Abstract
Flaviviral replication is believed to be exclusively cytoplasmic, occurring within virus-induced membrane-bound replication complexes in the host cytoplasm. Here we show that a significant proportion (20%) of the total RNA-dependent RNA polymerase (RdRp) activity from cells infected with West Nile virus, Japanese encephalitis virus (JEV), and dengue virus is resident within the nucleus. Consistent with this, the major replicase proteins NS3 and NS5 of JEV also localized within the nucleus. NS5 was found distributed throughout the nucleoplasm, but NS3 was present at sites of active flaviviral RNA synthesis, colocalizing with NS5, and visible as distinct foci along the inner periphery of the nucleus by confocal and immunoelectron microscopy. Both these viral replicase proteins were also present in the nuclear matrix, colocalizing with the peripheral lamina, and revealed a well-entrenched nuclear location for the viral replication complex. In keeping with this observation, antibodies to either NS3 or NS5 coimmunoprecipitated the other protein from isolated nuclei along with newly synthesized viral RNA. Taken together these data suggest an absolute requirement for both of the replicase proteins for nucleus-localized synthesis of flavivirus RNA. Thus, we conclusively demonstrate for the first time that the host cell nucleus functions as an additional site for the presence of functionally active flaviviral replicase complex.
Several members of the genus Flavivirus, which comprises viruses with a single-strand RNA of positive polarity, such as West Nile virus (WNV), dengue virus (DENV), Japanese encephalitis virus (JEV), yellow fever virus (YFV), Murray Valley encephalitis virus and tick-borne encephalitis virus, are pathogens of humans and animals. The 11-kb-long flaviviral genomes encode three structural and seven nonstructural proteins, derived by processing of the primary polyprotein translation product by host signalases and a viral protease, NS3. The NS3 protein, in addition, has helicase and NTPase functions in flaviviral replication. The viral RNA-dependent RNA polymerase (RdRp), the product of the viral NS5 gene, is responsible for replication of the viral genome within putative complexes comprising both viral and an as-yet-unidentified host protein(s). The replication complex (RC) uses the genomic RNA to generate a double-stranded replicative form (RF), and initiation of RNA synthesis on this template results in formation of replicative intermediates (RI) that resolve, upon completion of strand synthesis, to produce a single-stranded viral RNA (vRNA) and RF. This model of flavivirus replication by an asymmetric and semiconservative mode was first proposed for Kunjin virus (KUNV) (8) and has been subsequently confirmed for DENV (9) and JEV (49).
The RdRp activity of several flaviviruses including KUNV, DENV, JEV, and WNV has been established to be tightly associated with intracellular cytosolic membranes in numerous studies through biochemical (7, 15, 16, 45, 46, 51, 54) and ultrastructural (52) analyses. This has led most investigators to use postnuclear supernatant fractions as a source of membrane-bound flaviviral RC in in vitro RdRp assays. However, a few early studies employing in vivo labeling of JEV and WNV RNA have suggested that, in addition to cytoplasm, the viral RNA species is also found associated with the nuclear fraction of infected cells (16, 45, 54). Consequently, the outer nuclear envelope membrane (ONEM) and its extensions into the endoplasmic reticulum (ER) were also proposed to harbor the flavivirus RC. A single study on Saint Louis encephalitis virus reported nuclear localization of vRNA synthesis (3). However, the methods used in these studies did not clearly differentiate vRNA species from those of the host, nor did these studies validate the identity of the subcellular fractions using appropriate biochemical markers.
Many positive- and negative-strand RNA viruses that use the host cytoplasm as the primary site of replication engage either the nucleus or its components by sequestering nuclear factors and by altering nuclear-cytoplasmic trafficking to promote viral replication (for a review, see reference 21). Flaviviruses have been demonstrated to involve host nuclei in their replication, albeit indirectly. Treatment of host cells with either actinomycin D or alpha-amanitin before virus infection compromised viral replication and subsequently viral titers (49, 54). Enucleation of cells during the latent period of virus infection also inhibited JEV replication (29). Taken together, these results pointed to a requirement for a factor(s) coded for by the host DNA and, therefore, a functional nucleus, especially during the early phase of JEV replication. Thus, while the role of nuclear involvement in flaviviral replication is established, the question of whether active viral RNA synthesis occurs inside the nucleus remained unaddressed.
There is compelling evidence for the translocation of flaviviral and picornaviral RdRp into the nucleus (28, 50). In the case of two flaviviruses, DENV and YFV, a fraction of NS5 has been shown to migrate into the nucleus, consistent with the presence of nuclear localization signals (NLS) in the NS5 protein (4, 5, 28). Although the exact function(s) carried out by flaviviral NS5 in the nucleus is yet unknown, a recent demonstration that recombinant DENV NS5 alone was sufficient to carry out synthesis of negative strands by transcription of exogenously added subgenomic positive-strand RNA templates with a specificity similar to that obtained using infected cell-derived RC (1) raised the possibility that flaviviral RNA synthesis within infected cell nuclei might be achieved solely by NS5. Hence, it is likely that the nucleus of infected cells may function as an additional site for flaviviral RNA synthesis.
We have therefore examined in detail, using a combination of biochemical, confocal microscopic, and ultrastructural analyses, the role of the host nucleus in flaviviral replication using JEV, WNV, and DENV as model systems. Our results provide evidence that nuclei in flavivirus-infected cells harbor active flaviviral RC, establishing that flaviviral replication is not restricted to the cytoplasm but occurs in the host nucleus as well. Thus, effective therapy for flaviviral infections would need to target both sites of viral RNA synthesis.
MATERIALS AND METHODS
Viruses and cells.
WNV strain E101, JEV strain P20778, and DENV-2 strain TR 1751 (National Institute of Virology, Pune, India) were propagated in the Aedes albopictus cell line C6/36 (National Centre for Cell Science, Pune, India) as described earlier (49). The porcine kidney cell line PS (National Centre for Cell Science) infected with WNV, JEV, or DENV at a multiplicity of infection of 10 was used as a source of viral RC at 22 h postinfection (p.i.) for WNV and JEV and at 48 h p.i. for DENV.
Subcellular fractionation of infected cells and preparation of flaviviral RC.
Flavivirus-infected PS cells were harvested at various time points p.i. and disrupted as described previously (8, 49). Briefly, cell pellets were resuspended in TNMg buffer (10 mM Tris, pH 8.0, 10 mM sodium acetate, 1.5 mM MgCl2) at a density of 5 × 106 cells per ml and allowed to swell on ice for 10 min before being disrupted by sequential passage through 21- and 29-gauge needles 20 times each. The homogenate obtained was centrifuged at 800 × g for 7 min to obtain a nuclear (N) pellet fraction and a postnuclear supernatant (PNS). The latter was further centrifuged at 16,000 × g in a refrigerated microcentrifuge to obtain a heavy membrane pellet fraction (P16) and a microsomal supernatant fraction (S16). Nuclear fractions were resuspended in TNMg buffer containing 10% sucrose and sedimented in a refrigerated swing-out centrifuge at 1,800 × g for 10 min through two volumes of a 30% sucrose cushion, followed by two washes with TNMg buffer to rid them of cellular debris. The membranes in the above sucrose supernatant from the centrifugation at 1,800 × g were sedimented at 1,000 × g following threefold dilution with TNMg buffer and combined with PNS for further analyses. The protein concentrations of the various fractions were determined as described earlier (44).
Nocodazole treatment.
