Abstract
Recently, we demonstrated that plant DNA virus replication was inhibited in planta by using an artificial zinc finger protein (AZP) and created AZP-based transgenic plants resistant to DNA virus infection. Here we apply the AZP technology to the inhibition of replication of a mammalian DNA virus, human papillomavirus type 18 (HPV-18). Two AZPs, designated AZPHPV-1 and AZPHPV-2, were designed by using our nondegenerate recognition code table and were constructed to block binding of the HPV-18 E2 replication protein to the replication origin. Both of the newly designed AZPs had much higher affinities towards the replication origin than did the E2 protein, and they efficiently blocked E2 binding in vitro. In transient replication assays, both AZPs inhibited viral DNA replication, especially AZPHPV-2, which reduced the replication level to approximately 10%. We also demonstrated in transient replication assays, using plasmids with mutant replication origins, that AZPHPV-2 could precisely recognize the replication origin in mammalian cells. Thus, it was demonstrated that the AZP technology could be applied not only to plant DNA viruses but also to mammalian DNA viruses.
The papillomaviruses are double-stranded DNA viruses that induce benign proliferative squamous epithelial and fibroepithelial lesions (warts and papillomas) in their natural hosts. They have been isolated from a variety of animal species, and >100 human papillomavirus (HPV) types have been identified and fully sequenced so far (reviewed in reference 13). A subgroup of HPVs classified as “high-risk” viruses, including HPV types 16, 18, 31, 35, 39, 45, 51, 52, 58, and 59, has been found to be associated with the development of cervical cancer (1, 28). Each year, about 500,000 such infections at the uterine cervix undergo malignant conversion, making cervical cancer the second most common malignancy in women worldwide (17). About 90% of such tumors contain high-risk HPVs, with HPV-16 and -18 being the most prevalent. The incidence shows no evidence of declining, and current treatment options are limited(http://www.boehringer-ingelheim.ca/research/res_area_humpap.asp).Therefore, effective antiviral therapies/treatments for this widespread and troublesome disease are clearly needed.
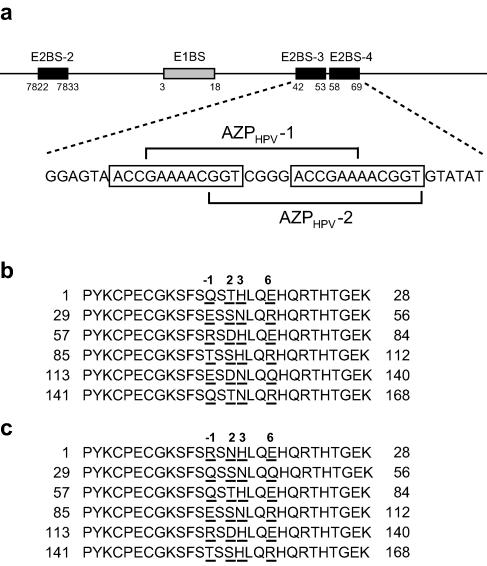
The papillomavirus proteins required for viral DNA replication are the viral E1 and E2 proteins (reviewed in reference 9). The E1 protein is a 70- to 80-kDa nuclear phosphoprotein possessing DNA helicase activity (reviewed in reference 29). Sequence-specific binding of E1 to the viral origin of replication is most likely mediated by the papillomavirus E2 protein (2, 6, 26, 30, 31); E2 bound to the origin recruits E1 to the origin, which results in initiation of the replication process. The 43- to 50-kDa E2 protein comprises two well-conserved functional domains linked by a hinge domain (reviewed in reference 14). The amino-terminal domain of E2 is necessary for direct association with the E1 protein. The carboxyl-terminal portion of E2 binds a 12-base-pair palindromic DNA sequence, 5′-ACCGNNNNCGGT-3′. This sequence is repeated four times near the viral replication origin. Systematic mutational analysis around the replication origin revealed that two E2-binding sites, designated E2BS-3 and E2BS-4 (see Fig. 1a), seem to be most important for replication (12, 20). Therefore, it is highly likely that one strategy for efficient inhibition of HPV replication is to block E2 binding to E2BS-3 and E2BS-4.
FIG. 1.
Organization of the HPV-18 replication origin and amino acid sequences of AZPs for inhibition of DNA replication. (a) DNA targets of AZPHPV-1 and AZPHPV-2 in the replication origin. The gray and black boxes indicate an E1-binding site (E1BS) and an E2-binding site (E2BS), respectively. The two open rectangles indicate the 12-bp DNA sequence recognized by the E2 protein. The numbers below the boxes indicate their locations (in nt) in the HPV-18 genome. The two 19-bp DNA targets were chosen for AZPHPV-1 and AZPHPV-2 to block E2 binding to the region containing E2BS-3 and E2BS-4, which is the most important cis element for DNA replication (12, 20). (b) Amino acid sequence of AZPHPV-1. AZPHPV-1 binding to the 19-bp DNA occurs through six zinc finger domains. The underlined amino acids in each finger domain show recognition amino acids at positions −1, 2, 3, and 6 in the α-helix of the finger domain. These amino acids were chosen from our recognition code table (23). (c) Amino acid sequence of AZPHPV-2.
Recently, we demonstrated that DNA replication of a plant geminivirus, beet severe curly top virus (BSCTV), was inhibited by an artificial zinc finger protein (AZP) that was designed to block binding of the replication protein (Rep) to its replication origin, and transgenic Arabidopsis thaliana plants expressing the AZP showed complete resistance to BSCTV infection (22). The six-finger AZP, which binds to the 19-bp DNA containing the entire Rep-binding site, was designed using our nondegenerate recognition code table (23).
Since we reported the characterization of the DNA-binding properties of AZPs designed for the inhibition of replication of plant DNA viruses (23), three other groups reported applications of zinc finger proteins to human viruses, such as human immunodeficiency virus type 1 and herpes simplex virus type 1 (10, 15, 19, 21). In these studies, inhibition of virus replication was attempted through repression of viral transcription. For this purpose, zinc finger proteins were fused with an effector domain, such as the Krüppel-associated box repressor domain. Zinc finger proteins alone were used as the controls, and fusion with an effector domain was required for efficient inhibition of virus replication.
