Abstract
The human cytomegalovirus UL99 open reading frame encodes a 190-amino-acid (aa) tegument protein, pp28, that is myristoylated and phosphorylated. pp28 is essential for assembly of infectious virus, and nonenveloped virions accumulate in the cytoplasm of cells infected with recombinant viruses with a UL99 deletion. pp28 is localized to the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) in transfected cells, while in infected cells, it is localized together with other virion proteins in a juxtanuclear compartment termed the assembly compartment (AC). We investigated the sequence requirements for pp28 trafficking to the AC and assembly of infectious virus. Our studies indicated that the first 30 to 35 aa were required for localization of pp28 to the ERGIC in transfected cells. Mutant forms of pp28 containing only the first 35 aa localized with other virion structural proteins to cytoplasmic compartments early in infection, but localization to the AC at late times required a minimum of 50 aa. In agreement with previous reports, we demonstrated that the deletion of a cluster of acidic amino acids (aa 44 to 59) prevented wild-type trafficking of pp28 and recovery of infectious virus. A recombinant virus expressing only the first 50 aa was replication competent, and this mutant, pp28, localized to the AC in cells infected with this virus. These findings argued that localization of pp28 to the AC was essential for assembly of infectious virus and raised the possibility that amino acids in the amino terminus of pp28 have additional roles in the envelopment and assembly of the virion other than simply localizing pp28 to the AC.
Human cytomegalovirus (HCMV) is the largest and most complex member of the family of human herpesviruses. The virion of HCMV consists of three distinct structures, a nucleocapsid containing a double-stranded linear DNA genome, an envelope including an as-yet-undefined number of viral glycoproteins, and a tegument layer located between the capsid and the envelope (27, 40, 44, 45). HCMV assembly is a multistage and poorly understood process. Although all proposed models include well-studied mechanisms of capsid assembly within the nucleus of infected cells, final tegumentation and envelopment in the cytoplasm of infected cells remain poorly understood (25). The assembly pathway and protein interactions that are required for formation of the tegument layer are not defined. As an example, the tegument protein ppUL69 is expressed only in the nucleus, whereas some tegument proteins such as pp150 (ppUL32) and pp28 (ppUL99) are expressed only in the cytoplasm during the replication of HCMV (33). Other tegument proteins such as ppUL53 and pp65 (ppUL83) are expressed in the nucleus of cells early in infection but are localized predominantly in the cytoplasm late in infection (33). Thus, it is unclear whether tegument proteins associate with the capsid in the nucleus or in the cytoplasmic assembly compartment at a later step (25, 33). Electron microscopic studies have revealed that both nuclear and cytoplasmic subviral particles have an additional electron-dense layer consistent with a tegument layer, suggesting that tegumentation takes place partially in the nucleus and is presumably completed within the cytoplasm.
The role of individual tegument proteins in the replication and assembly of infectious HCMV has not been completely elucidated. However, it has been shown that many of the tegument proteins regulate viral gene expression or modify host cell responses to HCMV infection and likely have less important or nonessential roles in the assembly of the virion. As examples, pp71 (ppUL82) has been shown to transactivate immediate-early viral promoters, target cellular Rb family members for degradation, and inhibit the degradation of incoming DNA; ppUL69 has been shown to restrict cell cycle progression; and pp65 has been shown to inhibit the expression of genes associated with the induction of interferon responses (1, 10, 11, 15, 18, 23). Deletion of viral genes encoding any of these tegument proteins results in various degrees of impaired replication, but none exhibit comparable null phenotype of viruses with deletions in structural proteins such as the envelope glycoproteins gB and gM or the tegument protein pp28 (7, 8, 16, 17, 24, 36).
HCMV pp28 is a 190-amino-acid (aa) tegument protein that is encoded by the UL99 open reading frame (ORF). It is a true late protein that is both myristoylated and phosphorylated (19, 26, 34). The pp28 protein is one of the most abundant constituents of the tegument layer and is highly immunogenic (26, 42). Our previous studies have approached the investigation of the envelopment and assembly of HCMV by studying the intracellular trafficking of this protein. We have determined that pp28 is expressed only in the cytoplasm and is localized to the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC) in the absence of other viral proteins, suggesting that viral functions are required for its localization to the cytoplasmic assembly compartment (AC) late in infection (33, 34). Because pp28 is essential for the assembly of infectious virus, its localization in the AC suggested that pp28 may be involved in late steps of viral morphogenesis such as final tegumentation or envelopment (7, 17, 36). The finding that a mutant virus lacking pp28 failed to spread as cell-free infectious virus and the demonstration of nonenveloped cytoplasmic virions in cells infected with this pp28 deletion mutant virus were consistent with a key role of this tegument protein in the assembly of an enveloped virus (7, 36).
In this study, we investigated sequence requirements for intracellular trafficking of pp28 as an initial attempt to define the function of pp28 in the infectious cycle of HCMV. Our previous studies as well as those of other investigators have determined that myristoylation at glycine 2 is required for localization of pp28 in the ERGIC in the absence of other viral proteins as well as for the production of infectious virus (7, 17, 34, 36). More recently, Jones and Lee argued that an acidic cluster (aa 44 to 57) in the amino terminus of pp28 was required for the cytoplasmic localization of pp28 in virus-infected cells and for replication of infectious virus (17). Those authors also reported that the carboxyl-terminal two-thirds (aa 58 to 190) of pp28 were not essential for virus replication (17). To investigate sequence requirements for the trafficking and function of pp28, we created a panel of C-terminal deletion mutants after each 30 aa of pp28 and produced recombinant viruses expressing this series of C-terminal deletions by the use of a lambda phage-based linear recombination system. In addition, to define the possible functions of the acidic cluster domain for trafficking of pp28, we made two mutants, an acidic cluster (aa 44 to 59) deletion mutant and an acidic cluster deletion and insertion mutant in which the acidic cluster (aa 44 to 59) was transplanted to the C terminus of pp28. Our data indicated that in addition to myristoylation at glycine 2, the first 30 to 35 aa were required for localization of pp28 in the ERGIC in the absence of virus infection. Furthermore, the first 35 aa were sufficient for the cytoplasmic trafficking of pp28 with other virion structural proteins early in infection, but this mutant expressing only the first 35 aa of pp28 as well as other pp28 mutants that contained less than the first 50 aa of pp28 failed to accumulate mutant forms of pp28 in the AC late in infection. In addition, we also found that the first 50 aa were sufficient for the production of infectious virus and for wild-type (WT) trafficking of pp28 late in infection. Finally, our findings suggested that the sequences between aa 35 and 60 of pp28, especially the acidic cluster (aa 44 to 59), may function in a context-dependent fashion in protein interactions required for the final envelopment and assembly of virus within the infected cell.
MATERIALS AND METHODS
Cells, viruses, plasmids, and antibodies.
Primary human foreskin fibroblasts (HF) were prepared, propagated, and infected as previously described (9). HCMV strain AD169 was used for all experiments. Infectious stock was prepared from supernatants of infected HF cells which exhibited a 100% cytopathic effect and were titered as described previously (9).
For transient expression assays, a panel of carboxyl-terminal deletions was generated by the insertion of a stop codon into the gene encoding pp28 after each 30th amino acid (cloning vector pEF1; Invitrogen, San Diego, Calif.) (Fig. 1). To test the function of an acidic cluster domain in the trafficking of pp28, we also made two mutants, an acidic cluster (aa 44 to 59) deletion mutant (pp28Δac) and an acidic cluster insertion mutant (pp28Δac-CtTR), in which the acidic cluster (aa 44 to 59) was deleted and then transplanted to the carboxyl terminus (between aa 189 and 190) (Fig. 1). In addition, these truncated forms of pp28 (expressing 10, 14, 20, 25, 30, 35, 40, 50, 61, 80, 110, or 145 aa from the N terminus) were fused to enhanced green fluorescent protein (EGFP) (cloning vector EGFP-N2; Clontech, Palo Alto, Calif.). All pp28 deletion mutants were constructed using standard cloning techniques and PCR-based mutagenesis, and each mutant was sequenced prior to use in the experiments described in this report. Each truncated gene was transiently expressed in Cos-7 cells or 293T cells following calcium phosphate-mediated transfection (34). For transient expression/infection assays, pp28 mutants were transfected into HF cells, followed by infection with HCMV. In some experiments, cellular proteins such as ERGIC53 (a recycling ERGIC protein), mannosidase II (Man II) (a Golgi protein), and galactosyl transferase (Gal T) (a trans-Golgi protein) were fused to EGFP and served as markers for compartments of the secretory system. EGFP-Gal T and EGFP-Man II were kindly provided by Brian Storrie (University of Arkansas Medical Center, Little Rock, AR) (39).
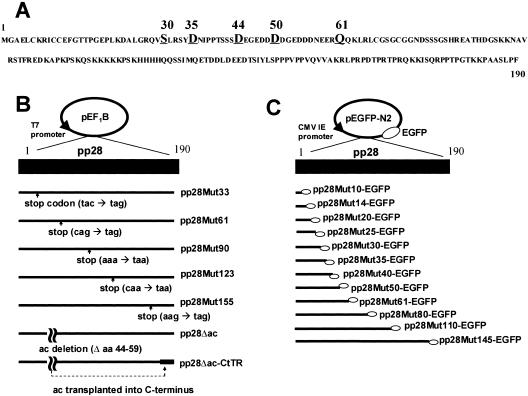
FIG. 1.