Nocodazole (Sigma-Aldrich) at a concentration of 6 μg per ml was added at 16 h p.i. for JEV and WNV and at 42 h p.i. for DENV; treatment was carried out for a period of 6 h prior to harvesting cells. Nocodazole treatment of Kunjin virus-infected cells 18 h p.i. did not affect viral titers or the localization of viral proteins (35). Nocodazole treatment has also been reported to have no effect on viral maturation and secretion or on viral titer in a variety of cell lines infected with WNV (24). We found that nocodazole treatment under our conditions had no effect on the total RdRp activity in the infected cell homogenates, nor did it alter the distribution of RdRp activity in the subcellular fractions reported above (data not shown).
In vitro RdRp assay.
The in vitro RdRp assays and subsequent extraction and analysis of labeled RNA products by partially denaturing 7 M urea-3% polyacrylamide gel electrophoresis (PAGE) as well as computation of enzyme activity using Fuji MacBAS V2.4 software were as described earlier (49).
Detergent treatment of nuclear fractions from flavivirus-infected cells.
All detergent treatments of sucrose-purified nuclear fractions used a protein concentration of 2 mg/ml on ice for 1 h. The ONEM was solubilized by using 1% Triton X-100 (TX100) or a premixed combination of 0.5% sodium deoxycholate and 1% Tween 80 (referred to as double detergent [DD]), which was reported to be more efficient for this purpose (22, 42). The treated samples were centrifuged at 800 × g for 10 min at 4°C to obtain the soluble supernatant and an insoluble nuclear pellet fraction.
Organelle-specific marker proteins and enzymes.
Absence of nuclear contamination of the PNS and P16 was confirmed by blotting with monoclonal antibody LA2B3 specific to the A-type lamins A and C, which are alternatively spliced products of the lamin A gene, LMNA (a gift from V. Parnaik, Centre for Cellular and Molecular Biology, Hyderabad, India). Conversely, cytosolic contamination in nuclear fractions was ruled out based on absence of the cytosolic enzyme lactate dehydrogenase (LDH; Roche Applied Science, Germany), and nuclei were determined to be greater than 97% pure. Complete removal from the nuclei of the outer nuclear membrane by DD treatment was ascertained based on the absence of the ONEM marker enzyme mannose-6-phosphatase (M6P) (14, 47), as described earlier (14, 23). The inorganic phosphate liberated by M6P was estimated by a modified molybdate-malachite green method (32), with the absorbance read at 650 nm. More than 96% of the M6P activity was routinely removed following the detergent treatments mentioned above. Absence of the ER marker protein calnexin in DD-treated nuclei as determined by confocal microscopy following indirect immunofluorescence analysis with a specific antibody (Stressgen Biotechnologies Corporation) also confirmed total removal of the ONEM by this treatment. We further confirmed removal of the ER-derived ONEM by electron microscopic analyses of resin-embedded (see below), DD-treated nuclear fractions. Nuclease-free molecular biology grade TX100, sodium deoxycholate, and Tween 80 were obtained from Sigma-Aldrich.
Metabolic labeling of viral proteins.
Mock- and JEV-, WNV-, or DENV-infected PS cells were labeled using [35S]methionine-cysteine (EXPRE35S35S; 1175.0 Ci/mmol; PerkinElmer Life Sciences), fractionated as described above, electrophoresed, and subjected to fluorography as described previously (48). The identities of labeled proteins of JEV were confirmed using antisera specific to the NS3, NS5, NS1, and envelope proteins.
Indirect immunofluorescence of flavivirus-infected cells.
JEV-infected PS cells grown on coverslips were washed with phosphate-buffered saline (PBS) at 16 h p.i. and fixed with freshly prepared 3.7% paraformaldehyde in PBS for 10 min. Nuclei obtained from JEV-infected cells and purified by centrifugation through sucrose as described above were treated with DD for 1 h at 4°C to remove the ONEM and then fixed as mentioned above. After three washes with PBS containing 0.1 M glycine to quench unreacted aldehyde groups, the nuclei were deposited on poly-l-lysine (Mr, 150,000 to 300,000; Sigma, St. Louis, Mo.)-treated coverslips. The nuclei were fixed again as above for 5 min, washed as before, and permeabilized with 100% acetone for 10 min. Nuclear matrix preparations were obtained by treating DD-extracted nuclei deposited on coverslips with DNase I, followed by sequential extraction with 0.25 M ammonium sulfate and 2 M sodium chloride as previously described (38, 40). After a blocking step with PBS containing 0.1% bovine serum albumin (PBS-BSA, fraction V; Sigma) and normal goat serum (1:1,000 dilution; Vector Laboratories), immunostaining was performed with rabbit anti-JEVNS3, mouse anti-JEVNS5, mouse anti-lamin A/C monoclonal LA2B3, or mouse anti-calnexin antibodies (1:2,000) in PBS-BSA, followed by fluorescein isothiocyanate (FITC)-conjugated appropriate goat immunoglobulin G (IgG; 1:100) (Bangalore Genei Pvt. Ltd., Bangalore, Karnataka, India) for 30 min. In experiments aimed at simultaneous detection of NS3 and NS5, the former was detected using mouse anti-JEVNS3 antibodies followed by affinity-purified goat anti-mouse IgG-tetramethylrhodamine isothiocyanate (TRITC; Invitrogen). For colocalization of NS3 and lamin, the former was detected with TRITC- and the latter with FITC-conjugated secondary reagents. The samples were finally washed twice before being mounted on slides with antifade reagent [100% glycerol containing 5% n-propyl gallate, 0.25% 1,4-diazobicyclo-(2, 2, 2)-octane, and 0.0025 g of para-phenylenediamine; Sigma] and viewed in a Leica DMIRB inverted confocal laser scanning microscope with the pinhole set at 1.5. Images were processed with Adobe Photoshop, version 7.0. Antibodies to JEVNS3 and JEVNS5 were raised in rabbit and mouse, respectively, immunized with recombinant proteins expressed in Escherichia coli from the pET3d vector carrying DNA inserts obtained by reverse transcription-PCR of RNA from JEV-infected PS cells using appropriate primers (30).
For localizing nascent RNA, sucrose-purified nuclei from flavivirus-infected cells were subjected to RdRp assays in vitro as described above but with a mix of 100 μM biotin-16-UTP (Roche Applied Science, Germany), 5 μM UTP, 100 μM GTP, 100 μM CTP, and 100 μM ATP for 5 min; samples were then washed and treated as described above. Biotinylated RNA was detected using streptavidin-FITC (1:100 in PBS-BSA; BD Biosciences) at room temperature for 30 min after blocking with PBS-BSA containing 2% blocking reagent (Roche Applied Science, Germany) and visualized as above. In colocalization experiments, rabbit anti-JEVNS3 antibodies were detected using affinity-purified goat anti-rabbit IgG-TRITC (Invitrogen).
Western blot analysis.
Proteins in the nuclear matrix and P16 fractions obtained from noninfected and JEV-infected PS cells were electrophoresed on sodium dodecyl sulfate (SDS)-10% polyacrylamide gels. Proteins transferred onto nitrocellulose membranes were detected using rabbit anti-NS3 serum and mouse anti-lamin A/C monoclonal LA2B3 antibody.
Coimmunoprecipitation of viral NS3, NS5, and nascent viral RNA.
JEV-infected cells were metabolically labeled with [35S]methionine-cysteine (EXPRE35S35S; 1175.0 Ci/mmol; NEN) as described above. Cells were harvested at 18 h p.i., and nuclei were purified as described above. Nuclei were incubated briefly for 5 min in RdRp assay buffer containing 10 μCi of [32P]UTP (specific activity of 6,000 Ci/mmol) to label newly synthesized viral RNA. These nuclei were then extracted exhaustively with DD to remove the ONEM and associated ER membranes, which we verified both by a biochemical assay for the absence of M6P activity and by confocal microscopy for the absence of immunoreactive calnexin. These nuclear preparations were treated with RNase-free DNase I and then solubilized in immunoprecipitation buffer (150 mM sodium chloride, 10 mM Tris, pH 7.5, 0.1% sodium lauryl sulfate, 1.0% deoxycholate, and 1% Nonidet P-40), and immunoprecipitation was carried out using rabbit anti-JEVNS3 or mouse anti-JEVNS5. The precipitates were divided into two fractions, one of which was processed for SDS-PAGE followed by fluorography and the other of which was processed for RNA isolation and partially denaturing 3% PAGE to visualize viral RNA species as described above.