For this study, we applied the AZP technology to HPV type 18 (HPV-18), one of the high-risk HPVs. In order to block E2 binding to E2BS-3 and E2BS-4, two AZPs were designed and constructed. We examined whether the AZPs are able to inhibit HPV-18 DNA replication in transient replication assays.
MATERIALS AND METHODS
Plasmid construction.
Two AZPs for the inhibition of E2 binding, designated AZPHPV-1 and AZPHPV-2, were designed using our nondegenerate recognition code table, and each of the Escherichia coli expression plasmids was constructed with pET-21a (Novagen), as described previously (23). The control AZP, designated AZPAla, was produced by mutation of all recognition amino acids to alanine. An E. coli expression plasmid encoding HPV-18 E2 was prepared by cloning the E2 open reading frame (ORF) (HPV-18 nucleotides [nt] 2817 to 3914) from pHPV-18 (American Type Culture Collection) into the EcoRI/HindIII sites of pET-21a. Three plasmids used for transient replication assays (described below), named pRL-E1, pRL-E2, and pUC-Ori177, were prepared. pRL-E1 and pRL-E2 were constructed by cloning the E1 and E2 ORFs (HPV-18 nt 904 to 2887 and 2817 to 3914, respectively), respectively, into the NheI/XbaI sites of a pRL-SV40 mammalian expression plasmid (Promega), and pUC-Ori177 was constructed by cloning the 177-bp AluI/BamHI fragment (HPV-18 nt 7800 to 7857 and 1 to 119), designated Ori177 and including the HPV-18 replication origin, into the HincII/BamHI sites of pUC-19. The E1 and E2 ORFs and Ori177 were amplified from pHPV-18 by PCR. Mammalian expression plasmids for AZP derivatives, designated pCMV-AZPHPV-1, pCMV-AZPHPV-2, and pCMV-AZPAla, were prepared by cloning each AZP ORF into modified pcDNA3.1 (Invitrogen). The modified plasmid contains an N-terminal T7 tag, a nuclear localization signal from the simian virus 40 large T antigen, and a multicloning site for AZPs.
Protein overexpression and purification of AZPs and E2.
Three AZPs, AZPHPV-1, AZPHPV-2, and AZPAla, were overexpressed in E. coli and purified as previously described (23). These AZPs were >95% homogeneous, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Each protein concentration was determined using an ESL protein assay kit (Roche Molecular Biochemicals). The HPV-18 E2 protein was also overexpressed in E. coli and purified essentially as previously described (23), except that the protein was eluted with 500 mM NaCl buffer. In our procedure, the expression level of E2 was very low and was monitored only by Western blot analysis. Therefore, the concentration of purified E2 protein was determined in Western blot analysis by comparison with AZPAla, whose concentration was determined using an ESL protein assay kit.
DNA-binding assays.
A 56-bp synthetic DNA duplex consisting of the sequence 5′-(TA)4GGAGTAACCGAAAACGGTCGGGACCGAAAACGGTGTATAT(TA)4-3′, including two E2-binding sites (underlined) (see Fig. 1a), was labeled by a Klenow fill-in reaction with [α-32P]dATP and [α-32P]dTTP. Purified AZP was incubated on ice for 1 h in 10 mM Tris-HCl (pH 7.5)-100 mM NaCl-5 mM MgCl2-0.1 mM ZnCl2-0.05% bovine serum albumin-10% glycerol containing the labeled probe (0.03 fmol per 10 μl of buffer) and 1 μg of poly(dA-dT)2. A shorter 32P-labeled (40-bp) probe with the sequence 5′-(TA)4GGAGTA ACCGAAAACGGTCGGGAC(TA)4-3′, containing a single E2-binding site (underlined), was used only for E2-binding experiments. In competition binding experiments, a mixture of an AZP (i.e., AZPHPV-1 or AZPHPV-2) and E2 protein was added to the above binding buffer containing the 32P-labeled 56-bp probe. The concentrations of AZPs and E2 protein used are described in detail in the figure legends. The probe-protein complex and the free probe were resolved in 6% nondenaturing polyacrylamide gels in 45 mM Tris-borate buffer by electrophoresis at 140 V for 2 h at 4°C. The radioactive signals were recorded on X-ray films.
Transient replication assays.
A total of 8 × 105 cells of the human cell line 293H (Invitrogen) were plated onto a BioCoat poly-d-lysine 12-well plate (Becton Dickinson) and maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 0.1 mM nonessential amino acids and 10% fetal bovine serum (Invitrogen). Three plasmids necessary for transient replication, i.e., pRL-E1 (1.5 μg), pRL-E2 (0.17 μg), and pUC-Ori177 (0.17 μg), were cotransfected with pCMV-AZPHPV-1, pCMV-AZPHPV-2, pCMV-AZPAla, or pcDNA3.1 (0.17 μg) by using Lipofectamine 2000 (Invitrogen) according to the protocol accompanying the reagent. Three days after transfection, low-molecular-weight DNA was isolated by Hirt extraction (7). The samples were first treated with HindIII to linearize them. To distinguish between replicated and unreplicated DNAs, one-half of each sample was then treated with an excess of DpnI to remove the unreplicated, methylated input DNA (16). DpnI resistance has been used to demonstrate DNA replication in studies with mammalian cells, including studies with HPVs (3, 4). One percent of the remaining half of each linearized sample was used to confirm that equal amounts of the plasmids used for each transient replication assay were introduced into 293H cells. The DNA samples were separated by electrophoresis (in 0.8% agarose gels with 0.5× Tris-borate-EDTA buffer), followed by Southern blot hybridization.
A 200-bp digoxigenin (DIG)-labeled probe specific to an ampicillin resistance gene was prepared from pUC-19 by PCR using DIG-11-dUTP (Roche Molecular Biology) and the primer set 5′-CGGCATCAGAGCAGATTGTACTGAGAGTGC-3′and 5′-TACCCAACTTAATCGCCTTGCAGCACATCC-3′. Because pRL-E1, pRL-E2, pCMV-AZP, and pUC-Ori177 contain the ampicillin resistance gene as a selection marker, all of these plasmids could be detected by using the DIG-labeled probe. DNAs were resolved in a 0.8% agarose gel and transferred onto a Nytran SuPerCharge membrane by use of a TurboBlotter (Schleicher & Schuell). After hybridizing with the DIG-labeled probe, DNA bands corresponding to the plasmids used for transient replication assays were recorded on X-ray films, using anti-DIG-AP and CDP-Star according to the accompanying protocols (Roche Molecular Biology). DNA band intensities on X-ray films were digitized and quantitated by UN-SCAN-IT (Silk Scientific, Inc.). Average DNA band intensities of replicated pUC-Ori derivatives were calculated from four independent experiments and normalized with DNA band intensities of input pUC-Ori derivatives.