Generation of pp28 mutants and pp28 EGFP fusion proteins. (A) Amino acid sequences of 190-aa pp28. Amino acids that are relevant to this study are identified by larger font, and positions are listed above the sequence. (B) pp28 deletion mutants. In mutants pp28Mut33 to pp28Mut155, a stop codon was inserted into the nucleotide sequence following the codon designated in the mutant. In the pp28Δac mutant, the stretch of acidic amino acids (aa 44 to 59) was deleted internally, leaving the wild-type reading frame of the remainder of the molecule intact. The pp28Δac-CtTR mutant was generated by transplanting the acidic cluster (ac) (aa 44 to 59) into the C terminus of the pp28Δac mutant. The mutants were cloned into the pEF1B vector or pCDNA vector for transient expression assays. (C) Generation of the pp28 deletion mutants fused with EGFP. pp28 truncation mutants (Mut10 to Mut145) were fused with EGPF by cloning into the pEGFP-N2 vector. The mutants are numbered such that the final amino acid of the wild-type pp28 sequence that is expressed is designated in the mutant. CMV IE promoter, cytomegalovirus immediate-early promoter.
HCMV-encoded proteins were detected with monoclonal antibodies (MAbs) as previously described (33). MAbs used in this study included those specific for IE-1 (UL123) (MAb P63-27), pp150 (UL32) (MAb 36-14), pp28 (UL99) (MAb 41-18), gB (UL55) (MAb 7-17), and the gM/gN complex (UL100/UL73) (MAb 14-16A) (8, 24, 33). The antibodies that were reactive with cellular markers included a MAb specific for ERGIC53 (generously provided by Peter Hauri, University of Basel, Basel, Switzerland) and a MAb specific for p115 (purchased from Transduction Laboratories, Lexington, KY) as recycling ERGIC proteins, a rabbit antiserum specific for mannosidase II (kindly provided by Marilyn Farquhar, University of California, San Diego) and a rabbit antiserum against GM130 (purchased from Transduction Laboratories, Lexington, KY) as probes for Golgi proteins, a rabbit antiserum against TGN46 (purchased from Serotec Ltd., Oxford, United Kingdom) as a trans-Golgi protein, and a rabbit antiserum specific for calreticulin as an ER-resident protein (purchased from Affinity BioReagents, Golden, Colo.). A rabbit antiserum against the c-Myc epitope tag was purchased from Affinity BioReagents, Golden, Colo. Fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG) subclass-specific antibodies, FITC-conjugated goat anti-rabbit IgG antibodies, Texas Red-conjugated goat anti-mouse IgG subclass-specific antibodies, Texas Red-conjugated goat anti-rabbit IgG antibodies, tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-mouse immunoglobulin G subclass-specific antibodies, and TRITC-conjugated goat anti-rabbit IgG antibodies were purchased from Southern Biotechnology Associates, Birmingham, Ala.
Generation of recombinant viruses.
Recombinant viruses were constructed utilizing a two-step strategy for the introduction of point mutations into the HCMV genome maintained as an infectious bacterial artificial chromosome (BAC) in Escherichia coli as previously described (7). The BAC-containing HCMV strain AD169 (HB-5) was provided by Martin Messerle and Ulrich Koszinowski (University of Munich, Germany) (6). In the first step, the UL99 ORF was deleted from a BAC-maintained HCMV genome by replacement with an Ampr lacZ cassette. Growth in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) enabled the identification of HCMV recombinants by visual inspection for blue bacterial colonies. The mutagenesis was performed with RED locus-mediated recombination to delete sequences between positions 145310 and 145795 of the AD169 genome. A primer set amplifying sequences from positions 145210 (forward primer) and 146025 (reverse primer) was used to produce a recombination cassette of approximately 2.0 kbp that carried the Ampr lacZ cassette together with approximately 100 to 200 bp of viral sequence flanking the site of the deletion within the HCMV genome. The cassette was recombined into the AD169 BAC by RED locus-mediated recombination using the protocol described previously by Lee et al., resulting in the production of Ampr blue colonies (7, 20). Insertion of the cassette resulted in the deletion of the entire pp28 ORF except for 100 bp of the 3′ end (recombinant designated 99KO BAC). To ensure the correct targeting of the recombination cassette into the desired genomic location, Southern blot analysis was performed. The recombinant BAC DNA was digested with HindIII, electrophoresed in agarose gels, transferred onto nitrocellulose membranes, and hybridized with a 32P-labeled probe specific for the Ampr lacZ cassette or the UL99 ORF. Nucleotide sequence analysis of PCR products amplified from these recombinant BACs confirmed the correct insertion of the Ampr lacZ cassette and replacement of the pp28 ORF. In the second step, RED recombination was used to replace the pp28 deletion in 99KO BAC with either the wild-type pp28 ORF sequence or the pp28 sequence containing the desired mutations by using a linear DNA fragment. Recombination removed the Ampr lacZ cassette from the 99KO BAC, and repaired or mutagenized recombinants were identified by visual inspection for white colonies. The same primer set described above was used to prepare linear DNA fragments containing the desired mutations by PCR from a pp28 template as previously described (7). This methodology allowed the insertion of single nucleotide changes in the native UL99 ORF to create stop codons and maintain the transcription program and the normal regulation of the expression of this region of the wild-type genome.
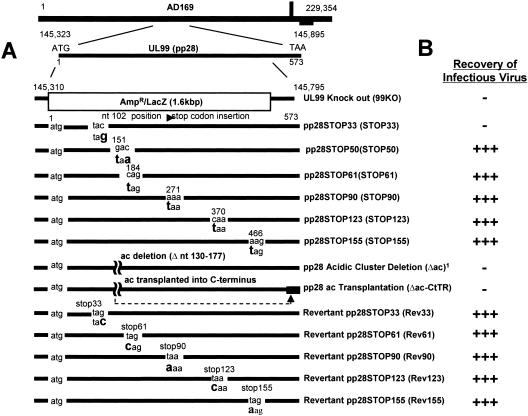
In the two-step strategy described above, we produced mutated BACs that contained mutations in the pp28 ORF that resulted in lethal mutations. Using a similar recombination strategy, the mutated BACs could also be repaired by using single-stranded oligonucleotides that contained a repaired stop codon and approximately 20 nucleotides flanking each side of the repair codon. The repair oligonucleotides were electroporated into E. coli containing the mutated HCMV BAC and recombined, and following expansion of this mixed population of E. coli (mutant and repaired BACs), BAC DNA was purified and electroporated together with a plasmid encoding pp71 into HF cells. Finally, 14 recombinant BAC DNAs (UL99KO; a series of pp28 deletion mutants expressing the first 33 [pp28STOP33], 50 [pp28STOP50], 61 [pp28STOP61], 90 [pp28STOP90], 123 [pp28STOP123], or 155 aa [pp28STOP155] of pp28; an acidic cluster deletion mutant in pp28 [pp28Δac]; an acidic deletion mutant in which the acidic cluster was deleted and transplanted to the C terminus [pp28Δac-CtTR]; and a series of revertant BAC DNAs repaired from the pp28 mutants [pp28Rev33, pp28Rev61, pp28Rev 90, pp28Rev123, and pp28Rev155]) were generated for this study (see Fig. 7).
FIG. 7.
Construction of UL99 recombinant BACs containing mutations in pp28. (A) Recombinant BACs containing pp28 deletion mutants were generated by the insertion of single nucleotide (nt) changes in the UL99 coding sequence that altered wild-type codons to translational stops. Mutations were confirmed by sequence analysis. Revertant or repaired BACs were made by oligonucleotide-directed repair of the single nucleotide mutations. Recombinant viruses were recovered by electroporation of BAC DNA into HF cells. (B) Successful recovery of infectious viruses is listed on the right. In all cases, at least three independent attempts were made to recover infectious virus. The acidic cluster (ac) deletion BAC was made by the deletion of nucleotides 130 to 177 in the UL99 coding sequence. A virus was recovered from this pp28Δac BAC, but it formed small and very slowly expanding plaques. We could not isolate cell-free infectious virus from these cultures, nor did the plaques expand through the monolayer.
To determine whether the recombinant pp28 gene was essential for virus infectivity in HF cells, DNA was purified from the BAC-containing E. coli cells and electroporated into HF cells. One microgram of an expression plasmid encoding pp71 was included to enhance the recovery of infectious virus. Infectivity was monitored by observing the production of visible plaques. In assays for virus replication, virus titers were determined by a fluorescent virus infectivity assay of HF cells infected with WT or recombinant viruses at the indicated times after infection (2).
SDS-PAGE and immunoblotting.
Sodium dodecyl sulfate (SDS)-polyacrylamide electrophoresis (PAGE) under reducing conditions and immunoblotting were carried out as described previously (4). Virus-infected cell proteins were extracted from wild-type virus- or recombinant virus-infected HF cells grown in 35-mm-diameter tissue culture dishes. Following washing in phosphate-buffered isotonic saline (PBS) (pH 7.4), the cells were lysed in sample buffer containing 5% 2-mercaptoethanol and 2% SDS and heated to 100°C. The solubilized proteins were then subjected to SDS-PAGE and transferred onto nitrocellulose membranes. Murine MAbs or in some cases a 1:100 dilution of the IgG fraction of the rabbit anti-c-Myc serum was used to detect specific proteins. Antibody binding was detected by 125I-protein A followed by autoradiography.
Immunofluorescence microscopy.