Immunoelectron microscopy of flavivirus-infected cells and nuclei.
Cell permeabilization and labeling of nascent RNA with biotin-16-UTP was carried out essentially as described earlier (33). Briefly, JEV- and mock-infected PS cells at 16 h p.i. were treated for 1 h with 10 μg of actinomycin D per ml to inhibit cellular RNA synthesis, washed three times with serum-free medium, and treated with 200 μg of lysolecithin per ml for 1 min on ice. The permeabilized cells were then incubated for 10 min in a transcription mix containing 0.5 mM biotin-16-UTP (Roche Applied Science, Germany) to label de novo synthesized viral RNA. Labeling was terminated by fixation of the cells. Sample fixation, resin embedding, ultrathin sectioning, and immunostaining for electron microscopy were carried out essentially as described previously (48). Biotinylated RNA was detected using streptavidin conjugated to 15-nm gold particles (1:100 dilution; Sigma-Aldrich), and JEVNS3 was localized using rabbit anti-JEVNS3 polyclonal antibodies (1:500) followed by detection with anti-rabbit IgG (heavy plus light chains) antibodies coupled to 10-nm gold particles (Ted Pella Inc.). In colocalization experiments JEVNS3 and biotinylated RNA were detected using appropriate antibodies conjugated to 5- and 15-nm gold particles, respectively. After being stained with uranyl acetate and lead citrate, the samples were visualized in a JEOL JEM-100CXII electron microscope operated at 80 kV.
RESULTS
Nuclear association of viral RC in flavivirus-infected cells.
We initially addressed the question of nuclear localization of flaviviral RC using in vitro RdRp assays and visualization of all viral RNA species using the 7 M urea-3% PAGE system for three different flaviviruses (JEV, WNV, and DENV). At appropriate times postinfection, RdRp activity in each of the fractions (N, PNS, P16, and S16) from flavivirus-infected cells measured as radioactivity incorporated in the three viral RNA species (see Materials and Methods) was compared. The P16 fractions (heavy membranes) from all three virus-infected cells harbored ∼60% of the total activity (Fig. 1A, lanes 3, 7, and 11), while the S16 fraction recorded negligible incorporation (Fig. 1A, lanes 4, 8, and 12). Interestingly, ∼30 to 40% of the RdRp activity was associated with the nuclear fractions in all three flaviviruses studied (Fig. 1A, lanes 1, 5, and 9), as reported earlier for JEV (45, 54). Preparations of these nuclei were ascertained to be free of cytosolic contamination and more than 97% pure based on the absence of LDH (data not shown).
FIG. 1.
Flaviviral replication complexes are associated with the nuclear fraction. (A) In vitro RdRp activity associated with cellular fractions. Sucrose-purified nuclear fractions (N), postnuclear supernatants (PNS), heavy membrane (P16), and its supernatant (S16) fractions from cells infected with JEV (lanes 1 to 4), WNV (lanes 5 to 8), and DENV (lanes 9 to 12) were subjected to in vitro RdRp assays using [α-32P]GTP. The labeled RNA products generated were resolved by a partially denaturing 7 M urea-3% polyacrylamide gel electrophoresis. Values above lane numbers indicate the proportion of total RdRp activity seen in each fraction of flavivirus-infected cells. The sum of the values in the N and PNS fractions is equal to 1. The arrowheads in panel A denote the position of the three viral RNA species RI, vRNA, and RF. (B) Protein profile in cellular fractions of flavivirus-infected cells. N, PNS, P16, and S16 fractions from actinomycin D-treated cells metabolically labeled with [35S]methionine-cysteine and mock infected (lanes 1 to 4) or infected with JEV (lanes 5 to 8), WNV (lanes 9 to 12), and DENV (lanes 13 to 16) were electrophoresed on SDS-10% polyacrylamide gels followed by autoradiography. The asterisks indicate the locations of flavivirus-specific NS5 and NS3. The positions of standard molecular mass markers are indicated on the left.
Since the cytosolic membranous compartments which host flaviviral RC are localized in close apposition with nuclear membranes, we treated virus-infected cells with the microtubule-depolymerizing drug nocodazole, which disrupts attachment of microtubules with the nuclei and nucleus-associated structures (36), thus achieving better separation of the cytoskeletal and cytoplasmic membrane network from the nuclear membrane. The proportions of RdRp activity in the various fractions obtained from nocodazole-treated cells were similar to those obtained from untreated cells reported above (data not shown), confirming the genuine nuclear location of the RdRp activity observed in that fraction. Metabolic labeling of proteins in cells infected with JEV, WNV, and DENV in the presence of actinomycin D revealed the presence of both NS3 and NS5 in the sucrose-purified nuclear fraction for all three viruses (Fig. 1B, lanes 5, 9, and 13), consistent with the presence of substantial RdRp activity in this compartment. Curiously, a sizeable proportion of NS5 of all three flaviviruses was found in the S16 fraction (Fig. 1B, lanes 8, 12, and 16), which, however, was devoid of RdRp activity (Fig. 1A, lanes 4, 8, and 12), probably due to either the absence of viral RNA template or essential viral/host factors in this fraction.
Nonionic detergent treatment reveals association of flaviviral RC with the host nucleus.
The presence of RdRp activity as well as major replicase proteins in nuclear fractions could be attributed to the association of flaviviral RC with the ONEM and the continuous ER, as has been reported previously (45, 46, 54). In our initial analyses we labeled the viral RNAs in an in vitro assay using purified nuclear fractions and then studied the effect of detergents (such as TX100 or DD, which specifically remove the ONEM and solubilize the ER) on the localization of the newly synthesized labeled RNA species. Remarkably, ∼50% of the labeled viral RNA species remained associated with the nuclear pellet even after extensive detergent treatment (Fig. 2A). Nearly complete removal of M6P, a specific marker enzyme for the ONEM and ER (14, 23) (Table 1), as well as quantitative removal of calnexin, an ER marker (see Fig. 4C), ruled out contamination of detergent-treated nuclei by the ONEM and validated our fractionation procedure. These data suggested that the flaviviral RCs were, in fact, associated with the nucleus in addition to the ONEM. The earlier reported absence of labeled viral RNA species in TX100-washed nuclei (54) was probably due to nuclease contamination in the detergent used, leading to degradation of viral RNA species associated with the nucleus.
FIG. 2.
Distribution of nuclear fraction-associated flaviviral RdRp activity and in vitro labeled RNA following DD treatment. (A) The nuclear fractions from cells infected with JEV (lanes 1 to 3), WNV (lanes 4 to 6), and DENV (lanes 7 to 9) were subjected to an in vitro RdRp assay using [α-32P]GTP and processed as depicted in the flowchart to obtain RNA from total (T), pellet (P), and supernatant (S) fractions. (B) Purified nuclear fractions from WNV-infected cells were processed as shown in the flowchart after treatment with DD for 1 h on ice to obtain total (T), pellet (P), and supernatant (S) fractions and then subjected to in vitro RdRp assay using [α-32P]GTP. The activity in control nuclear fractions not treated with detergent (N) is shown in lane 1. The labeled RNA obtained from each of these fractions was resolved by urea-PAGE. Values above the lane numbers in panels A and B indicate total radioactivity incorporated by all three viral RNA species as a proportion of that detected in appropriate control assays shown in lanes T. The value of 0.45 in brackets in panel B, below lane 2, represents the proportion of radioactivity incorporated as a proportion of that detected in lane 1. The arrowheads in panels A and B indicate the positions of the three viral RNA species RI, vRNA, and RF. (C) Proteins from total (T) and DD-treated pellet (P) and supernatant (S) fractions of purified nuclei from metabolically labeled WNV-infected cells were analyzed by SDS-10% PAGE. The positions of the standard molecular weight size markers are indicated at left.