Immunoblotting analysis of AZPs.
Three days after transfection, carried out as described above, 293H cells were washed with phosphate-buffered saline once and lysed in 1× passive lysis buffer (Promega). The protein concentration of each lysis sample cleared by centrifugation was determined by using an ESL protein assay kit. Equal amounts (2.4 μg) of the protein samples were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, electroblotted onto a polyvinylidene difluoride membrane, and probed with a T7 tag antibody conjugated with horseradish peroxidase (Novagen) by standard methods. For chemiluminescence detection, ECL Plus (Amersham) was used. The resulting chemiluminescent signals were recorded on X-ray films.
RESULTS AND DISCUSSION
Design of AZPs to block E2 binding.
The replication origin of HPV-18 contains four E2-binding sites (E2BS). Systematic mutational analysis around the replication origin revealed that two E2-binding sites, designated E2BS-3 and E2BS-4 (Fig. 1a), seem to be most important for replication (12, 20). Therefore, a region containing E2BS-3 and E2BS-4 was chosen as an AZP target for the inhibition of virus replication. As described in our previous report (23), AZPs containing N (N ≥ 3) fingers recognize (3N + 1)-bp DNA sequences, and functional AZPs recognize DNA targets containing more than three guanines (at any position) per 9 bp. Accordingly, the following two 19-bp DNAs were chosen as six-finger AZP targets to block the binding of E2 to E2BS-3 and E2BS-4: 5′-GAAAACGGTCGGGACCGAA-3′ and 5′-GGTCGGGACCGAAAACGGT-3′ (Fig. 1a). The AZP designed to recognize the former 19-bp sequence was designated AZPHPV-1, and the AZP designed to recognize the latter 19-bp sequence was designated AZPHPV-2 (Fig. 1a). Each AZP gene was designed and constructed using our nondegenerate recognition code table as previously described (23). The amino acid sequences of AZPHPV-1 and AZPHPV-2 are listed in Fig. 1b and c, respectively. An additional AZP, designated AZPAla, in which all recognition amino acids at positions −1, 2, 3, and 6 are replaced with alanine, was also generated as the control protein.
In vitro binding properties of AZPs.
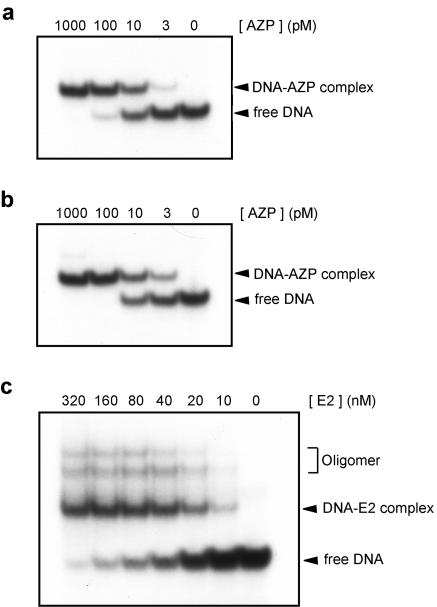
Each of the AZPHPV-1 and AZPHPV-2 ORFs was cloned into an E. coli expression plasmid under the control of a bacteriophage T7 promoter, and each AZP was overexpressed in E. coli expressing T7 RNA polymerase and purified to >95% homogeneity (data not shown). The purified AZPs were used to examine binding properties in vitro by gel shift assays. As shown in Fig. 2a and b, protein concentrations for half-maximal binding are about 10 pM for both AZPHPV-1 and AZPHPV-2. By comparison of band shifting at protein concentrations of 3 and 100 pM, AZPHPV-2 seemed to have a slightly higher affinity for the given DNA target than did AZPHPV-1. Under the same conditions [e.g., in the presence of an excess amount of cold poly(dA-dT)2], the apparent dissociation constant (Kd) of the E2 protein, where half-maximal binding was observed, was approximately 40 nM, as shown in Fig. 2c. Thus, both AZPHPV-1 and AZPHPV-2 have approximately 4,000-fold higher affinities for DNA than that of E2.
FIG. 2.
DNA-binding assays with AZPHPV-1 (a), AZPHPV-2 (b), and the E2 protein (c). A 32P-labeled 56-bp probe including E2BS-3 and E2BS-4 was used for AZPHPV-1 and AZPHPV-2. A 32P-labeled 40-bp probe containing a single E2BS was used for the E2 protein. The protein concentrations used in each assay are indicated above the lanes. As indicated in panel c (Oligomer), additional complexes migrating more slowly than the DNA-E2 complex were also observed. The same event, presumably generated by oligomerization of DNA-E2 complexes (8), has been reported for E2 proteins.
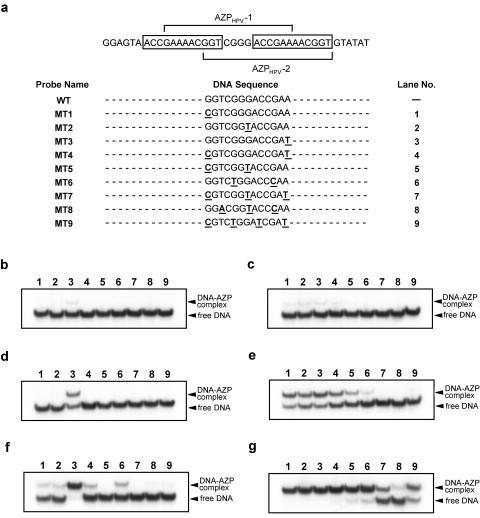
Next, the DNA-binding specificities of AZPHPV-1 and AZPHPV-2 were examined using various mutant probes. The mutant probes contained one to four mutations in a region overlapped by the DNA targets of AZPHPV-1 and AZPHPV-2 (Fig. 3a). In the presence of 10 pM AZP, where half-maximal binding to the wild-type probe was observed (Fig. 2a and b), very faint shifted bands were observed only with MT3 for AZPHPV-1 (Fig. 3b, lane 3) and with MT1 to MT4 for AZPHPV-2 (Fig. 3c, lanes 1 to 4), indicating that both AZPs recognize a 1-bp difference. AZPHPV-2 shifted approximately 50% or more with MT1 to MT4, but not with MT7 to MT9, which contain more than three mutations, at a protein concentration of 100 pM (Fig. 3e). In the presence of 1 nM AZPHPV-2, MT8 did not show significant band shifting (Fig. 3g). Furthermore, AZPHPV-1 showed greater specificity (Fig. 3d and f). Even at a protein concentration of 1 nM, AZPHPV-1 did not show a significant affinity for any mutant probe except MT3 (Fig. 3f).