HF cells were grown in 24-well tissue culture plates containing a 13-mm-diameter coverslip. After the cells were 90% confluent, they were infected with HCMV strain AD169 derived from the HB-5 BAC or from mutant viruses generated from the HB-5 BAC for 1 to 2 h, washed once, and incubated for the indicated time. The coverslips were harvested by first washing the cells with PBS and then fixing the cells for 45 min at room temperature in 3% paraformaldehyde freshly prepared in PBS. The coverslips were washed in PBS and permeabilized with 0.05% Triton X-100 and 0.001% SDS in PBS for 7 min. The coverslips were then blocked with PBS containing 10% normal goat serum for 20 min at room temperature, followed by the addition of primary antibody, and incubated for 60 min at 37°C. Following washing (three times) with 0.2% Tween in PBS, the coverslips were incubated with FITC-conjugated and/or Texas Red-conjugated or TRITC secondary antibody diluted in PBS-Tween containing 2.5% normal goat serum for 45 min at 37°C. The coverslips were washed three times and then rinsed once in PBS, mounted with SlowFade antifade reagent (Molecular Probes, Eugene, Oreg.), and viewed using a Leitz Diavert fluorescence microscope or an Olympus confocal microscope. The images were captured with a digital camera (Photometrics, Tucson, Ariz.) using the Leitz epifluorescence microscope at a magnification of ×60. In some cases, images were processed with Image Pro software (Media Cybernetics, Silver Spring, Md.). Deconvolution was accomplished with Hazebuster (Vaytek, Fairfield, Iowa).
Cos-7 cells grown on coverslips were transfected with an expression vector containing the truncated pp28 sequence or a vector encoding a pp28-EGFP fusion protein (pp28EGFP). Transfected cells were fixed 36 to 48 h posttransfection, and cells expressing pp28 were reacted with MAb 41-18 followed by Texas Red-conjugated goat anti-mouse IgG antibody as described above. In a transient expression/infection assay, HF cells were electroporated with approximately 5 μg of DNA from an expression vector containing the truncated pp28 sequence, a vector encoding a pp28-EGFP fusion protein, or a vector encoding EGFP fused to markers for cellular compartments. Forty-eight hours later, the cells were infected with HCMV and then washed once and incubated for the indicated times. The infected cells were fixed and stained as described above.
Subcellular fractionation.
A 75-cm2 flask of HF cells was electroporated with 5 μg of an expression vector encoding pp28Mut40EGFP, pp28Mut50EGFP, or pp28ΔacEGFP. Twenty-four hours later, the cells were washed and infected with HCMV at a multiplicity of infection (MOI) of 0.2. The HF cells were harvested on day 6 postinfection by trypsinization, and the cell pellet was washed twice with cold PBS and then resuspended in 1 ml of homogenization buffer (0.25 M sucrose, 10 mM HEPES, pH 7.4, 1 mM EDTA). The cell suspension was repeatedly passed through a 23-gauge needle until there were no intact cells in the suspension, as determined by light microscopy, and a postnuclear supernatant was collected following centrifugation at 1,000 × g for 10 min. Subcellular fractionation was performed using a density gradient prepared from iodixanol (Optiprep; Sigma, St. Louis, Mo.) and ultracentrifugation using a modification of protocols described previously (37, 43). A discontinuous gradient was prepared using 30%, 25%, 20%, 15%, and 10% (vol/vol) Optiprep solution. The gradient was allowed to equilibrate vertically for 30 min at room temperature. The postnuclear supernatant was overlaid onto the discontinuous gradient and centrifuged at 100,000 × g in an SW41 rotor for 3 h at 4°C. Equal fractions were collected from the top of the gradient, and individual fractions were assayed for viral and host cell proteins by immunoblotting.
RESULTS
Sequence requirements for trafficking of transiently expressed pp28 to the ERGIC.
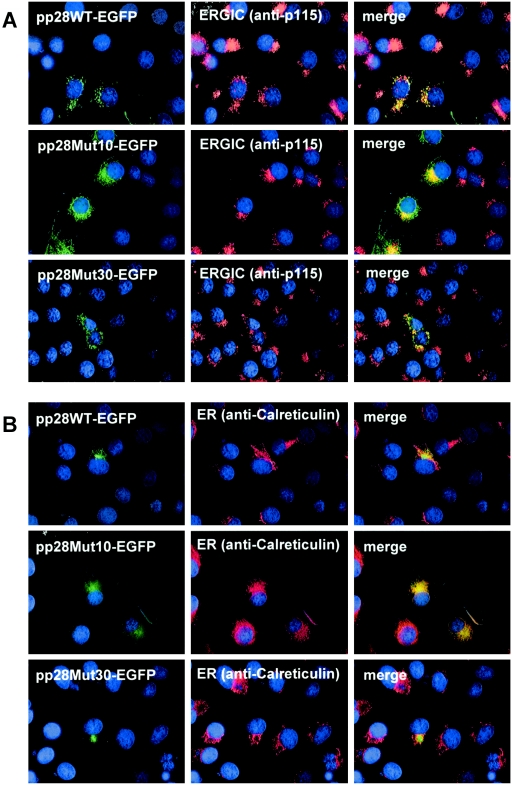
Initially, we determined sequences within pp28 that were required for its localization in the ERGIC. Previously, we have shown that pp28 is localized in the ERGIC and does not traffic to more distal compartments of the Golgi/trans-Golgi network (TGN) in cells transiently expressing pp28 in the absence of other viral proteins (34). To define specific domains required for the localization of this protein in the ERGIC, we transfected Cos-7 cells with plasmids encoding a series of pp28 C-terminal deletion mutants and determined their intracellular localization. This panel of pp28 mutants was generated by the insertion of stop codons into the coding sequence of the wild-type gene or by the generation of C-terminal truncations of pp28 followed by a fusion of the remaining coding sequence with EGFP, as described in the legend of Fig. 1. In addition, we also created two mutants in which the acidic domain located between aa 44 and 59 was either deleted or excised and transplanted to the C terminus of the molecule, as detailed in the legend of Fig. 1. These latter two mutants were constructed because it has been previously reported that sequences within the first acidic domain of pp28 (aa 44 to 57) are required for its localization within infected cells (17). All mutations were confirmed by nucleotide sequencing, and expression of the mutant form of pp28 was demonstrated by Western blotting of pp28 mutants following transient expression in 293T cells (data not shown). The intracellular trafficking of pp28 deletion mutants was monitored by localizing their intracellular expression using antibodies that are reactive with proteins specific for cellular compartments (Fig. 2). Surprisingly, the trafficking of most pp28 deletion mutants was indistinguishable from that of wild-type pp28 and remained localized within the ERGIC (Fig. 2). Mutants expressing polypeptides of the first 33, 61, 90, 123, or 155 aa of pp28 all appeared to have intracellular localization similar to that of wild-type pp28 (Table 1). Colocalization with a protein marker of the ERGIC, p115, was also observed following expression of C-terminal truncation pp28 mutants that encoded the first 30, 35, 40, 50, 61, 80, 110, or 145 aa of pp28 fused to EGFP (Table 1). The two acidic cluster mutants, pp28Δac (aa 44 to 59) and pp28Δac-CtTR, also colocalized with markers of the ERGIC (Table 1). An example of this pattern of localization is shown by the distribution of pp28Mut30EGFP following transient expression in Cos-7 cells (Fig. 2). We noted that the vesicular distribution of this pp28 mutant protein closely resembles that of wild-type pp28, and there was partial overlap with the protein marker of the ERGIC, p115 (Fig. 2). In contrast, pp28 mutants that contained less than the first 30 aa failed to colocalize with markers of the ERGIC, and their intracellular localization differed from that of wild-type pp28. This is illustrated by the mutant pp28Mut10EGFP, which was distributed in a pattern most consistent with ER-resident proteins, such as calreticulin, when transiently expressed in Cos-7 cells (Fig. 2). Each of these pp28 mutants contained a glycine codon at position 2 and was therefore presumably myristoylated, and thus, mislocalization cannot be ascribed to a lack of membrane association. These results indicated that the first 30 aa of pp28 together with the myristoylation modification at amino acid position 2 were required for the localization of this pp28 protein in the ERGIC when pp28 was expressed in the absence of virus infection.
FIG. 2.
Localization of transiently expressed pp28 deletion mutants in the ERGIC. pp28 deletion mutants were transfected into Cos-7 cells. At day 2 posttransfection, the cells were fixed with 3% paraformaldehyde and examined by fluorescence microscopy as described in Materials and Methods. (A) Wild-type pp28EGFP (pp28WTEGFP) or pp28MutEGFP fluorescence is green, and organelle (p115, ERGIC marker) fluorescence is red (Texas Red). (B) Wild-type pp28EGFP (pp28WTEGFP) or pp28MutEGFP fluorescence is green, and organelle (calreticulin, ER marker) fluorescence is red (Texas Red). Nuclei are pseudocolored in blue following staining with Hoechst dye.
TABLE 1.
pp28 sequence requirements for localization in the ERGIC
| Proteinb | ERGICa | ERa |
|---|---|---|
| pp28Mut33 | ++ | |
| pp28Mut61 | ++ | |
| pp28Mut90 | ++ | |
| pp28Mut123 | ++ | |
| pp28Mut155 | ++ | |
| pp28Δac | ++ | |
| pp28Δac-CtTR | ++ | |
| pp28Mut10-EGFP2 | − | ++ |
| pp28Mut14-EGFP | − | ++ |
| pp28Mut20-EGFP | + | ++ |
| pp28Mut25-EGFP | + | ++ |
| pp28Mut30-EGFP | ++ | − |
| pp28Mut35-EGFP | ++ | − |
| pp28Mut40-EGFP | ++ | − |
| pp28Mut50-EGFP | ++ | − |
| pp28Mut61-EGFP | ++ | − |
| pp28Mut80-EGFP | ++ | − |
| pp28Mut110-EGFP | ++ | − |
| pp28Mut145-EGFP | ++ | − |
| pp28WT-EGFP | ++ | − |
Localization in the ERGIC was determined by colocalization of the pp28 mutant with ERGIC53 (ERGIC) or p115, and localization to the ER was determined by colocalization with calreticulin (ER). The intensity of the signal based on colocalization with the marker was scored as partially (+), primarily (++), or not (−) detected in the organelle.
pp28 mutants were designated by the last remaining amino acid from the amino terminus of pp28.