TABLE 1.
Marker enzyme activitya
| Fraction | M6P activityb
|
LDH activityc
|
||
|---|---|---|---|---|
| Total | Specific | Total | Specific | |
| Homogenate | 570.73 (100) | 265.46 | 2049.4 (100) | 953.2 |
| Nuclei | 119.4 (20.92) | 306.9 | 48.6 (2.37) | 124.94 |
| DD-extracted nuclei | 1.22 (0.21) | 3.97 | 3.17 (0.15) | 10.3 |
Total activities of M6P and LDH are given for fractions obtained from 10 million PS cells, with a total protein content of 2.15 mg. Values in parentheses are percentages.
Total M6P activity is expressed as nmol of Pi produced/min, while specific activity is expressed as nmol/min/mg of protein.
Total LDH activity is expressed as nmol of reduced NADH/min, while specific activity is expressed as nmol/min/mg of protein.
FIG. 4.
JEVNS3 and JEVNS5 colocalize within virus-infected cells and along the rim of DD-treated nuclei. Distribution of NS5 (A) and NS3 (B) within DD-treated nuclei by indirect immunofluorescence using mouse and rabbit polyclonal antibodies, respectively, followed by appropriate secondary reagents conjugated to FITC. NS5 is present throughout the nucleoplasm and along the rim (A, frame ii), while NS3 is present only along the inner membrane of the nucleus (B, frame iv). Frames i and iii show bright-field images of the same nuclei. (B) Quantitative loss of calnexin from DD-treated nuclei (compare frames i and ii) confirms complete removal of the ONEM. Such nuclei, however, retained NS3 signals (frame iv). (C) Colocalization of NS3 and NS5 in JEV-infected cells and in DD-treated nuclei. Frame i reveals colocalization of the two proteins in the perinuclear region, while frame ii reveals their combined presence along the inner nuclear membrane of DD-treated nuclei. NS5 and NS3 were stained by mouse and rabbit sera, respectively, and detected by FITC- and TRITC-conjugated secondary reagents, respectively. Additional nucleoplasmic distribution of NS5 is evident in both frames i and ii.
In the above-mentioned experiments, the fractionation of nuclei using detergents was carried out after the RdRp assay. This approach was necessary to localize newly synthesized RNA in the nuclear fraction for JEV where RdRp activity was totally inactivated following detergent treatment (48, 49). In the case of WNV, however, 45% of the total nuclear RdRp activity was retained even after DD treatment (Fig. 2B, compare lanes 1 and 2), a feature also observed with DENV (data not shown). Following DD treatment, the bulk of this residual nucleus-associated WNV RdRp activity fractionated into the DD extracts of nuclei (Fig. 2B, compare lanes 3 and 4). The weak residual activity in the nuclear pellet fraction resulted in incorporation of label into viral RI species (Fig. 2B, lane 3), the species of viral RNA into which label first incorporates as shown for both Kunjin virus (8) and JEV (49). These, however, failed to elongate to completion and resolve into RF and vRNA, reflecting the damage inflicted by the detergent on the replicase complex. This loss of RdRp activity from the DD-treated nuclear pellet fraction could most probably be due to solubilization and subsequent loss upon centrifugation of vital host or viral factors such as NS1 (Fig. 2C and see below) from the associated RC.
We next analyzed the protein profile of nuclei from metabolically labeled WNV-infected cells. Fractionation of such nuclei after DD treatment revealed that the major replicase proteins, NS5 and NS3, were distributed in both the nuclear pellet and the supernatant fractions (Fig. 2C, compare lanes 2 and 3). Notably, this treatment caused dissociation of NS1 from replicase proteins associated with the nuclear pellet, as evidenced by its near complete extraction into the supernatant fraction (Fig. 2C, lane 3), probably accounting for the loss of RdRp activity from the detergent-treated nuclear pellet. On the other hand, we surprisingly found reproducible association of significant amounts of the viral envelope protein with detergent-extracted nuclei (Fig. 2C, lane 2). Thus, our data proved that both flaviviral RNA and replicase proteins NS5 and NS3 that constitute the RC are associated with the nucleus in addition to the ONEM.
Immunofluorescence studies to determine intracellular localization of flaviviral replicase proteins.
In order to confirm the association of the replicase component with the nucleus, we carried out indirect immunofluorescence analysis of JEV-infected PS cells with polyclonal antisera to the JEV helicase NS3 and polymerase NS5. As has been previously reported for JEV and other flaviviruses (46), we observed intense staining for NS3 in the perinuclear region and emanating into the cytoplasm (Fig. 3A, frame i). Since the perinuclear staining pattern of NS3 made it difficult for us to establish its association with the nucleus, we again used nocodazole treatment to distinguish the cytoplasmic NS3 signal from that associated with the nucleus. Nocodazole treatment caused the reticular NS3 fluorescence in the cytoplasm to coalesce into intense large foci (Fig. 3A, frames ii and iii, arrowheads). More importantly, a clear and intense ring of NS3 staining associated with the nuclear periphery was now evident (Fig. 3A, frames ii and iii, arrows) in addition to its abundant presence in the cytoplasm. Careful observation under the microscope also revealed that the outer nuclear envelope in these cells had withdrawn from the rest of the nucleus, a feature that was difficult to capture and visualize clearly in the pictures.
FIG. 3.
Subcellular localization of the major replicase proteins NS3 and NS5 in JEV-infected cells. (A) Intracellular distribution of NS3 by indirect immunofluorescence using rabbit polyclonal antibodies to JEVNS3. Frame i shows the perinuclear and reticular staining of NS3 in infected cells while frames ii and iii show its localization after nocodazole treatment. Solid arrowheads point to the coalesced intense foci of NS3 signals and arrows indicate those present along the rim of the nucleus. Mock-infected cells processed and stained as described for frame i are shown in frame iv. (B) Intracellular distribution of NS5 by indirect immunofluorescence using rabbit polyclonal antibodies to JEVNS5. Frame i shows the cytoplasmic (arrows) as well as the intense and punctate perinuclear (solid arrowheads) staining pattern of JEVNS5 detected as described in Materials and Methods. Frame ii shows mock-infected cells processed and stained as in frame i and reveals nonspecific staining of nucleoli by the NS5-specific antiserum referred to in the Results section. The infected cells after appropriate processing and treatments were detected with anti-rabbit IgG conjugated to FITC. Appropriate scale bars are shown in each frame.
In order to unambiguously determine whether NS3 was indeed localized in the nucleus, immunofluorescence studies were carried out with sucrose-purified nuclei that were extracted with DD to solubilize and remove the ONEM and other associated membranes. Quantitative loss of calnexin from the detergent-extracted nuclei again confirmed the complete absence of any ER-derived membranes in the nuclei analyzed (Fig. 4B, compare frames i and ii). Calnexin signals were unaffected by JEV infection (data not shown). DD-treated nuclei, especially from virus-infected cells, were fragile and somewhat damaged, often with debris clinging to them. The rim of such nuclei stained positive for JEVNS3, clearly revealing the presence of NS3 along the periphery of host nuclei (Fig. 4B, frame iv) and corroborating the protein profile obtained above. The resolution obtained here, however, made it difficult to distinguish whether this signal was on the inner or outer side of the rim of DD-treated nuclei.