FIG. 3.
DNA binding of AZPHPV-1 and AZPHPV-2 with mutant DNA probes. (a) DNA sequences of mutant probes. Mutations were introduced into a DNA region overlapped by the DNA targets of the AZPs. Underlining represents mutation. The two open rectangles indicate E2BS-3 and E2BS-4 (see Fig. 1a). The lane numbers correspond to those shown above panels a to g. (b) Gel shift with 10 pM AZPHPV-1. (c) Gel shift with 10 pM AZPHPV-2. (d) Gel shift with 100 pM AZPHPV-1. (e) Gel shift with 100 pM AZPHPV-2. (f) Gel shift with 1 nM AZPHPV-1. (g) Gel shift with 1 nM AZPHPV-2.
Finally, the control protein, AZPAla, was unable to shift the target DNA even at 1 μM in the presence of an excess amount of cold poly(dA-dT)2 competitor DNA, whereas AZPHPV-1 caused a single shifted species (data not shown), showing that the specific recognition amino acids in AZPHPV-1 and AZPHPV-2 are required to recognize the 19-bp DNA targets.
Inhibition of HPV-18 DNA replication by AZPs.
HPV DNA replication can be transiently reconstituted in mammalian cells by using mammalian expression plasmids for the viral E1 and E2 proteins and a plasmid containing an HPV replication origin (HPV ori plasmid), and the DNA replication mechanism has been extensively investigated by using transient replication assays (for example, see references 3, 4, and 18). These transient experiments revealed that the E2 protein plays a critical role in replication by recruiting the E1 helicase to the replication origin and that the ratio of the E1 expression plasmid to the E2 expression plasmid significantly affects the DNA replication efficiency. Previous reports also showed that the absence of an E2 expression plasmid resulted in no replication or a significant loss of replication but that a higher ratio of E2 expression plasmid reduced the replication efficiency, with ratios ranging from 3:1 to 60:1 showing optimal replication (12, 24, 25, 27). We first examined the replication efficiencies of ratios of E1 expression plasmid (i.e., pRL-E1) to E2 expression plasmid (i.e., pRL-E2) of 3:1, 7:1, 9:1, and 15:1, where pUC-Ori177, an HPV-18 ori plasmid, was used in an amount equal to that of pRL-E2. The plasmid pUC-Ori177 includes the 177-bp HPV-18 replication origin (nt 7800 to 7857 and 1 to 119), containing one E1BS and three E2BSs (Fig. 1a), and has been demonstrated to be sufficient for maximal DNA replication (18, 24, 25). From these experiments, we found that ratios higher than 9:1 did not improve the replication efficiency (data not shown). Thus, the ratio was fixed to 9:1 in all of the following transient replication assays.
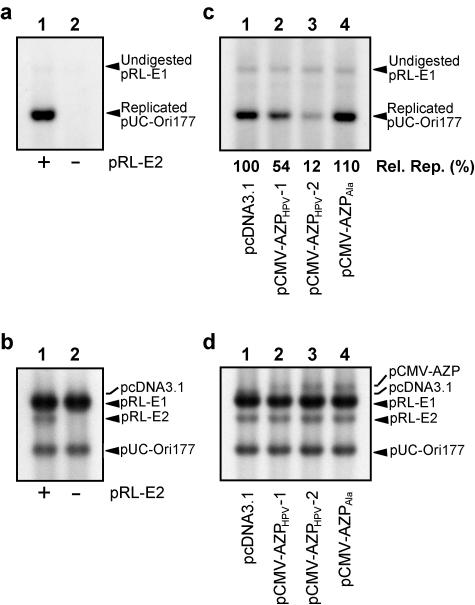
Under the above conditions, we next confirmed the significance of the E2 protein for HPV-18 DNA replication in transient replication assays. Specifically, we compared the replication efficiency in the presence of pRL-E2 with that in the absence of pRL-E2. As shown in Fig. 4a, when Hirt-extracted DNA samples were treated with DpnI to eliminate unreplicated input plasmids, no replicated pUC-Ori177 was observed in the absence of pRL-E2 (lane 2), corresponding to previous reports (3, 12, 24), although it has been reported that the E1 protein alone promoted a low level of origin-independent replication (11). In the transient replication assay without pRL-E2, the same amounts of plasmids as those used in that with pRL-E2 were introduced into 293H cells, as judged from Southern blot analysis with Hirt-extracted DNA samples untreated with DpnI (Fig. 4b). Therefore, the significance of the E2 protein for HPV-18 DNA replication was also confirmed in the transient replication assay.
FIG. 4.