Localization of pp28 to the assembly compartment of virus-infected cells requires a late viral function.
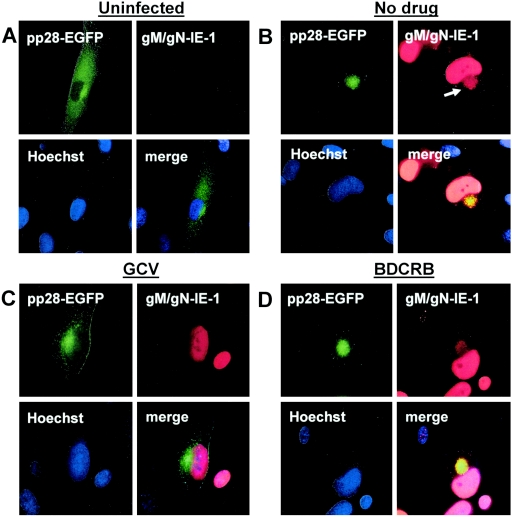
In virus-infected cells, pp28 was localized to a membranous cytoplasmic compartment that we have designated the virus AC based on its morphological appearance, the isolation of this membrane-bound compartment by cell fractionation, and the localization of a number of structural tegument and envelope proteins in this compartment late in infection (33). Because pp28 remained in the ERGIC when transiently expressed in the absence of other viral proteins, the trafficking pathway of pp28 to the assembly compartment likely required the expression of an undefined viral function. To formally demonstrate this possibility, we developed an assay based on transient expression of EGFP- or epitope-tagged pp28 or mutant pp28 followed by virus infection to supply the virus function in trans (transient expression/infection assay). In this assay, the EGFP-tagged pp28 protein was expressed in HF cells following electroporation of a plasmid encoding the pp28 molecule fused at its C terminus to EGFP. Electroporated HF cells were infected with HCMV 48 h later at an MOI of 0.1. In uninfected cells, the pp28EGFP protein was expressed throughout the cytoplasm of the cell in a punctuate distribution that was similar to the distribution of ERGIC markers in uninfected HF cells (Fig. 3). Shortly after infection of electroporated cells, the pp28EGFP fusion protein was expressed in the cytoplasm of infected cells in a distribution similar to that of uninfected cells (data not shown). However, late in infection, the transiently expressed pp28EGFP protein could be colocalized with the gM/gN glycoprotein complex in the AC as illustrated by its expression in infected cells expressing IE-1 (pp72) in the nucleus and the gM/gN complex in the cytoplasm (Fig. 3). In contrast to these findings, when monolayers of pp28EGFP-electroporated/infected cells were treated with ganciclovir to block the expression of late viral proteins, the pp28EGFP fusion protein no longer localized to the AC at late times in infection but was expressed diffusely in the cytoplasm (Fig. 3). The transiently expressed pp28EGFP fusion protein was localized to the AC when these cultures of electroporated/infected cells were treated with the antiviral compound BDCRB (2-bromo-5,6-dichloro-1-β-d-ribofuranosyl benzimidazole), a compound that inhibits late steps of virus assembly, cleavage, and packaging of viral DNA but not late protein synthesis (Fig. 3) (41). These results demonstrated that authentic localization of pp28 to the AC required the expression of a late viral function and that localization of pp28 to the AC was not inhibited by antiviral compounds that specifically blocked nuclear events of virus assembly but not late protein synthesis.
FIG. 3.
pp28 localization in the assembly compartment requires late gene expression. HF cells (3 × 106 cells) were electroporated with approximately 5 μg of pp28EGFP and were either left uninfected (A) or infected with HCMV 48 h later at an MOI of 0.2 (B to D). Individual wells were treated with medium control (no drug, control) (B), ganciclovir (GCV) (C), or BDCRB (D). Cells were fixed 120 h postinfection, stained with anti-gM/gN and anti-IE-1 MAbs, and developed with TRITC anti-mouse IgG secondary antibodies. Nuclei were stained with Hoechst dye (blue) and IE-1 (red) in infected cells. The arrow in B demarcates the assembly compartment as indicated by gM/gN staining. Note that pp28EGFP was present in the assembly compartment of >90% of cells in control wells compared to <10% of cells in GCV-treated cultures.
Sequence requirements for localization of pp28 to the AC in virus-infected cells.
Having demonstrated that wild-type pp28EGFP localized to the AC in cells transfected with pp28EGFP and infected with HCMV, we utilized this assay to investigate the intracellular localization of transiently expressed mutant forms of pp28 to identify sequence requirements for the localization of pp28 to the AC. The EGFP plasmids expressing the first 30, 35, 40, 50, and 61 aa of pp28 (pp28Mut30EGFP, pp28Mut35EGFP, pp28Mut40EGFP, pp28Mut50EGFP, and pp28Mut61EGFP, respectively) were used in this transient expression/infection assay. In addition, the localization of the acidic cluster mutant pp28Δac (aa 44 to 59) fused with EGFP (pp28ΔacEGFP) and the pp28 mutant in which the acidic cluster (aa 44 to 59) was deleted and transplanted tothecarboxyl terminus and tagged with the Myc epitope (pp28ΔacCtTRMyc) were also studied. After electroporation of plasmids expressing pp28EGFP, pp28Mut30EGFP, pp28Mut35EGFP, pp28Mut40EGFP, pp28Mut50EGFP, pp28Mut61EGFP, pp28ΔacEGFP, and pp28ΔacCtTRMyc, we observed comparable intracellular distributions within 48 h after electroporation (data not shown). The electroporated cells were then infected with HCMV at an MOI of 0.2, and the localization of pp28EGFP or mutant forms of pp28 in infected cells (IE-1-expressing cells) was assayed. Interestingly, all of the pp28 mutants, with the exception of pp28Mut30EGFP, were expressed in a perinuclear compartment early after infection (day 4 postinfection) (data not shown). However, the efficiency of localization of wild-type and mutant forms of pp28 in this compartment varied between mutants such that the relative efficiency of localization to the perinuclear compartment early in infection depended on the length of the pp28 mutant. The shorter mutants, such as pp28Mut35EGFP, localized to this perinuclear compartment less efficiently than the longer mutants, such as pp28Mut50EGFP. These results suggested that only the first 35 aa of pp28 were required for trafficking to a perinuclear compartment that appeared to form shortly after virus infection, although the efficiency of localization to the this compartment was decreased for the shorter pp28 mutants.
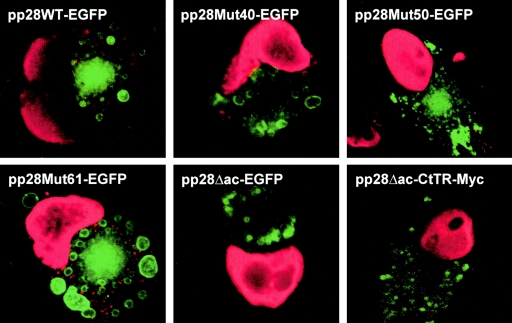
In marked contrast to findings early in infection, the localization of mutant forms of pp28 on day 7 postinfection revealed qualitative differences between several mutants and wild-type pp28EGFP. The mutants pp28Mut40EGFP, pp28ΔacEGFP, and pp28ΔacCtTR-Myc failed to localized to the AC and were distributed peripherally to the AC compared to wild-type pp28EGFP (Table 2 and Fig. 4). As shown in some panels of Fig. 4, we costained cells with anti-IE-1 MAb and anti-GM130 MAb to allow the detection of infected cell nuclei and the Golgi compartment to help further define the intracellular localization of pp28 mutants. In the experiments in which we successfully visualized GM130, we noted that this Golgi compartment marker was displaced peripherally from the AC (Fig. 4). This finding was further explored in a series of experiments (see below). Interestingly, pp28Mut50EGFP was localized to the AC in only about 50% of EGFP-positive cells at 7 days postinfection (Table 2 and Fig. 4). pp28Mut61EGFP was concentrated in the AC with an efficiency comparable to that of wild-type pp28EGFP (Table 2). The differences in the localization of pp28 mutant forms in the AC at late times during infection suggested that sequences between aa 35 and 61 of pp28, particularly the N-terminal acidic cluster (aa 44 to 59), might function through specific interactions with other viral proteins or cellular proteins for the final localization of pp28 to the AC. Interestingly, pp28Mut50EGFP, containing one-half of the acidic cluster of amino acids, appeared to have an intermediate phenotype. These findings were consistent with the replication-defective phenotype of the previously reported recombinant viruses expressing less than 57 aa of the amino terminus of pp28 (17). Together, these findings argued that a defect in the localization or concentration of pp28 mutant proteins in the mature AC late but not early in infection was limiting in the process of the envelopment and assembly of infectious virus.
TABLE 2.
pp28 sequence requirements for localization to the AC following transient expression and infection of HF cells
| Proteinb | ACa |
|---|---|
| pp28Mut10-EGFP | NT |
| pp28Mut14-EGFP | NT |
| pp28Mut20-EGFP | NT |
| pp28Mut25-EGFP | NT |
| pp28Mut30-EGFP | − |
| pp28Mut35-EGFP | − |
| pp28Mut40-EGFP | − |
| pp28Δac-EGFP (Δaa 44-59) | − |
| pp28Δac-CtTR-Myc | − |
| pp28Mut50-EGFP | + |
| pp28Mut61-EGFP | ++ |
| pp28Mut80-EGFP | ++ |
| pp28Mut110-EGFP | ++ |
| pp28Mut145-EGFP | ++ |
| pp28WT-EGFP | ++ |
Localization to the AC was assayed by imaging of cells 6 to 7 days postinfection. Localization was scored as − if there was no localization in the compact AC, + if 50% of cells demonstrated localization to the AC, and ++ if >70% of cells exhibited localization of mutant pp28 or wild-type pp28 in the compact AC. NT, not tested in this experiment.
pp28 mutants were designated by the last remaining amino acid of the amino terminus of pp28.