For the viral polymerase NS5, cytoplasmic signals were intense in the perinuclear regions as distinct foci (Fig. 3B, frame i, arrowheads) and less intense but spread out in the cytoplasm (Fig. 3B, frame i, arrows). The former would probably represent NS5 associated with the membrane-bound RC predominantly localized in the perinuclear region, while the latter most likely represents the fraction of NS5 we detected in the replication-incompetent S16 fraction (Fig. 1B). Importantly, we were able to detect JEVNS5 for the first time inside the nucleus with signals seen throughout the nucleoplasm (Fig. 4A, frame ii, and C, frame ii). While this staining was weak, its absence from mock-infected cells (Fig. 4A, frame iv) suggested that it represented the genuine presence of NS5 in the nucleoplasm. Our NS5-specific antiserum consistently stained nucleoli, including those from mock-infected cells, which led us to conclude that this was nonspecific (Fig. 3B, frame ii, and 4A, frame iv). Indirect immunofluorescence of DD-treated nuclei for NS5 revealed signals along the rim as well as diffuse signals throughout the nucleoplasm (Fig. 4A, frame ii, and C, frame ii). Again, nucleolar staining for NS5 in mock-infected cells revealed this to be nonspecific in nature (Fig. 4A, frame iv).
We also observed colocalization of the replicase proteins NS5 and NS3 within JEV-infected cells in both the cytoplasm and the perinuclear region (Fig. 4C, frame i). Dual labeling of DD-treated nuclei again demonstrated convincing colocalization of NS5 and NS3 only along the periphery of the nuclei, with nucleoplasm staining weakly for only NS5 (Fig. 4C, frame ii). To ascertain if this staining along the nuclear rim represented the inner nuclear membrane or a region within the nucleus just below the rim, we immunostained DD-treated nuclei with the monoclonal antibody LA2B3, which specifically detects the A-type lamins A and C in the peripheral nuclear lamina (38) (Fig. 5A, frame i), which is the filamentous network underlying the inner nuclear membrane. Colocalization of NS3 with lamin A/C was evident (Fig. 5A, frame iv), suggesting that the viral helicase was indeed present inside the nucleus, just below the inner membrane along the peripheral lamina.
FIG. 5.
JEV helicase NS3 and polymerase NS5 are present within nuclear matrix. (A) NS3 colocalizes with lamin A/C in DD-treated nuclei and within nuclear matrix preparations (C) from JEV-infected cells. Panel A shows the distribution of lamin A/C (i) and NS3 (ii) within nuclei by indirect immunofluorescence using mouse and rabbit antibodies followed by appropriate secondary reagents conjugated to FITC and TRITC, respectively. Frame iv shows colocalization of the two proteins. (B) Colocalization of JEVNS3 and JEVNS5 within nuclear matrix preparations of DD-treated nuclei. Panel B shows the distribution of NS3 (i) and NS5 (ii) and their colocalization (iv) in nuclear matrix preparations, using rabbit and mouse antibodies followed by secondary reagents conjugated to TRITC and FITC, respectively. (D) Western blot analysis of nuclear matrix (lanes 1 and 3) and P16 (lanes 2 and 4) fractions from JEV-infected (lanes 1 and 2) and noninfected (lanes 3 and 4) PS cells using anti-JEVNS3 and anti-lamin antibodies as described in Materials and Methods. The two A-type lamin species A and C detected by this antibody and the JEVNS3 are indicated by arrows. Molecular weight markers are indicated on the right.
To our surprise, we detected both NS3 and NS5 also in nuclear matrix preparations (Fig. 5B, frames i and ii, respectively) following digestion of DD-treated nuclei with DNase and salt extraction, which leaves behind only the core filaments. As shown earlier in Fig. 4C (frame ii), these viral replicase proteins colocalized with each other (Fig. 5B, frame iv). NS3 was again colocalized with lamin A/C in the nuclear matrix (Fig. 5C, frame iv) along the nuclear lamina, a location in such matrix preparations where these A-type lamins have been previously detected (38). The nuclei in these nuclear matrix preparations invariably appeared collapsed and damaged due to removal of their contents.
We further confirmed the presence of JEVNS3 in nuclear matrix preparations obtained from infected PS cells by Western blotting to NS3-specific serum. JEVNS3 was detected not only in P16 fractions from infected PS cells (Fig. 5D, lane 2), as expected, but also in nuclear matrix preparations (Fig. 5D, lane 1). The presence of the two characteristic nuclear lamin A and C proteins of 70 and 62 kDa, respectively (38), in the nuclear matrix alone but not in the P16 fractions of infected and control cells (Fig. 5D, lanes 1 and 3) was used to validate these fractions.
Viral polymerase complex proteins and freshly synthesized viral RNA colocalize along the rim of the nucleus.
In order to determine if the NS3 and NS5 proteins found along the rim of nuclei from infected cells were, in fact, involved in viral RNA synthesis, we next attempted to localize de novo synthesized biotinylated viral RNA in purified, DD-treated nuclei. We resorted to using biotinylated UTP rather than brominated UTP or bromouridine in order to enhance the signals associated with the nucleus. To rule out the possibility that the nucleus-localized flaviviral RNA was synthesized in the cytoplasm and then translocated to the nucleus, labeling was carried out for a short duration of 5 min. RNA synthesized de novo in purified and DD-extracted nuclei from JEV- and WNV-infected cells showed fluorescence that was localized to the periphery of the nucleus (Fig. 6A, frames i and v). Distinct and punctate regions of intense fluorescence were seen spread along the boundary of the nucleus. Control samples, which used flavivirus-infected nuclei in RdRp assays without biotinylated UTP, did not produce signals under identical conditions (Fig. 6A, frames iii and vii). No signals were visible in the interior parts or in the nucleoli (Fig. 6A, frames i and v). In addition, we also successfully demonstrated colocalization of the newly synthesized RNA present along the periphery of JEV-infected nuclei with JEVNS3 (Fig. 6B, frame iii). Similarly treated nuclei from mock-infected cells did not incorporate biotinylated UTP, proving the absence of cellular RNA synthesis following treatment with actinomycin D (Fig. 6B, frames v and vi). Optical sectioning along the z axis of the dually stained nuclei further confirmed the peripheral location of both RNA and JEVNS3 in the nuclei (Fig. 6C). Collectively, these results indicate that nuclear synthesis of viral RNA requires both polymerase and helicase functions of the viral RC.
FIG. 6.
Flaviviral RNA and helicase protein NS3 colocalize along the inner membrane of DD-treated nuclei. (A) Localization of nascent flaviviral RNA synthesis inside nuclei. Purified nuclei from JEV (frames i to iv) and WNV (frames v to viii) were subjected to an in vitro RdRp assay with (frames i and ii and frames v and vi) or without (frames iii and iv and frames vii and viii) biotin-16-UTP. The nuclei after the assay were treated with DD, and nascent biotinylated RNA was detected using streptavidin-FITC. Distinct and punctate foci of biotinylated RNA in the periphery of the nuclei were observed. (B) Dual labeling of nascent biotinylated RNA and JEVNS3. Nuclei were processed as described in panel A. Anti-rabbit IgG conjugated to FITC was used for immunolabeling JEVNS3 (i), and the RNA was detected with streptavidin-phycoerythrin (ii). Apparent coincidence of RNA and NS3 in merged images can be seen in frame iii. Frame v shows nuclei from mock-infected cells processed as described for frame i. Phase-contrast images are also shown (iv and vi). (C) Merged images of optical sections of the dually labeled nuclei treated as described for panel B show signals only along the rim, suggesting the localization of the active replicase to the inner nuclear membrane.
Coimmunoprecipitation of viral NS3, NS5, and nascent viral RNA.