Transient replication analysis of HPV-18 DNA with AZPs. (a) Importance of E2 for replication. A transient replication assay was performed with pRL-E1 for E1 expression and pUC-Ori177 as the HPV-18 ori plasmid in the presence of the E2 expression plasmid pRL-E2 (lane 1) or in the absence of pRL-E2 (lane 2). Hirt-extracted samples were digested with DpnI to eliminate unreplicated input plasmids (see Materials and Methods). DNA replication efficiencies were judged by comparison of relative amounts of pUC-Ori177. (b) Examination of amounts of plasmids introduced into 293H cells for panel a. This panel shows Southern blot hybridization with Hirt-extracted DNA samples which were not treated with DpnI, indicating relative amounts of plasmids introduced into 293H cells. (c) Inhibition of HPV-18 DNA replication by AZPs. Transient replication assays were performed with pRL-E1, pRL-E2, and pUC-Ori177 along with the AZP expression plasmid described below each lane. Hirt-extracted samples were digested with DpnI. Inhibition efficiencies of DNA replication by AZPs were judged by comparison of relative amounts of pUC-Ori177. The amount of replicated pUC-Ori177 in the presence of each AZP derivative was compared with that in the absence of AZP (100%). The average data [Rel. Rep. (%)] from four independent assays are indicated below each lane. Under our conditions, input pRL-E1 could not be eliminated completely by DpnI digestion. (d) Examination of amounts of plasmids introduced into 293H cells for panel c. This panel shows Southern blot hybridization with Hirt-extracted DNA samples which were not treated with DpnI, indicating relative amounts of plasmids introduced into 293H cells.
We then examined whether both designed AZPs, AZPHPV-1 and AZPHPV-2, can inhibit HPV-18 DNA replication. Each of the AZPHPV-1 and AZPHPV-2 ORFs was cloned into a mammalian expression plasmid under the control of a cytomegalovirus promoter. The resulting plasmids were designated pCMV-AZPHPV-1 and pCMV-AZPHPV-2, respectively. In transient replication assays, the ratio of pRL-E1, pRL-E2, pUC-Ori177, and the expression plasmid for each AZP derivative was 9:1:1:1. This experiment was repeated independently four times. The results are shown in Fig. 4c, where Hirt-extracted DNA samples were treated with DpnI to eliminate unreplicated input plasmids. Both AZPHPV-1 and AZPHPV-2 inhibited DNA replication. AZPHPV-2, especially, reduced the replication level to 12% ± 4%. In the transient replication assay with each pCMV-AZP derivative, the same amounts of plasmids as those used in that with pcDNA3.1 were introduced into 293H cells, as judged from Southern blot analysis with Hirt-extracted DNA samples untreated with DpnI (Fig. 4d).
Under the same conditions, the control protein, AZPAla, was unable to inhibit DNA replication (Fig. 4c, lane 4), showing that specific recognition amino acids in AZPHPV-1 and AZPHPV-2 are required for inhibition.
AZPHPV-2, with a greater ability to inhibit HPV-18 DNA replication, inhibits E2 binding more efficiently in vitro.
In the aforementioned transient replication assays, AZPHPV-2 inhibited DNA replication of pUC-Ori177 more efficiently than AZPHPV-1 did, although these AZPs had similar binding affinities towards the DNA probe containing E2BS-3 and E2BS-4 (Fig. 2a and b). One possible cause of this result is that higher expression of AZPHPV-2 may result in more efficient inhibition of DNA replication. To examine this possibility, the expression level of each AZP was investigated by Western blot analysis with an anti-T7-tag antibody because AZPs expressed from pCMV-AZP derivatives contain a T7 tag at the N terminus. After transfection of pRL-E1, pRL-E2, pUC-Ori177, and a pCMV-AZP derivative (i.e., pCMV-AZPHPV-1, pCMV-AZPHPV-2, pCMV-AZPAla, or pcDNA3.1 as the control), as performed in transient replication assays, the transfected 293H cells were lysed, and each lysis sample was cleared by centrifugation. Using the same amount (i.e., total protein content of 2.4 μg) of each lysis sample, the protein expression level of each AZP was then analyzed by immunoblotting with an anti-T7-tag antibody. As shown in Fig. 5a, the expression level of AZPHPV-2 was lower than that of AZPHPV-1. The control AZP, AZPAla, showing no inhibition of DNA replication, was expressed most abundantly.
FIG. 5.
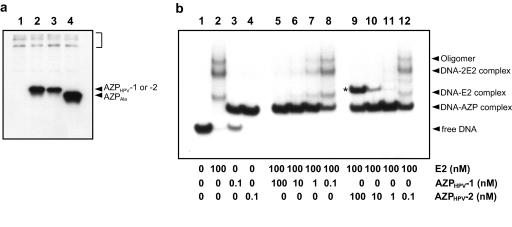
Identification of the cause of more efficient inhibition of HPV-18 DNA replication by AZPHPV-2. (a) Western blot analysis of AZPs. The immunoblot was obtained by using protein extracts derived from the transient replication assays performed for Fig. 4c. The AZP expression plasmids used were pCMV-AZPHPV-1 (lane 2), pCMV-AZPHPV-2 (lane 3), and pCMV-AZPAla (lane 4). When pcDNA3.1 (lane 1) was used as the control, there was no signal except for faint artifact signals derived from endogenous proteins cross-reactive to the anti-T7-tag antibody (shown by the square bracket). (b) Competitive binding experiment with E2 and AZP. DNA-binding assays were performed with the 32P-labeled 56-bp probe containing E2BS-3 and E2BS-4 along with a constant level of E2 protein (100 nM) in the presence of increasing concentrations of AZP (nM), from lane 8 to lane 5 for AZPHPV-1 and from lane 12 to lane 9 for AZPHPV-2. Lanes 1 to 4 indicate band positions of free DNA, DNA bound to E2, DNA bound to AZPHPV-1, and DNA bound to AZPHPV-2, respectively, which are used as markers for lanes 5 to 12. The concentrations of AZP and E2 protein used are indicated below each lane. The identities of the shifted bands are described in Results and Discussion.