FIG. 4.
Localization of pp28 deletion mutants to the assembly compartment late in infection in HCMV-infected HF cells. HF cells were electroporated with approximately 5 μg of expression plasmids encoding wild-type pp28EGFP (pp28WT-EGFP), pp28Mut40EGFP, pp28Mut50EGFP, pp28Mut61EGFP, pp28ΔacEGFP, or pp28ΔacCtTR-Myc and infected 2 days later with HCMV at an MOI of 0.2. The cells were harvested at day 7 postinfection, fixed with 3% paraformaldehyde, stained with anti-IE-1 MAbs combined with anti-GM130 MAbs (pp28EGFP, pp28Mut40EGFP, pp28Mut50EGFP, pp28Mut61EGFP, and pp28ΔacEGFP) to localize the AC to a secretory compartment or with anti-IE-1 MAbs only (pp28ΔacCtTR-Myc) followed by TRITC-labeled anti-mouse IgG to identify infected cells (red nuclei) and examined by confocal microscopy. The expression of the pp28ΔacCtTR-Myc-tagged mutant was detected with an anti-Myc MAb followed by FITC anti-mouse IgG (green fluorescence). The AC can be seen in the juxtanuclear position in cells expressing pp28EGFP, pp28Mut61EGFP, and pp28Mut50EGFP. Note the weak staining from the GM130 MAb staining of the Golgi compartment that is surrounding the AC in cells expressing wild-type pp28EGFP, pp28Mut40EGFP, pp28Mut50EGFP, and pp28Mut61EGFP. The GM130 reactivity in infected cells expressing pp28ΔacEGFP cannot be appreciated in this photograph.
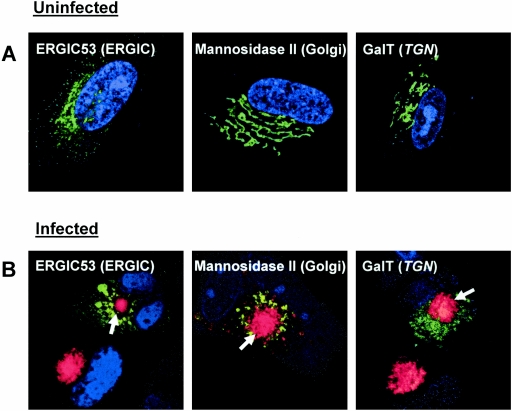
The striking differences between the localization of wild-type pp28 in the AC and the distribution of pp28 mutants such as pp28Mut40 on the periphery of the AC suggested that pp28 mutants such as pp28Mut40 were restricted to more proximal compartments of the secretory pathway that were reorganized during HCMV infection. This possibility was consistent with previous imaging findings that demonstrated the exclusion of protein markers of the Golgi compartment and TGN from the AC (33). To explore the possibility that morphological reorganization of the host cell secretory compartment took place during virus infection, we investigated the distribution of markers of the secretory compartment, including ERGIC53-EGFP, Man II-EGFP, and Gal T-EGFP, in transient expression/infection assays. These markers of the secretory system redistributed into perinuclear spherical structures that also contained viral proteins, such as gB, and pp28 early in infection (<4 days) (data not shown). We then utilized the same transfection/infection assays to determine the resident distribution proteins of compartments of the secretory system late in infection. The analysis was done at late times during infection for several reasons, including that characteristics of the AC have been defined in cells late in infection, pp28 is expressed with true late kinetics, and virus production and presumably virus assembly are maximal late in infection. By day 7 postinfection, signals from these protein markers of the secretory compartment revealed a dramatic morphological reorganization of the host cell secretory system and demonstrated that the secretory compartment had been reorganized into layers that ringed the pp28-containing, juxtanuclear AC (Fig. 5). Moreover, we observed that late in infection, the AC was a compact structure compared to the more diffuse and spherical perinuclear structure early in infection, and in contrast to the colocalization with markers of the secretory pathway early in infection, the AC containing pp28 was devoid of markers of the Golgi compartment or TGN (Fig. 5). These findings were consistent with the maturation of the AC during the course of infection in an individual cell, a mechanism consistent with results from previous studies (33). Previously, we demonstrated that several viral proteins were localized in the AC late in infection, but interestingly, in some experiments utilizing these markers of the host secretory pathway, gB could also be detected both in the center of the AC and partially overlapped with the signal from the Golgi compartment (data not shown). Thus, it appeared that the AC represented a site of viral protein accumulation within a morphologically altered secretory compartment displaced to the periphery of the AC. Some viral glycoproteins such as gB could be colocalized with the Golgi compartment and TGN as well in the more compacted AC late in infection, suggesting the possibility that this molecule and other viral proteins trafficked from the Golgi/TGN into the AC during virus assembly.
FIG. 5.
Altered morphology of the secretory compartment following HCMV infection. HF cells were electroporated with approximately 5 μg of plasmids encoding ERGIC53 (ERGIC marker), mannosidase II (Golgi marker), and Gal T (TGN marker) proteins fused to EGFP and were either (A) left uninfected or (B) infected with HCMV at an MOI of 0.2 36 h after electroporation. The infected cells were incubated for 7 days prior to fixation as described in Materials and Methods. Cells that were fixed 7 days postinfection were reacted with anti-pp28 MAb followed by TRITC anti-mouse IgG to show the assembly compartment. Nuclei were stained (blue) with Hoechst dye. The arrow demarcates the assembly compartment.
Subcellular localization of wild-type pp28 and pp28 mutants.
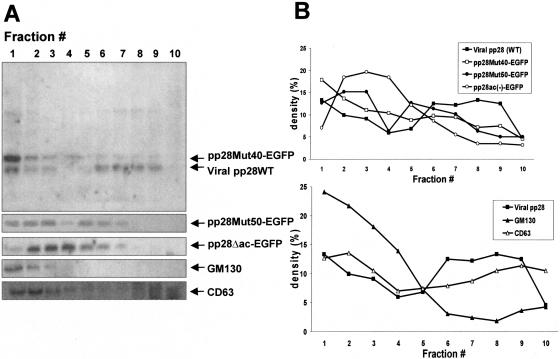
To further investigate the nature of the AC containing the pp28 WT protein and to compare the intracellular distribution of wild-type pp28 with mutant forms of pp28 late in infection, we used centrifugation through density gradients to analyze the distribution of transiently expressed viral proteins in cells transfected with pp28Mut40EGFP, pp28Mut50EGFP, or pp28ΔacEGFP and then infected with HCMV. Postnuclear supernatants were prepared from cells harvested late in infection (>6 days) and subjected to centrifugation through discontinuous iodixanol (Optiprep) density gradients, and individual fractions were analyzed by Western blotting for the presence of virus-encoded wild-type pp28, the mutant pp28EGFP proteins, and host cell proteins GM130 (Golgi compartment) and CD63 (late endosomes). The amount of protein detected in each fraction was quantified by densitometry and is presented as a fraction of the total amount of the specific protein recovered from the entire gradient. The wild-type pp28 viral protein was distributed into two broad peaks, with one being associated with the first three fractions and a second broad peak migrating further into the gradient (fractions 6 to 9) (Fig. 6). The mutant pp28 protein pp28Mut40EGFP exhibited a very different distribution in the gradient, with the majority of this protein migrating in the first three fractions of the gradient (Fig. 6). Similarly, the pp28 mutant protein pp28ΔacEGFP also partitioned primarily in the top fractions of the gradient as a single broad peak (Fig. 6). Interestingly, the pp28 mutant pp28Mut50EGFP was found in two broad peaks in a distribution that was most similar to that of wild-type pp28 (Fig. 6).
FIG. 6.
Iodixanol density gradient fractionation of HCMV-infected HF cells transfected with pp28 mutants. HF cells were electroporated with approximately 5 μg of the designated pp28MutEGFP expression plasmids and infected 2 days later with HCMV at an MOI of 0.2. The cells were harvested at day 6 postinfection and fractionated by centrifugation through iodixanol (Optiprep; Sigma Co., St. Louis, Mo.) gradients as described in Materials and Methods. The gradient was fractionated by removing 1-ml fractions from the top; thus, fraction 1 represents the top of the gradient, and fraction 10 represents the bottom of the gradient. (A) Gradient fractions were analyzed by Western blot. Proteins were detected with specific antibodies, anti-pp28 MAb for WT pp28 and pp28Mut40EGFP, pp28Mut50EGFP, and pp28ΔacEGFP and developed with 125I-protein A. MAbs reactive with cellular proteins specific for compartments of the secretory pathway, anti-GM130 and anti-CD63, were used to localize pp28 and pp28 mutants to different intracellular compartments. (B) Results of panel A were analyzed by densitometry [density (%) = peak density of each fraction/total density of signal from protein in all gradient fractions × 100]. The top of B is a graphic comparison of the pattern of fractionation of virus-encoded pp28 to those of pp28Mut40EGFP, pp28Mut50EGFP, and pp28ΔacEGFP. Viral pp28 (WT) (▪), pp28Mut40EGFP (□), pp28Mut50EGFP (•), and pp28ΔacEGFP (○) are depicted. The bottom of B represents a comparison of the pattern of fractionation from viral pp28 to those of subcellular organelles (GM130, Golgi; CD63, late endosome). Viral pp28 (WT) (▪), GM130 (▴), and CD63 (▵) are depicted.