We then attempted to demonstrate biochemically the functional interaction of the nucleus-localized viral polymerase complex proteins NS5 and NS3 with nascent viral RNA. We carried out radioimmunoprecipitation assays of purified DD-extracted nuclei isolated from JEV-infected cells metabolically labeled with [35S]methionine, with antisera to JEVNS3 and JEVNS5 after a 5-min labeling with [32P]UTP to label both viral proteins and nascent viral RNA (see Materials and Methods for details). Antiserum to green fluorescent protein (GFP) served as the control. The immunoprecipitates were either subjected to SDS-PAGE followed by fluorography to visualize the JEV replicase proteins (Fig. 7A) or processed for RNA (Fig. 7B). Antibodies to either NS3 or NS5 specifically coimmunoprecipitated the other protein (Fig. 7A, lanes 3 and 4) along with newly synthesized viral RNA (Fig. 7B, lanes 3 and 4), which was not seen with the control serum (Fig. 7A and B, lanes 2). These results unambiguously demonstrated the interaction of JEV NS3, NS5, and newly synthesized viral RNA. While the viral RNA was also labeled, it appears not to have survived the subsequent manipulations and exposure to serum samples that could not be rid of RNase contamination (Fig. 7B, compare lane 1 with lanes 3 and 4).
FIG. 7.
Coimmunoprecipitation of JEV replicase proteins NS3 and NS5 with nascent viral RNA from DD-treated nuclei. Sucrose-purified nuclei isolated from [35S]methionine-cysteine-labeled JEV-infected cells were incubated in RdRp assay buffer containing [32P]UTP for a brief period of 5 min to label nascent viral RNA. The nuclei were subsequently treated with DD to remove cytoplasmic membranes and solubilized prior to immunoprecipitation with either NS3-specific or NS5-specific polyclonal sera. (A) SDS-10% polyacrylamide gels of immunoprecipitates developed for fluorography as described in Materials and Methods. Antibodies to both NS3 (lane 3) and NS5 (lane 4) coprecipitate viral RNA and the other protein. The specificity of the reaction is confirmed by absence of signals using GFP-specific serum (lane 2). JEVNS1 is also precipitated by both specific sera. (B) Partially denaturing 3% polyacrylamide gel of RNA extracted from the immunoprecipitates. Newly synthesized viral RF is precipitated by both NS3-specific (lane 3) and NS5-specific (lane 4) sera but not by nonspecific GFP-specific serum (lane 2). While the labeled vRNA is clearly visible in the labeled RNA extracted from DD-treated nuclei (Con, lane 1), it appears to degrade upon exposure to sera.
Electron microscopy detects viral helicase protein and viral RNA within nuclei.
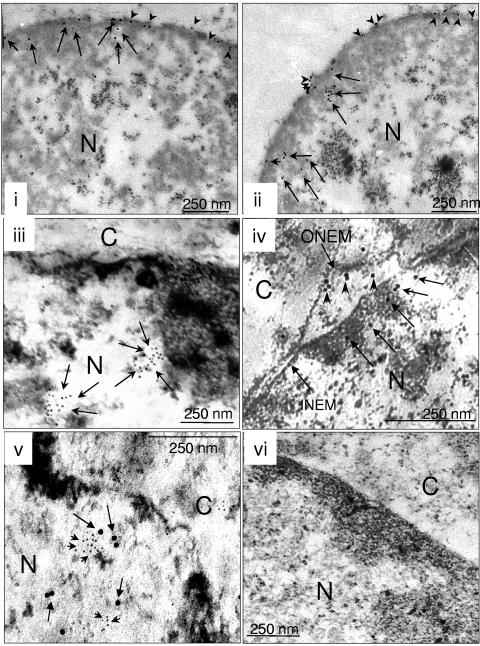
In order to conclusively demonstrate the nuclear localization of functional flaviviral replication complexes, we resorted to immunoelectron microscopy. In DD-treated nuclei from JEV-infected cells (Fig. 8, frames i and ii) and in intact JEV-infected cells (Fig. 8, frame iii), we observed a concentration of NS3 signal along the inner nuclear envelope membrane on both the inner and the outer surfaces of the nuclei (frames i and ii, arrowheads) as well as clusters of NS3 signals inside the nucleus but close to the periphery (frames i to iii, arrows), consistent with results from confocal microscopy. We also carried out electron microscopy of JEV-infected cells that were permeabilized for de novo incorporation of biotin-16-UTP in the presence of actinomycin D (see Materials and Methods). The biotinylated JEV RNA that was detected using streptavidin conjugated to 15-nm gold particles could be clearly seen within the nucleus and close to the periphery (Fig. 8, frame iv, arrows) in addition to the ONEM (Fig. 8, frame iv, arrowheads). Thus, RdRp activity appeared to localize only to those parts of nuclei that harbored both the replicase proteins NS3 and NS5. Colocalization of NS3 (Fig. 8, frame v, short arrows) and de novo biotinylated RNA (Fig. 8, frame v, long arrows) close to the periphery of nuclei of JEV-infected cells was also evident, with clusters of biotinylated RNA located so as to corroborate the punctate signals obtained in confocal analyses. The sensitivity of the JEVNS5-specific antiserum to the fixation conditions required for immunoelectron microscopy precluded its use in this analysis. Noninfected cells processed as above and probed with streptavidin gold along with anti-rabbit gold antibody showed no signals (Fig. 8, frame vi). Taken together, our results demonstrate for the first time that nuclei of infected cells represent a second site for the presence of functionally competent flaviviral RC, in addition to and distinct from host cytoplasmic membranes.
FIG. 8.
Immunoelectron microscopy detects NS3 and de novo synthesized RNA inside JEV-infected cells and purified DD-treated nuclei. Sucrose-purified nuclei obtained from JEV-infected PS cells and treated with DD (i and ii) and intact JEV-infected cells (iii) were processed for immunoelectron microscopy. Ultrathin sections were cut and probed with antibodies to JEVNS3 and detected with secondary antibody conjugated to 10-nm gold particles. The ONEMs were clearly absent in these nuclei. There was selective enrichment of JEVNS3 along both the outer and inner surface (frames i and ii, arrowheads) of the inner nuclear membrane. In addition, impressive clusters of NS3 signals were located close to the nuclear inner membrane but sufficiently removed from it inside the nucleus (frames i to iii, arrows). Actinomycin D-treated JEV-infected PS cells were permeabilized at 16 h postinfection with lysolecithin, treated with biotin-16-UTP, fixed, and processed as above (frames iv and v). Ultrathin sections were stained with streptavidin conjugated to 15-nm gold particles to detect de novo synthesized biotinylated RNA, which could be located both along the ONEM (iv, arrowheads) and also inside the nucleus as clusters (v, arrows). In frame v, similarly treated sections from JEV-infected cells were stained with both anti-NS3 antibodies (5-nm gold particles; short arrows) and RNA as above (15-nm gold particles; long arrows), revealing clusters of NS3 signals colocalized with biotinylated RNA. Frame vi shows ultrathin sections obtained from mock-infected cells and treated as described for frame v. N, nucleus; C, cytoplasm; INEM, inner nuclear envelope membrane. Bar, 250 nm.
DISCUSSION
The nucleus of the infected cell has been conventionally recognized as the abode of retroviruses, many DNA viruses, some negative-strand RNA viruses, and insect rhabdoviruses (53). Positive-strand RNA viruses such as flaviviruses have been considered to be cytoplasmic, since the major part of the viral life cycle, including translation, replication, and virion morphogenesis, has been believed to be confined exclusively to the host cytoplasm. Several positive- and negative-strand RNA viruses, which replicate in the cytosol, nevertheless interact with the host nucleus or its components. Avoiding host cell responses appears to be a major factor contributing to the successful outcome of a viral infection. Viruses achieve this by sequestering nuclear factors as well as by use of virally encoded nucleoproteins that alter host gene expression, thereby attenuating host responses that serve to counter viral infection (21). For example, the vesicular stomatitis virus M protein, which is vital for assembly and budding of the virus, inhibits export of U snRNA, rRNAs, and mRNAs across the nuclear envelope into the cytosol (20), while the polioviral 3C protease gains access to the nucleus and degrades basal transcription factors, shutting down the host transcription machinery (50). Thus, so far the role of the nucleus in the life cycle of positive-strand RNA viruses has been presumed to be one that aids viral replication in the cytoplasm. Specifically, the nucleus has not yet been reported to harbor active RC of positive-strand RNA viruses. In this report we offer conclusive evidence for a nucleus-localized replication component of flaviviruses.