Another possible cause of our results is a differential inhibition of E2 binding by these AZPs. To examine whether AZPHPV-2 inhibits E2 binding more efficiently than does AZPHPV-1, competitive binding experiments with AZP and E2 protein were performed with a 32P-labeled 56-bp probe containing E2BS-3 and E2BS-4 (for the sequence, see Materials and Methods). In gel shift assays, mixtures of AZP (final concentration, 0.1, 1, 10, or 100 nM) and E2 protein (final concentration, 100 nM) were added to the binding buffer containing the DNA probe. After incubation at 4°C for 1 h, DNA-protein complexes and free DNA were resolved in a 6% nondenaturing polyacrylamide gel. In the presence of E2 alone (Fig. 5b, lane 2), two DNA-protein complexes (DNA-2E2 and DNA-E2 complexes) were observed. The main, DNA-2E2 complex represents E2 binding to both E2BS-3 and E2BS-4, while the minor, DNA-E2 complex is formed between E2 and one E2BS (either E2BS-3 or E2BS-4). An additional complex migrating more slowly than the DNA-2E2 complex was also observed. This complex (oligomer), presumably generated by oligomerization of DNA-E2 complexes (8), has been reported for E2 proteins. Formation of these E2 complexes was inhibited by AZPHPV-2 at a protein concentration lower than that of AZPHPV-1. As shown in lane 11 of Fig. 5b, 1 nM AZPHPV-2 inhibited E2 binding completely. On the other hand, AZPHPV-1 showed the same level of inhibition at a protein concentration of 10 nM (see lane 6 of Fig. 5b). Therefore, since AZPHPV-2 inhibits E2 binding more efficiently than AZPHPV-1, it is likely that AZPHPV-2 inhibits HPV-18 DNA replication more effectively than AZPHPV-1. This result also indicates that the location of an AZP-targeting site is more important than the binding specificity of AZP for efficient inhibition of E2 binding.
In the competition assay with AZPHPV-2, we observed an additional complex (indicated with an asterisk in Fig. 5b) migrating more slowly than the DNA-AZP complex at protein concentrations of ≥10 nM (see lanes 9 and 10 of Fig. 5b). This complex was also clearly observed in gel shift assays with AZPHPV-2 alone at protein concentrations of ≥10 nM (data not shown). While we have generated many six-finger AZPs so far, such an additional complex has not been detected, even at protein concentrations of 1 μM (as an example, see lane 5 of Fig. 5b).
Specific binding of AZPHPV-2 to HPV-18 DNA replication origin causes inhibition of DNA replication.
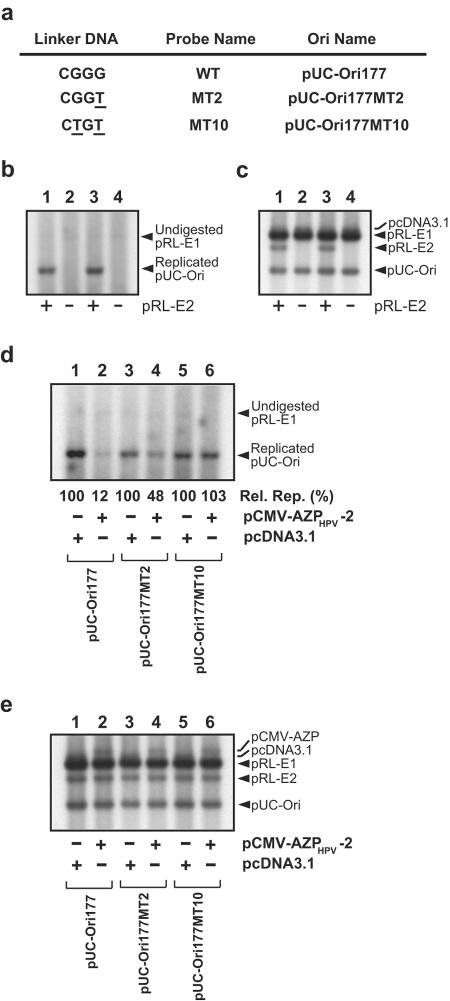
The results described above suggest that specific binding of AZPs to the HPV-18 replication origin inhibits DNA replication. To examine this further, we conducted transient replication assays by using plasmids with mutant replication origins and AZPHPV-2 because of its greater inhibition ability. If no repression of DNA replication of mutant ori plasmids, whose mutations do not ideally affect E2 binding but do impair AZPHPV-2 binding, was observed in the presence of AZPHPV-2, this would demonstrate that the inhibition of DNA replication of the wild-type HPV-18 ori shown in Fig. 4c was caused by specific binding of the designed AZPHPV-2 to the replication origin; however, if effective inhibition of mutant ori replication was observed even in the presence of AZPHPV-2, then the cause of the inhibition of DNA replication of the wild-type HPV-18 ori shown in Fig. 4c was an unknown nonspecific binding of AZPHPV-2. For this experiment, two mutant ori plasmids were prepared (Fig. 6a). These mutant ori plasmids, pUC-Ori177MT2 and pUC-Ori177MT10,contained one and two mutations, respectively, in a spacer region (i.e., 5′-CGGG-3′) (Fig. 1a) between E2BS3 and E2BS4.
FIG. 6.
Transient replication of HPV-18 mutant ori plasmids with AZPHPV-2. (a) Mutant probes and mutant ori plasmids. The mutations were introduced into the 4-bp spacer DNA (i.e., 5′-CGGG-3′) located between E2BS-3 and E2BS-4 to minimize any negative effect on HPV-18 DNA replication due to the mutations. (b) Importance of E2 for mutant ori replication. A transient replication assay was performed with pRL-E1 for E1 expression and pUC-Ori177M2 (lanes 1 and 2) or pUC-Ori177M10 (lanes 3 and 4) as a mutant ori plasmid in the presence of the E2 expression plasmid pRL-E2 (lanes 1 and 3) or in the absence of pRL-E2 (lanes 2 and 4). Hirt-extracted samples were digested with DpnI to eliminate unreplicated input plasmids. (c) Examination of amounts of plasmids introduced into 293H cells for panel b. This panel shows Southern blot hybridization with Hirt-extracted DNA samples which were not treated with DpnI, indicating relative amounts of plasmids introduced into 293H cells. (d) Replication of HPV-18 mutant ori plasmids in the presence of pCMV-AZPHPV-2. Transient replication was assayed with pRL-E1, pRL-E2, and a pUC-Ori177 derivative along with pCMV-AZPHPV-2 or pcDNA 3.1, as indicated below each lane. Hirt-extracted samples were digested with DpnI. Efficiencies of inhibition of DNA replication by AZPs were judged by comparison with relative amounts of each pUC-Ori177 derivative. The amount of each replicated pUC-Ori177 derivative in the presence of AZPHPV-2 was compared with that in the absence of AZP (100%). The average data [Rel. Rep. (%)] from four independent assays are indicated below each lane. (e) Examination of amounts of plasmids introduced into 293H cells for panel d. This panel shows Southern blot hybridization with Hirt-extracted DNA samples which were not treated with DpnI, indicating relative amounts of plasmids introduced into 293H cells.
