- 56.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson, J. F., L. S. Hayes, and D. B. Lloyd. 1991. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene 103:171-177. [DOI] [PubMed] [Google Scholar]
- 59.Valente, L., and K. Nishikura. 2005. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 79:299-338. [DOI] [PubMed] [Google Scholar]
- 60.Wang, W., and R. M. Krug. 1996. The RNA-binding and effector domains of the viral NS1 protein are conserved to different extents among influenza A and B viruses. Virology 223:41-50. [DOI] [PubMed] [Google Scholar]
- 61.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. Garcia-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams, B. R. 2001. Signal integration via PKR. Sci. STKE 2001:RE2. [DOI] [PubMed] [Google Scholar]
- 64.Xiang, Y., R. C. Condit, S. Vijaysri, B. Jacobs, B. R. Williams, and R. H. Silverman. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 76:5251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 66.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 67.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 68.Yoneyama, M., W. Suhara, and T. Fujita. 2002. Control of IRF-3 activation by phosphorylation. J. Interferon Cytokine Res. 22:73-76. [DOI] [PubMed] [Google Scholar]
- 69.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan, W., J. M. Aramini, G. T. Montelione, and R. M. Krug. 2002. Structural basis for ubiquitin-like ISG 15 protein binding to the NS1 protein of influenza B virus: a protein-protein interaction function that is not shared by the corresponding N-terminal domain of the NS1 protein of influenza A virus. Virology 304:291-301. [DOI] [PubMed] [Google Scholar]
- 71.Yuan, W., and R. M. Krug. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao, C., C. Denison, J. M. Huibregtse, S. Gygi, and R. M. Krug. 2005. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. USA 102:10200-10205. [DOI] [PMC free article] [PubMed] [Google Scholar]