Our data and those of others (39) clearly show that ∼60% of the RdRp activity is associated with cytoplasmic membranes. Out of the remaining RdRp activity, ∼20% was associated with membranes that formed the cytoplasmic extensions of the host ONEM in WNV-infected cells (16). The study by Grun et al., however, did not ascertain that the RdRp activity would remain with the nuclear pellet devoid of ONEM. Earlier studies carried out on JEV (45, 54), despite demonstrating ONEM-associated RdRp activity, lacked the validation of identity and purity of each membrane fraction as determined by using marker enzymes. Thus far, sufficient analyses have not been performed to locate, without ambiguity, the remaining viral RdRp activity. We demonstrate that the final 20% of RdRp activity for three different flaviviruses resides wholly within the nucleus, an observation reinforced by the lack of LDH, M6P activity, and calnexin in the detergent-treated nuclear fraction. A comparable proportion of the in vivo labeled total 3H-labeled KUNV RNA species has also been known to associate with nuclei that were DD treated to remove cytoplasmic membrane contamination (39).
We observed that DD treatment of WNV-infected nuclei extracted most of the RdRp activity from the nucleus as has been reported for KUNV (7), suggesting that the flaviviral RdRp is associated with membranes that could be solubilized by detergents and not with the nucleus. In contrast, however, when radiolabeling was performed prior to DD extraction, up to 50% of the radiolabeled viral RNA remained associated with nuclei. This clearly demonstrated that (i) RdRp activity exists within the nucleus and (ii) this activity is detergent sensitive. Investigation of the protein profile revealed extraction from the nuclear pellet of nearly all the WNVNS1-sized protein by the detergent (Fig. 2C, compare lanes 2 and 3). While the role for this extracted NS1 in the nuclear RdRp activity and, hence, nuclear viral RNA synthesis remains to be confirmed, one is tempted to hypothesize a role for NS1 in nuclear viral RNA synthesis, thereby ascribing the detergent-sensitive character of nucleus-associated RdRp activity to solubilization and subsequent loss of NS1 from the nuclear RC. NS1 was previously reported within the nuclei of flavivirus-infected cells (6), and a role for NS1 in negative-strand RNA synthesis has been proposed in YFV (34). Furthermore, our data also revealed coimmunoprecipitation of a 45-kDa protein, which is presumably NS1, by antibodies to NS3 or NS5 (Fig. 7), suggesting interaction of the nuclear NS1 with NS3, NS5, and nascent nuclear viral RNA.
Earlier investigators (45, 46) had also observed an association of JEVNS3 and JEVNS5, the viral replicase proteins, with the nuclear fraction by SDS-PAGE analyses similar to our demonstration in this report. Our analyses showed the presence of metabolically labeled NS3 in detergent-treated nuclei (Fig. 2C, lane 2), together with its location along the rim of the nuclei in nocodazole-treated JEV-infected cells. Analysis of detergent-treated nuclei by confocal as well as immunoelectron microscopy confirmed the association of NS3 with the nucleus. Furthermore, that this nuclear association was well entrenched and not superficial was revealed by the presence of both NS3 and NS5 in nuclear matrix preparations, where they colocalized with the peripheral lamina (Fig. 5). To the best of our knowledge, this is the first report demonstrating migration of a flaviviral NS3 into the nucleus. The mechanism by which NS3 gains entry into the nucleus can only be speculated upon. Proteins larger than 45 kDa require active transport into the nucleus (27), suggesting that the 68-kDa NS3 may be interacting with nuclear import factors or “piggybacking” into the nucleus using an interacting partner, as shown for the coronavirus N protein interacting with nucleolin (21). In this regard, a similar stretch of basic amino acid residues forming a putative bipartite NLS present in the linker region of DENV NS5 that was sufficient to confer nuclear location to a fused heterologous protein (12) can also be seen in JEVNS3 (Fig. 9, amino acid residues 185 to 203). In fact, another region of JEVNS3 (amino acids 381 to 400) contains a stretch of basic amino acid residues similar to those found in bipartite NLS from the yeast transcription factor SW15 and the human cytokine interleukin-5 (Fig. 9). It remains to be seen if these regions actually play a role in nuclear translocation of NS3. Interestingly, poliovirus 3C protease and the protease-helicase (nsP2) of the alphavirus Semliki Forest virus have both been shown to localize inside the nucleus (43). Hence, nuclear localization of this class of proteins is suggestive of a common strategy followed by positive-strand RNA viruses in modulating virus-host interactions. It should be noted, however, that while JEVNS3 localized solely to the rim of nuclei devoid of ONEM, Semliki Forest virus nsP2 was localized exclusively to nucleoli (43).
FIG. 9.
Comparison of known nuclear localization signals with two putative regions in JEVNS3 sequence. The sequence of amino acid residues 185 to 203 and 381 to 400 of JEVNS3 (GenBank accession number AFO82051) and the known bipartite NLS of nucleoplasmin (NPM) (26), the yeast transcription factor SW15 (27), human retinoblastoma protein (HuRb) (10), the cytokine interleukin-5 (HuIL-5) (25), and DENV NS5 (12) are shown. The important basic residues are in bold type, and the regions of similarity are boxed. The numbers on the right represent positions of the last amino acids depicted in their respective proteins.
It was intriguing that the WNV envelope protein was found in the nuclear fraction even after extraction with DD (Fig. 2C). The reproducible association of the E protein with the DD-treated nuclear pellet that we observed corroborates earlier documentation of this phenomenon in JEV- and WNV-infected cells (17, 31). However, in contrast to E of YFV, which could be coimmunoprecipitated by monoclonal antibody GJL-1, specific to NS5, from detergent-extracted nuclei of infected cells (5), JEV envelope protein did not coimmunoprecipitate with the replicase proteins NS3 and NS5 and RNA (Fig. 7). It is to be emphasized here that we have used three cellular markers—LDH, M6P, and the ER-resident protein calnexin—to prove beyond doubt that the nuclei are free of any cytoplasmic contamination. The presence of flaviviral structural proteins E (this study) and capsid (37, 51) in nuclei points to a need to reevaluate our understanding of the role of nuclear localization of viral structural proteins, especially in light of the demonstrated alteration in virulence and neuroinvasiveness of JEV carrying a mutant capsid with a nuclear localization defect (37).
The NS5 protein of JEV has so far not been visualized in the nucleus of infected cells despite sufficient evidence for its association with the nucleus from protein profile analyses of nuclear fractions (45). Despite the presence of NS5 throughout the nucleoplasm and nuclear periphery, only that localized in the periphery along with NS3 appeared to be involved in viral RNA synthesis, as evidenced by the localization of nascent RNA and the coimmunoprecipitation of these two proteins with nascent viral RNA. Thus, despite the demonstrated proficiency of recombinant purified DENV NS5 in specific viral negative-strand RNA synthesis in vitro (1), in vivo synthesis of viral RNA from endogenous templates in nuclei of JEV-infected cells appears to have an absolute requirement for both NS5 and NS3. The exact role of the fraction of nuclear NS5 not colocalizing with NS3 is presently unclear, but it is possible that it may influence the host response to viral infections, as has been proposed for many viral proteins that localize to the nucleus (21).