First, we examined the in vitro binding properties of AZPHPV-2 towards DNA probes corresponding to these mutations. Since half-maximal binding to a mutant probe, MT1, containing one mutation was observed at a protein concentration of 100 pM (Fig. 3e, lane 2), gel shift assays with the mutant probe MT10 were performed. Half-maximal binding of AZPHPV-2 to MT10 was observed at 140 pM (data not shown).
We next confirmed the significance of the E2 protein for mutant ori replication, as performed for Fig. 4a. The efficiency of each mutant ori plasmid's replication in the presence of pRL-E2 was compared with that in the absence of pRL-E2. As shown in Fig. 6b, when Hirt-extracted DNA samples were treated with DpnI to eliminate unreplicated input plasmids, no replicated pUC-Ori177 was observed in the absence of pRL-E2 (see lanes 2 and 4). In the transient replication assay without pRL-E2, the same amounts of plasmids as those used in that with pRL-E2 were introduced into 293H cells, as judged by Southern blot analysis with Hirt-extracted DNA samples untreated with DpnI (Fig. 6c). In the mutant ori plasmids, mutations were introduced into the spacer region between E2BS-3 and E2BS-4 to minimize the effect of these mutations on E2 binding, whereas replication levels of these mutant ori plasmids in the absence of AZPHPV-2 were lower than that of the wild-type ori, pUC-Ori177 (compare lanes 3 and 5 with lane 1 in Fig. 6d). It has been reported that mutation of the spacer region also significantly reduces the replication level (12).
Transient replication assays with the two mutant ori plasmids (together with the wild-type ori plasmid) were then carried out according to the procedure described for Fig. 4. As shown in Fig. 6d, lanes 3 and 4, one mutation reduced the inhibition level by AZPHPV-2 to 48% ± 4%, indicating that AZPHPV-2 recognizes a 1-bp difference in 293H cells as well. Moreover, two mutations increased the replication level in the presence of AZPHPV-2 (Fig. 6d, lane 6) to that of the control (Fig. 6d, lane 5). The same results were also obtained with an additional mutant ori plasmid containing a 4-bp mutation, from 5′-CGGG-3′ to 5′-ATAT-3′ (data not shown). These results demonstrate that AZPHPV-2 discriminates differences of >2 bp in 293H cells. In the transient replication assay with pCMVAZPHPV-2 for each mutant ori plasmid, the same amounts of plasmids as those used in that with pcDNA3.1 were introduced into 293H cells, as shown in Fig. 6e (for example, compare lane 3 with lane 4). Because half-maximal binding to the mutant probes was observed at concentrations of AZPHPV-2 that were 10- to 14-fold higher than that for the wild-type probe, the loss of inhibition of replication of these mutant ori plasmids indicates that specific binding of AZPHPV-2 to the HPV-18 replication origin inhibited DNA replication.
Before conducting this experiment with mutant ori plasmids, we considered that repression of E2 expression by AZP might also cause the inhibition of wild-type ori replication. If this happened in the transient replication assays with the wild-type ori plasmid, then the same results (i.e., inhibition of replication in the presence of AZP) should have been obtained in replication assays with the mutant ori plasmids because AZP should have repressed E2 expression in the mutant ori replication assays as well (the same E2 expression plasmid, pRL-E2, was used in both transient replication assays). However, as shown in lanes 5 and 6 of Fig. 6d, a significant reduction of replicated pUC-Ori177 was not observed in the presence of AZP. Therefore, it is unlikely that repression of E2 expression by AZP caused the inhibition of wild-type ori replication.
AZP technology for antiviral therapies.
Recently, we demonstrated in planta that blocking the binding of a viral replication protein of BSCTV, a plant DNA virus, to its replication origin by use of an artificial zinc finger protein led to inhibition of virus replication and that transgenic plants expressing the AZP were completely resistant to BSCTV infection (22). Because the binding of a viral protein(s) to its replication origin is also important for genome replication of animal DNA viruses, except in a few instances (e.g., parvoviruses) (reviewed in reference 5), the success of the strategy in planta prompted us to examine the effectiveness of the strategy against animal viruses. The results of this study demonstrate that our AZP technology for inhibiting replication can also be applied to animal viruses. One of the advantages of our strategy is the very low or nonexistent risk of the emergence of resistant viruses. Conventional antiviral drugs are designed to inactivate one component (usually viral DNA polymerase) of a viral protein. Therefore, viruses can easily escape the attack of an antiviral drug by mutation of the viral protein without a significant loss of their original activity. In contrast, our strategy targets E2 binding. In order to escape the AZP attack without sacrificing replication efficiency, viruses need to mutate a DNA base(s) in a region recognized by the AZP (so the AZP cannot bind to the site) and additionally to mutate the viral E2 protein at the same time so that the mutant E2 protein can bind to the mutated origin; mutation of an AZP binding site alone is unfavorable to DNA viruses because the native E2 protein cannot bind to the mutated replication origin either. The probability of the synchronized double mutation, which is equal to the probability of mutation in the viral replication origin multiplied by that of mutation in the E2 protein, should be extremely low. Therefore, it is likely that viruses resistant to our approach will emerge less frequently than those resistant to conventional antiviral drugs. We hope that this work will lead to the development of AZP-based antiviral therapies/treatments with much lower probabilities of emergence of resistant viruses.
Acknowledgments
We thank Tadayuki Imanaka and Haruyuki Atomi for their help with DNA sequencing of constructed plasmids and Takuya Kanamori for help with transfection experiments.
This work was supported in part by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (no. 17550154 to T.S.).