To determine the distribution of wild-type pp28 and mutant forms of pp28 in different compartments of the cellular secretory pathway, we assayed the distribution of two host cell proteins within these same gradient fractions. Our results were similar to those reported previously by Sims et al. (37). The Golgi compartment-specific protein GM130 could be localized to fractions 1 to 3, whereas the late endosomal marker CD63 was distributed over the gradient in two broad peaks, one of which coincided with the distribution of viral pp28 in fractions 6 to 9 (Fig. 6). This broad migration of endosomes has been previously observed by other investigators using similar conditions for density gradient separation of cellular organelles and is thought to be secondary to the heterogeneity of the density of endosomes as a result of differences in cargos (38). The results were also consistent with our imaging findings that suggested that mutant forms of pp28 localized outside of the AC and could be colocalized with proteins such as mannosidase II, a marker of the Golgi compartment, and not with the late endosomal marker CD63 (data not shown). Furthermore, these data also demonstrated that wild-type pp28 and the pp28Mut50EGFP mutant partitioned similarly in this gradient and, perhaps most importantly, could be found in fractions containing Golgi protein as well as fractions containing the late endosomal marker protein CD63. In contrast, the distribution of both pp28Mut40EGFP and pp28ΔacEGFP mutants were found most abundantly in fractions from the top of the gradient along with a cellular marker protein of the Golgi compartment. Together with data from our imaging studies, these findings provided additional evidence that mutant forms of pp28 that failed to localize to the AC late in infection were distributed in different cellular compartments compared to either wild-type pp28 or a mutant form of pp28, pp28Mut50. Lastly and most importantly, these data were also consistent with the capacity of the mutant protein pp28Mut50 to support virus replication when recombined into the viral genome, in that the pp28Mut50 protein partitioned similarly to the wild-type pp28 protein in these gradients and localized to the AC, as determined by image analysis (see below).
Mislocalization of pp28 prevents assembly of infectious progeny virus.
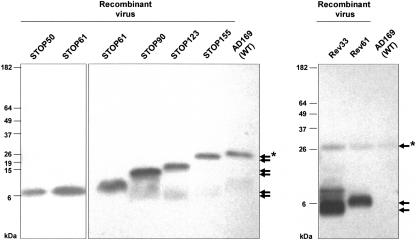
To examine the effect of mislocalization of pp28 mutants on the assembly of infectious virus, we generated a panel of recombinant viruses with stop codons inserted into the pp28 coding sequence using a linear recombination system previously described (7). Restriction fragment patterns of recombinant BAC DNAs containing mutations in UL99 were identical to that of the wild-type parent virus and differed from that of the UL99KO BAC utilized for replacement mutagenesis (data not shown). Southern blot analysis using a 32P-labeled probe generated by nick translation of the UL99 ORF revealed the presence of the 6-kbp HindIII R fragment in wild-type and mutant pp28 BAC DNA and the approximately 8-kbp HindIII R fragment that contained the 2-kbp Amp LacZ cassette in the UL99KO BAC (data not shown). Nucleotide sequencing of BAC DNAs that were mutated by the introduction of point mutations encoding translational stop codons verified the introduction of a stop codon into the predicted location and the insertion of a translational stop at the predicted location in the amino acid sequence. After electroporation of HF cells with DNAs from these recombinant BACs, infectious virus was recovered from pp28 deletion mutants expressing the first 50, 61, 90, 123, or 155 aa (STOP50, STOP61, STOP90, STOP123, and STOP155, respectively) of pp28 (Fig. 7). As reported previously, infectious virus was not recovered following electroporation of HCMV BAC containing a Gly→Ala mutation at codon 2 and the insertion of a stop codon at codon 4 (7). Infectious virus was also not recovered from a mutant BAC in which a stop codon had been inserted at codon 34 (STOP33), nor was infectious virus recovered from a recombinant BAC in which amino acids 45 to 59 were deleted (pp28Δac) (Fig. 7). Furthermore, we also failed to recover infectious virus from a recombinant BAC in which sequences encoding aa 45 to 59 were deleted and then transplanted to the C terminus of pp28 (pp28Δac-CtTR) (Fig. 7). Revertants of some of the BACs that failed to produce infectious virus were generated, and infectious virus could be recovered from these mutant HCMV BACs (Fig. 7). In the case of the pp28STOP33 mutant, the revertant was generated by linear recombination using a single defined oligonucleotide (Rev33) to repair the stop codon at position 33. Thus, the loss of infectivity in the pp28STOP33 mutant was secondary to the engineered mutation and not to additional mutations introduced by the mutagenesis procedure. In the case of the acidic cluster deletion mutant in pp28 (pp28Δac), small plaques were observed shortly after electroporation of HF cells with the BAC DNA, but they failed to increase in size, and infectious virus was not recovered from the supernatant of these cultures (data not shown). To confirm expression of the truncated pp28 proteins from the replication-competent recombinant viruses, we performed Western blot analysis of recombinant virus-infected HF cells at 5 days postinfection (Fig. 8). As expected, truncated pp28 was expressed from each recombinant virus. Because we repaired mutations in the coding sequence of pp28 by linear recombination and electroporation of a mixture of both repaired and mutant BAC DNA, revertants, including Rev33, generated by recombination with a repair oligonucleotide expressed both wild-type pp28 and truncated pp28. This result suggested that at least in the case of pp28STOP mutants, a replication-defective genome was maintained in these stocks of replicating virus that had been passaged through at least three serial passages.
FIG. 8.
Expression of truncated pp28 in HF cells infected with UL99 recombinant virus. HF cells were infected with recombinant virus at an MOI of 0.1. The cells were harvested at day 5 postinfection, lysed, and detected with anti-pp28 MAb by Western blot analysis as described in Materials and Methods. Arrows indicate the mass predicted (kilodaltons) from the amino acid sequence of truncated pp28. An asterisk (*) indicates the migration of wild-type pp28.
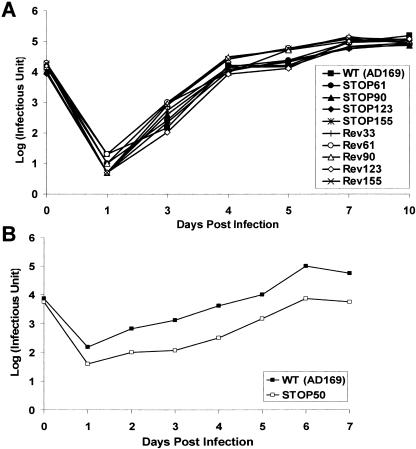
The phenotypes of the recombinant viruses generated from this panel of mutant HCMV BACs were characterized. As an example, image analysis showed that intracellular localizations of mutant pp28 from the recombinant virus containing the pp28Mut50 mutant and other viral proteins including gB, gM/gN, and pp150 in infected cells were similar to that of the wild-type parental virus (data not shown). This result indicated that a pp28 deletion mutant virus expressing only the first 50 aa of pp28 contained sufficient targeting information for authentic intracellular localizations of this mutant tegument protein with other viral proteins required for virus replication. To determine the kinetics of replication for viruses recovered from the mutant BACs, the replication kinetics of each mutant virus and revertants of mutants that failed to support virus replication were compared to those of wild-type HCMV in single-step growth assays. The growth curves for all recombinant viruses, with the exception of pp28STOP50, were similar to that of the wild-type parental virus (Fig. 9). The replication kinetics of the pp28STOP50 virus were delayed in comparison to wild-type virus and the pp28STOP61 virus, and the virus yield after 7 days in culture was approximately 10-fold less than that of the wild type (Fig. 9). The decreased yield was even more obvious at lower MOIs (data not shown). These results indicated that the first 50 aa were sufficient for the production of infectious virus and for normal trafficking of pp28 in the infected cell but that wild-type virus replication kinetics required the first 61 aa.
FIG. 9.
Replication kinetics of wild-type and recombinant viruses. Virus titer was quantified by a fluorescence-based virus infectivity assay on HF cells infected with WT or recombinant viruses at the indicated times after an initial infection using an MOI of 0.1. Results are expressed as log10 copies/milliliter (A) wild-type (▪) versus STOP61 (•), STOP90 (▴), STOP123 (⧫), STOP155 (*), Rev33 (+), Rev61 (○), Rev90 (▵), Rev123 (⋄), or Rev155 (×) virus. (B) Wild-type (▪) versus STOP50 (□) virus.
DISCUSSION
Although there is a consensus that HCMV virion assembly, more specifically, virion envelopment, takes place in the cytoplasm of infected cells, the trafficking of protein constituents of the virion tegument and envelope to cytoplasmic sites of assembly is not well understood. Because localization of tegumented capsids, tegument proteins, and envelopment proteins to the site of virion envelopment is a prerequisite for the assembly of the infectious particle, characterizations of intracellular trafficking of essential virion structural proteins should offer insights into the assembly of this large virus. In this study, we investigated the sequence requirements for localization of the essential tegument protein pp28 to a cytoplasmic site of virus assembly in HF cells, the AC. Our findings indicated that the first 50 amino acids of pp28 were required for localization to the AC and that, more importantly, a mutant virus encoding only the first 50 aa was replication competent. This finding was similar to the results of previous studies by Jones and Lee that described a mutant virus expressing only the first 57 aa of pp28 that was replication competent (17). Carboxyl-terminal deletions of the pp28 sequence at aa 44 resulted in a null phenotype of a recombinant virus, leading those investigators to postulate that acidic amino acids between positions 44 and 57 were responsible for the localization of pp28 to sites of assembly, perhaps through interactions with cellular adaptor proteins (17). We also found that deletion of the acidic cluster (aa 44 to 59) in pp28 resulted in a pp28 mutant virus that was defective in the production of infectious virus; however, our mutant virus was constructed differently than the C-terminal deletion mutants (aa 57 and aa 43) described in the previous study mentioned above in that this stretch of amino acids was deleted by the excision of this sequence from the full-length protein. Truncation mutants of pp28 that retain only the first 30 or 40 aa also exhibited a phenotype similar to that of the pp28 mutant containing the internal deletion of aa 44 to 59 in that they failed to localize to the AC and support virus replication. Thus, our studies utilizing both image and cell fractionation assays indicated that localization of pp28 to the AC late in infection was required for the assembly of infectious virus and that the minimal sequence requirements for localization of pp28 to the AC were contained within the first 50 aa of the molecule. It is unclear whether domains or sequences within these 50 aa are also responsible for other as-yet-undefined functions of pp28 in virion assembly after its localization to the AC.