REFERENCES
- 1.Bosch, F. X., M. M. Manos, N. Muñoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, and K. V. Shah. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) study group. J. Natl. Cancer Inst. 87:796-802. [DOI] [PubMed] [Google Scholar]
- 2.Chao, S.-F., W. J. Rocque, S. Daniel, L. E. Czyzyk, W. C. Phelps, and K. A. Alexander. 1999. Subunit affinities and stoichiometries of the human papillomavirus type 11 E1:E2:DNA complex. Biochemistry 38:4586-4594. [DOI] [PubMed] [Google Scholar]
- 3.Chiang, C.-M., M. Ustav, A. Stenlund, T. F. Ho, T. R. Broker, and L. T. Chow. 1992. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl. Acad. Sci. USA 89:5799-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Del Vecchio, A. M., H. Romanczuk, P. M. Howley, and C. C. Baker. 1992. Transient replication of human papillomavirus DNAs. J. Virol. 66:5949-5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiMaio, D., and D. M. Coen. 2001. Replication strategies of DNA viruses, p. 119-132. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 6.Frattini, M. G., and L. A. Laimins. 1994. Binding of the human papillomavirus E1 origin-recognition protein is regulated through complex formation with the E2 enhancer-binding protein. Proc. Natl. Acad. Sci. USA 91:12398-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 8.Hou, S. Y., S.-Y. Wu, and C.-M. Chiang. 2002. Transcriptional activity among high and low risk human papillomavirus E2 proteins correlates with E2 DNA binding. J. Biol. Chem. 277:45619-45629. [DOI] [PubMed] [Google Scholar]
- 9.Howley, P. M., and D. R. Lowy. 2001. Papillomaviruses and their replication, p. 2197-2229. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 10.Kim, Y.-S., J.-M. Kim, D.-L. Jung, J.-E. Kang, S. Lee, J. S. Kim, W. Seol, H.-C. Shin, H. S. Kwon, C. V. Lint, N. Hernandez, and M.-W. Hur. 2005. Artificial zinc-finger fusions targeting Sp-1 binding sites and the trans-activator-responsive element potently repress transcription and replication of HIV-1. J. Biol. Chem. 280:21545-21552. [DOI] [PubMed] [Google Scholar]
- 11.Kuo, S.-R., J.-S. Liu, T. R. Broker, and L. T. Chow. 1994. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J. Biol. Chem. 269:24058-24065. [PubMed] [Google Scholar]
- 12.Lee, D., H. Kim, Y. Lee, and J. Choe. 1997. Identification of sequence requirement for the origin of DNA replication in human papillomavirus type 18. Virus Res. 52:97-108. [DOI] [PubMed] [Google Scholar]
- 13.Lowy, D. R., and P. M. Howley. 2001. Papillomavirus, p. 2231-2264. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 14.McBride, A., and G. Myers. 1997. The E2 proteins, p. 37-53. In G. Myers, C. Baker, K. Münger, F. Sverdrup, A. McBride, and H.-U. Bernard (ed.), Human papillomavirus 1997. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 15.Papworth, M., M. Moore, M. Isalan, M. Minczuk, Y. Choo, and A. Klug. 2003. Inhibition of herpes simplex virus 1 gene expression by designer zinc-finger transcription factors. Proc. Natl. Acad. Sci. USA 100:1621-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peden, K. W. C., J. M. Pipas, S. Pearson-White, and D. Nathans. 1980. Isolation of mutants of an animal virus in bacteria. Science 209:1392-1396. [DOI] [PubMed] [Google Scholar]
- 17.Pisani, P., F. Bray, and D. M. Parkin. 2002. Estimates of the world-wide prevalence of cancer for 25 sites in the adult population. Int. J. Cancer 97:72-81. [DOI] [PubMed] [Google Scholar]
- 18.Remm, M., R. Brain, and J. R. Jenkins. 1992. The E2 binding sites determine the efficiency of replication for the origin of human papillomavirus type 18. Nucleic Acids Res. 20:6015-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynolds, L., C. Ullman, M. Moore, M. Isalan, M. J. West, P. Clapham, A. Klug, and Y. Choo. 2003. Repression of the HIV-1 5′ LTR promoter and inhibition of HIV-1 replication by using engineered zinc-finger transcription factors. Proc. Natl. Acad. Sci. USA 100:1615-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell, J., and M. R. Botchan. 1995. cis-Acting components of human papillomavirus (HPV) DNA replication: linker substitution analysis of the HPV type 11 origin. J. Virol. 69:651-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segal, D. J., J. Gonçalves, S. Eberhardy, C. H. Swan, B. E. Torbett, X. Li, and C. F. Barbas III. 2004. Attenuation of HIV-1 replication in primary human cells with a designed zinc-finger transcription factor. J. Biol. Chem. 279:14509-14519. [DOI] [PubMed] [Google Scholar]
- 22.Sera, T. 2005. Inhibition of virus DNA replication by artificial zinc finger proteins. J. Virol. 79:2614-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sera, T., and C. Uranga. 2002. Rational design of artificial zinc-finger proteins using a nondegenerate recognition code table. Biochemistry 41:7074-7081. [DOI] [PubMed] [Google Scholar]
- 24.Sverdrup, F., and S. A. Khan. 1994. Replication of human papillomavirus (HPV) DNAs supported by the HPV type 18 E1 and E2 proteins. J. Virol. 68:505-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sverdrup, F., and S. A. Khan. 1995. Two E2 binding sites alone are sufficient to function at the minimal origin of replication of human papillomavirus type 18 DNA. J. Virol. 69:1319-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Titolo, S., A. Pelletier, F. Sauvé, K. Brault, E. Wardrop, P. W. White, A. Amin, M. G. Cordingley, and J. Archambault. 1999. Role of the ATP-binding domain of the human papillomavirus type 11 E1 helicase in E2-dependent binding to the origin. J. Virol. 73:5282-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Horn, G., S. Sheikh, and S. A. Khan. 2001. Regulation of human papillomavirus type 1 replication by the viral E2 protein. Virology 287:214-224. [DOI] [PubMed] [Google Scholar]
- 28.Walboomers, J. M. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Stah, P. J. F. Snijders, J. Peto, C. J. L. Meijer, and N. Muñoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 29.Wilson, G. G., M. West, K. Woytek, and D. Rangasamy. 2002. Papillomavirus E1 proteins: form, function, and features. Virus Genes 24:275-290. [DOI] [PubMed] [Google Scholar]
- 30.Yasugi, T., J. D. Benson, H. Sakai, M. Vidal, and P. M. Howley. 1997. Mapping and characterization of the interaction domains of human papillomavirus type 16 E1 and E2 proteins. J. Virol. 71:891-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou, N., J.-S. Liu, S.-R. Kuo, T. R. Broker, and L. T. Chow. 1998. The carboxyl-terminal region of the human papillomavirus type 16 E1 protein determines E2 protein specificity during DNA replication. J. Virol. 72:3436-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]