Previously, we have shown that pp28 localizes in the ERGIC in the absence of other viral functions. This initial observation was confirmed by the finding that pp28 transiently expressed in virus-infected cells failed to localize in the assembly compartment in transfected/infected cells treated with ganciclovir, an inhibitor of late gene expression. Transiently expressed pp28 could localize in the assembly compartment in cells treated with BDCRB, a drug which has been shown to inhibit DNA packaging but not late protein synthesis. Thus, intracellular trafficking of pp28 differs from that of the alphaherpesvirus homologue, UL11, a virion tegument protein that has been reported to traffic to the Golgi compartment and TGN when expressed in the absence of virus infection (22). We also determined that localization of pp28 to the ERGIC required the expression of the first 30 aa based on findings that truncated forms of pp28 containing less than the first 30 aa colocalized with ER-resident proteins such as calreticulin. In contrast, deletion mutants expressing only the first 35 aa of pp28 trafficked similarly to wild-type pp28 and colocalized with ERGIC-localized proteins, suggesting that sequences within the first 35 aa were required for ERGIC localization. It is unclear if specific sequences within this stretch of amino acids target pp28 to the ERGIC. Although amino acid sequences that target specific proteins to the ERGIC have not been well defined, both a carboxyl-terminal dilysine ER retrieval signal and a phenylalanine ER export signal have been associated with localization to this compartment, presumably through the export from and retrieval to the ER (14, 21, 29, 30). Because this compartment is dynamic, localization of proteins within this compartment likely requires interactions with host cell proteins that partition within this compartment, such as ERGIC53 (3). Several well-studied ERGIC-localized proteins are either integral membrane proteins or membrane-associated proteins secondary to myristoylation and/or palmitoylation modifications. Interestingly, without the myristoylation modification, pp28 traffics throughout the cytoplasm and even enters the nucleus, suggesting that the membrane association of pp28 is necessary but not sufficient for its localization in the ERGIC (34). Whether other sequences within the first 35 aa of pp28 contribute to membrane association is not known, although preliminary studies have suggested that the protein is palmitoylated, and a consensus sequence for this posttranslational modification can be identified in the NH2 terminus of pp28 (data not shown). Even though our results are consistent with the interpretation that specific amino acid residues within the amino-terminal 35 aa of pp28 are required for ERGIC localization, other explanations include a loss of protein structure following deletions of these amino acids that results in the disruption of interactions with host proteins necessary for ERGIC localization.
The mechanism(s) that leads to pp28 trafficking from the ERGIC to the AC in virus-infected cells is a second and perhaps more interesting question. At least two possibilities can be proposed based on our findings. The first is that HCMV infection and late gene expression remodel the cellular secretory compartments, resulting in the approximation of the proximal compartments, such as the ERGIC, with more distal compartments, including the TGN. Thus, viral protein trafficking through the secretory pathway could be less compartmentalized and could occur following the mixing of closely approximated viral protein-containing vesicles. Alternatively, the dependence of pp28 localization to the AC on the expression of late gene products raises the possibility that interactions between pp28 and other viral proteins result in the redistribution of pp28 from the ERIGIC to more distal sites in the secretory pathway. This mechanism has been observed in assembly pathways of other viruses, including vaccinia virus and Mason-Pfizer monkey virus (32, 35). Preliminary findings have suggested that pp28 localizes to the distal compartments of the secretory pathway when transiently expressed with HCMV virion glycoproteins (data not shown), suggesting that pp28 could traffic with viral glycoproteins in virus-infected cells. This protein interaction could provide coordinated transport of this essential tegument protein with virion glycoproteins to the AC.
The defect in AC localization by the pp28 deletion mutants pp28Mut35 and pp28Mut40 and the internal deletion mutant pp28Δac (deletion of aa 44 to 59) was readily observed in transfected/infected cells on day 7 postinfection. The distribution of these pp28 mutants in the infected cell was clearly different than that of wild-type pp28 or mutant pp28Mut50 or pp28Mut61 in that deletion mutants at amino acid positions 35, 40, or 44 to 59 remained localized outside of the AC in a distribution that was consistent with proteins of the Golgi compartment or ERGIC. Imaging studies of cells coexpressing Myc-tagged pp28Mut35 and EGFP-ManII suggested that this mutant form of pp28 colocalizes with this Golgi protein late in infection, suggesting that this mutant form of pp28 entered the secretory pathway but was not transported to the AC (data not shown). The correlation between pp28 localization to the AC and the assembly of infectious virions strongly argued that virus replication and the production of infectious virus were dependent on pp28 localization within the AC. The finding that only the first 50 aa of this 191-aa protein were essential for the localization of pp28 to the AC and for the replication of infectious virus prompted questions about the function(s) of the remaining C-terminal 141 aa of pp28 that were nonessential for the replication of this virus in HF cells. Several possibilities could be considered, such as protein interactions with virion proteins that are not essential for infectivity in HF cells or, alternatively, interactions between these sequences and host cell proteins that are destined for incorporation into the virion. It is also of interest that pp28STOP50, which expressed only the first 50 aa of pp28, could be localized to the AC in infected cells and was incorporated into the virion even though the pp28STOP50 recombinant virus replicated less efficiently than the wild-type virus. This finding suggested that the replication defect in pp28STOP50 was unrelated to the authentic intracellular trafficking of pp28 but perhaps secondary to a loss or decrease in another function of pp28 such as interactions with virus-encoded or host cell proteins that contribute to the efficiency of virus assembly. Alternatively, the impairment in replication of pp28STOP50 could be secondary to a defect in the kinetics of localization of the mutant pp28 protein to the AC such that late in infection, a smaller fraction of pp28Mut50 localized to the AC and was available for the assembly of infectious virions.
The role of the cluster of acidic aa 44 to 59 in pp28 in the assembly of infectious virus has not been defined, but it has been suggested to be critical for localization to the AC through its interactions with cellular adaptor proteins such as PACS-1 (13, 17, 28). This potential function of the NH2-terminal acid cluster of pp28 has not been formally demonstrated and is based on the extrapolation of findings of previous studies of the alphaherpesvirus homolog UL11 (22). As noted previously, the UL11 protein of herpes simplex virus (HSV) is localized to the Golgi compartment and TGN in virus-infected cells and in cells transfected with an expression plasmid encoding UL11 (22). The deletion of the acidic cluster in the amino terminus of UL11 results in a protein that is expressed on the plasma membrane of transfected cells and also in the Golgi compartment and TGN (22). This finding together with the observation that the function of the acidic cluster of amino acids in the trafficking of UL11 in transient expression assays could be replaced with the PACS-1 binding consensus amino acid sequence from furin suggested a potential role of PACS-1 in the intracellular trafficking of UL11 (22). Those authors also noted that the targeting of HSV UL11 to the Golgi compartment appeared independent of the acidic cluster but required membrane association through fatty acid modifications in the extreme amino terminus of the molecule (22). Thus, the role of the cluster of acidic amino acids in the amino terminus of UL11 in the trafficking of this molecule remains unclear.
Although HSV UL11 and HCMV pp28 are presumed to be functional homologues and share some structural similarities, including a positionally conserved acid cluster of amino acids, several characteristics of the intracellular trafficking of HCMV pp28 appear to differ fundamentally from those of UL11. The first difference is that HCMV pp28 localizes in the ERGIC in the absence of other viral proteins and not in the Golgi compartment or TGN as has been previously reported for UL11 (34). In addition, unlike UL11, we have not detected the expression of pp28 or pp28 mutants on the plasma membrane in transfected cells. It should be noted, however, that we have detected expression on the plasma membrane in an occasional cell in transfection/infection assays following overexpression of pp28 very early during infection (data not shown), a finding that was reported previously by Jones and Lee using ectopic expression of pp28 from a strong, constitutive promoter (17). Furthermore, pp28 deletion mutants that fail to localize in the AC are not expressed on the plasma membrane in transfected/infected cells (data not shown). Thus, our findings suggest that the cluster of acidic amino acids in pp28 does not play an obvious role in retrieval from the plasma membrane as was observed in experiments with HSV UL11 (22). Secondly, it has been reported that the inhibition of PACS-1 expression with small interfering RNA resulted in only a two- to threefold decrease in virus production, whereas mutations in the UL99 ORF that alter intracellular trafficking of this viral protein result in the loss of infectious virus production (12). Thus, it could be argued that if pp28 interacts with PACS-1, then this interaction likely has only a limited role in the localization of pp28 to the AC. Alternatively, the acidic amino acid cluster between aa 44 and 59 could interact with cellular adaptor proteins other than PACS-1; however, this motif lacks other requisite signals such as adjacent dileucines that have been shown to function as signals recognized by cellular adaptor proteins responsible for trafficking between compartments of the secretory pathway (5, 31). Lastly, it was of interest that transplantation of the NH2-terminal acid cluster of pp28 (aa 44 to 59) to the carboxyl terminus of the molecule failed to direct the trafficking of this molecule to the AC, and when this UL99 mutant was incorporated into the viral genome, this recombinant molecule also failed to support virus replication. This result indicated that the role of the acid cluster in pp28 (aa 44 to 59) in localization to the AC and virus assembly was context dependent, a finding that is in contrast to the context independence of PACS-1 binding sequences and other trafficking motifs used to generate chimeric molecules for studies of intracellular trafficking. Thus, even though two independent studies have shown that the NH2-terminal acid cluster of pp28 plays a critical role in the localization of pp28 to the AC late in infection, the mechanism through which this sequence of amino acids functions to direct this molecule to the AC remains to be determined.
In a previous report, we noted that the AC was localized in a cellular compartment that failed to colocalize with markers of the Golgi compartment or the TGN and exhibited only minimal overlap with lysosomal markers such as LAMP-1 (33). In the current study, we further characterized the AC in transfection/infection assays in which EGFP-tagged markers of the Golgi compartment and trans-Golgi network were used to mark these secretory compartments. These experiments were carried out to further define the trafficking of pp28 and pp28 mutants to the AC, but unexpectedly, we found that the secretory pathway was morphologically remodeled during virus infection, resulting in the loss of ribbon-like stacks of the Golgi compartment and the TGN and the formation of a juxtanuclear, more spherically shaped organelle. The remodeling of the secretory pathway began within 72 h postinfection, and host cell markers such as mannosidase II and TGN38 (Gal T) colocalized with cytoplasmic structural virion proteins, including gB and gM/gN, as well as pp28 that was expressed transiently from an HCMV immediate-early promoter in cells transfected with this plasmid and infected with wild-type virus. However, at this relatively early time during infection, the characteristic compact AC containing virion structural proteins that has been seen late in infection was not present. The compact structure containing tegument proteins pp28 and pp150 was localized to the center of the AC only late in infection (>120 h in these experiments). These results suggested that the formation of the AC was associated the with remodeling of the secretory compartment with the eventual displacement of host cell proteins of the secretory compartment to the periphery of this structure, a finding consistent with previous studies (33). Furthermore, the dynamics of the changes in the morphology of the secretory compartment provided an explanation for results from experiments using transiently expressed pp28 mutants that demonstrated that pp28 mutants of more than 35 aa of the amino terminus of pp28 could localize in the ERGIC in transfected cells and also traffic to a perinuclear compartment early in infection in transfected/infected cells. Although this finding was initially interpreted as evidence that even the shortest pp28 truncation mutant, including several mutants that failed to support virus replication when recombined into the viral genome, could localize to a juxtanuclear compartment, we subsequently demonstrated that only those pp28 mutants that localized to the compact AC that formed late in infection supported virus replication. These observations were consistent with a morphological maturation of the AC during infection that can be described as a compaction of this virion protein-containing structure during maximal virus production in infected HF cells. Thus, it is possible that the characteristic compact appearance of the AC represents the concentration of virion structural proteins within this membranous compartment during the assembly of infectious particles and that the rate of production of infectious virus is governed by the concentration of structural proteins in this compartment. This pathway could in turn be dependent on viral protein trafficking to this compartment. Previous studies have demonstrated that virions lacking pp28 fail to become enveloped and can be found as individual nonenveloped particles in the cytoplasm of infected cells (36). Together with findings presented in the current study, those results suggested that the failure of pp28 mutants to localize in the AC at late times in infection could be expected to result in a loss of infectious virus production. An extension of this proposed function of pp28 during virus assembly would be that once pp28 is localized to the AC by targeting signals located between aa 44 and 50, other postlocalization functions such as interactions with other viral proteins and/or host cell proteins are required for virion envelopment and virus assembly.
Acknowledgments
This work was supported by a grant from the HHS, NIH, NIAID, to W.J.B. (R01 AI35602).
REFERENCES
- 1.Abate, D. A., S. Watanabe, and E. S. Mocarski. 2005. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 78:10995-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreoni, M., M. Faircloth, L. Vugler, and W. J. Britt. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J. Virol. Methods 23:157-167. [DOI] [PubMed] [Google Scholar]
- 3.Appenzeller, C., H. Andersson, F. Kappeler, and H. P. Hauri. 1999. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat. Cell Biol. 1:330-334. [DOI] [PubMed] [Google Scholar]
- 4.Billstrom, M. A., and W. J. Britt. 1995. Postoligomerization folding of human cytomegalovirus glycoprotein B: identification of folding intermediates and importance of disulfide bonding. J. Virol. 69:7015-7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonifacino, J. S., and L. M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395-447. [DOI] [PubMed] [Google Scholar]
- 6.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britt, W. J., M. Jarvis, J.-Y. Seo, D. Drummond, and J. Nelson. 2004. Rapid genetic engineering of human cytomegalovirus using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 78:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britt, W. J., M. A. Jarvis, D. D. Drummond, and M. Mach. 2005. Antigenic domain 1 is required for oligomerization of human cytomegalovirus glycoprotein B. J. Virol. 79:4066-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britt, W. J., and L. G. Vugler. 1992. Oligomerization of the human cytomegalovirus major envelope glycoprotein complex gB (gp55-116). J. Virol. 66:6747-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browne, E. P., and T. Shenk. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc. Natl. Acad. Sci. USA 100:11439-11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantrell, S. R., and W. A. Bresnahan. 2005. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J. Virol. 79:7792-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crump, C. M., C. H. Hung, L. Thomas, L. Wan, and G. Thomas. 2003. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J. Virol. 77:11105-11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crump, C. M., Y. Xiang, L. Thomas, F. Gu, C. Austin, S. A. Tooze, and G. Thomas. 2001. PACS-1 binding to adaptors is required for acidic cluster motif-mediated protein traffic. EMBO J. 20:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dogic, D., A. Dubois, B. de Chassey, Y. Lefkir, and F. Letourneur. 2001. ERGIC-53 KKAA signal mediates endoplasmic reticulum retrieval in yeast. Eur. J. Cell Biol. 80:151-155. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. USA 97:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, T. R., and S. W. Lee. 2004. An acidic cluster of human cytomegalovirus UL99 tegument protein is required for trafficking and function. J. Virol. 78:1488-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalejta, R. F., and T. Shenk. 2003. The human cytomegalovirus UL82 gene product (pp71) accelerates progression through the G1 phase of the cell cycle. J. Virology 77:3451-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerry, J. A., M. A. Priddy, C. P. Kohler, T. L. Staley, D. Weber, T. R. Jones, and R. M. Stenberg. 1997. Translational regulation of the human cytomegalovirus pp28 (UL99) late gene. J. Virol. 71:981-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 21.Lontok, E., E. Corse, and C. E. Machamer. 2004. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 78:5913-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, M., and T. Shenk. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mach, M., B. Kropff, M. Kryzaniak, and W. Britt. 2005. Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J. Virol. 79:2160-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer, H., A. Bankier, M. P. Landini, and R. Ruger. 1988. Identification and procaryotic expression of the gene coding for the highly immunogenic 28-kilodalton structural phosphoprotein (pp28) of human cytomegalovirus. J. Virol. 62:2243-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 28.Molloy, S. S., E. D. Anderson, F. Jean, and G. Thomas. 1999. Bi-cycling the furin pathway: from TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 9:28-35. [DOI] [PubMed] [Google Scholar]
- 29.Nufer, O., S. Guldbrandsen, M. Degen, F. Kappeler, J. P. Paccaud, K. Tani, and H. P. Hauri. 2002. Role of cytoplasmic C-terminal amino acids of membrane proteins in ER export. J. Cell Sci. 115:619-628. [DOI] [PubMed] [Google Scholar]
- 30.Nufer, O., F. Kappeler, S. Guldbrandsen, and H. P. Hauri. 2003. ER export of ERGIC-53 is controlled by cooperation of targeting determinants in all three of its domains. J. Cell Sci. 116:4429-4440. [DOI] [PubMed] [Google Scholar]
- 31.Ohno, H., R. C. Aguilar, D. Yeh, D. Taura, T. Saito, and J. S. Bonifacino. 1998. The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J. Biol. Chem. 273:25915-25921. [DOI] [PubMed] [Google Scholar]
- 32.Roper, R. L., and B. Moss. 1999. Envelope formation is blocked by mutation of a sequence related to the HKD phospholipid metabolism motif in the vaccinia virus F13L protein. J. Virol. 73:1108-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sfakianos, J. N., and E. Hunter. 2003. M-PMV capsid transport is mediated by Env/Gag interactions at the pericentriolar recycling endosome. Traffic 4:671-680. [DOI] [PubMed] [Google Scholar]
- 36.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sims, A. C., J. Ostermann, and M. R. Denison. 2000. Mouse hepatitis virus replicase proteins associate with two distinct populations of intracellular membranes. J. Virol. 74:5647-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stockli, J., and J. Rohrer. 2004. The palmitoyltransferase of the cation-dependent mannose 6-phosphate receptor cycles between the plasma membrane and endosomes. Mol. Biol. Cell 15:2617-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Storrie, B., J. White, S. Rottger, E. H. Stelzer, T. Suganuma, and T. Nilsson. 1998. Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 143:1505-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trus, B. L., W. Gibson, N. Cheng, and A. C. Steven. 1999. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites. J. Virol. 73:2181-2192. (Erratum, 73:4530, 1999.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Underwood, M. R., R. J. Harvey, S. C. Stanat, M. L. Hemphill, T. Miller, J. C. Drach, L. B. Townsend, and K. K. Biron. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wentz-Hunter, K., R. Kubota, X. Shen, and B. Y. Yue. 2004. Extracellular myocilin affects activity of human trabecular meshwork cells. J. Cell. Physiol. 200:45-52. [DOI] [PubMed] [Google Scholar]
- 44.Yu, X., S. Shah, I. Atanasov, P. Lo, F. Liu, W. J. Britt, and Z. H. Zhou. 2005. Three-dimensional localization of the smallest capsid protein in the human cytomegalovirus capsid. J. Virol. 79:1327-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 A. Science 288:877-880. [DOI] [PubMed] [Google Scholar]