The presence of viral RNA inside the nucleus has also been demonstrated in hepatitis C virus-infected hepatocytes by in situ hybridization of infected liver tissues (19, 41), although proof for their in situ generation within nuclei is lacking. Our analyses with purified nuclear preparations combined with short labeling periods, however, clearly demonstrated the presence of nascent viral RNA inside the nucleus and conclusively proved the existence of an active nuclear component of the flaviviral RC. While phosphorylation of NS5 influences its interaction with NS3 in dengue virus, this aspect may not influence the nuclear synthesis of viral RNA in flaviviruses, in light of our earlier demonstration that extremely small proportions of viral RC proteins suffice for viral RNA synthesis (49).
While our results beg for an explanation of the significance of nucleus-associated flaviviral RNA synthesis, any discussion on the unique purpose of the nucleus-localized RNA can at present only be speculative. However, in light of our previous demonstration of the need for the newly synthesized vRNA from cytoplasmic membrane-bound flaviviral RC to traverse a nonionic detergent-sensitive membrane for translation and viral morphogenesis (48), the nucleus-localized flaviviral RC could probably supply free viral RNA for these processes. It is pertinent to mention here that treatment of Dengue virus-infected cells with leptomycin B, a fungal metabolite that inhibits export of proteins from nucleus to cytoplasm, resulted in a log increase in virus titer (David Jans, Monash University, Australia, personal communication). The demonstration that a single amino acid change in the NLS of the alphaviral replicase component nsP2 protein affects the neurovirulence of Semliki Forest virus (11) has helped to validate the critical role played by nucleus-localized viral proteins in determining the outcome and severity of the infection process. In the extensive mutagenesis studies of DENV type 4 NS5 (18), it was mutants in the NLS and importin-β-binding domain of DENV type 4 NS5 more than those in the polymerase domain that drastically reduced viral titers in cultured cells. These mutants also displayed a striking reduction of mouse neurovirulence. We might therefore reasonably speculate that translocation of NS5 into the nucleus is essential to ensure viral fitness and virulence.
Our finding also impacts efforts to develop therapeutic agents against flaviviruses that are currently under way in several laboratories worldwide. The reported inability of RNA interference to target flavivirus RNA synthesis unless provided intracellularly prior to viral infection (2, 13) may very well be related to our earlier demonstration of the lack of accessibility for nucleases and proteases to the membrane-bound flaviviral RC (48). The tight association of viral replicase proteins with the nuclear matrix lends credence to the possibility that these proteins might deftly influence host gene expression in their favor within infected cells, further complicating our attempts and ability to control these viruses using anti-infective agents. Analysis of the architecture of nucleus-resident RC may help shed more light on additional aspects of flaviviral RNA synthesis, which in turn ought to not only enhance our understanding of virus-host interactions but also have far-reaching implications for developing antiviral therapeutic intervention strategies.
Acknowledgments
We gratefully acknowledge the gift of the LA2B3 monoclonal antibody from Veena Parnaik. We thank Priti Kumar for constant help and valuable discussions throughout the course of this investigation. Brijesh Narayan Bhatt is gratefully acknowledged for enthusiastic help with the colocalization of viral replicase proteins and nuclear lamin. We thank Akila Chandrashaker for her skilled assistance and knowledgeable contribution to the use of the confocal laser scanning microscope facility of the Indian Institute of Science. We also thank the staff of the electron microscope facility of the Department of Microbiology and Cell Biology. We thank Mridula Nandan and K. S. Ananda for excellent technical assistance.
This work was funded by a grant (SP/SO/BB-02/2000) from the Department of Science and Technology, Government of India.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Bai, F., T. Wang, U. Pal, F. Bao, L. Gould, and E. Fikrig. 2005. Apr 1. Use of RNA interference to prevent lethal murine West Nile virus infection. J. Infect. Dis. 191:1148-1154. [DOI] [PubMed] [Google Scholar]
- 3.Brawner, I. A., M. D. Trousdale, and D. W. Trent. 1979. Cellular localization of Saint Louis encephalitis virus replication. Acta Virol. 23:284-294. [PubMed] [Google Scholar]
- 4.Brooks, A. J., M. Johansson, A. V. John, Y. Xu, D. A. Jans, and S. G. Vasudevan. 2002. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin beta 1 and importin alpha/beta-recognized nuclear localization signals. J. Biol. Chem. 277:36399-36407. [DOI] [PubMed] [Google Scholar]
- 5.Buckley, A., S. Gaidamovich, A. Turchinskaya, and E. A. Gould. 1992. Monoclonal antibodies identify the NS5 yellow fever virus non-structural protein in the nuclei of infected cells. J. Gen. Virol. 73:1125-1130. [DOI] [PubMed] [Google Scholar]
- 6.Buckley, A., and E. A. Gould. 1988. Detection of virus-specific antigen in the nuclei or nucleoli of cells infected with Zika or Langat virus. J. Gen. Virol. 69:1913-1920. [DOI] [PubMed] [Google Scholar]
- 7.Chu, P. W., and E. G. Westaway. 1992. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch. Virol. 125:177-191. [DOI] [PubMed] [Google Scholar]
- 8.Chu, P. W., and E. G. Westaway. 1985. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology 140:68-79. [DOI] [PubMed] [Google Scholar]
- 9.Cleaves, G. R., T. E. Ryan, and R. W. Schlesinger. 1981. Identification and characterization of type 2 dengue virus replicative intermediate and replicative form RNAs. Virology 111:73-83. [DOI] [PubMed] [Google Scholar]
- 10.Efthymiadis, A., H. Shao, S. Hubner, and D. A. Jans. 1997. Kinetic characterization of the human retinoblastoma protein bipartite nuclear localization sequence (NLS) in vivo and in vitro. A comparison with the SV40 large T-antigen NLS. J. Biol. Chem. 272:22134-22139. [DOI] [PubMed] [Google Scholar]
- 11.Fazakerley, J. K., A. Boyd, M. L. Mikkola, and L. Kaariainen. 2002. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J. Virol. 76:392-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forwood, J. K., A. Brooks, L. J. Briggs, C. Y. Xiao, D. A. Jans, and S. G. Vasudevan. 1999. The 37-amino-acid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site. Biochem. Biophys. Res. Commun. 257:731-737. [DOI] [PubMed] [Google Scholar]
- 13.Geiss, B., T. Pierson, and M. Diamond. 2005. Actively replicating West Nile virus is resistant to cytoplasmic delivery of siRNA. Virol. J. 2:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilchrist, J. S., and G. N. Pierce. 1993. Identification and purification of a calcium-binding protein in hepatic nuclear membranes. J. Biol. Chem. 268:4291-4299. [PubMed] [Google Scholar]
- 15.Grun, J. B., and M. A. Brinton. 1987. Dissociation of NS5 from cell fractions containing West Nile virus-specific polymerase activity. J. Virol. 61:3641-3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grun, J. B., and M. A. Brinton. 1988. Separation of functional West Nile virus replication complexes from intracellular membrane fragments. J. Gen. Virol. 69:3121-3127. [DOI] [PubMed] [Google Scholar]
- 17.Gupta, A. K., M. M. Gore, V. J. Lad, and S. N. Ghosh. 1991. Nuclear immunofluorescence in porcine kidney cells infected with Japanese encephalitis virus. Acta Virol. 35:282-286. [PubMed] [Google Scholar]
- 18.Hanley, K. A., J. J. Lee, J. E. Blaney, Jr., B. R. Murphy, and S. S. Whitehead. 2002. Paired charge-to-alanine mutagenesis of dengue virus type 4 NS5 generates mutants with temperature-sensitive, host range, and mouse attenuation phenotypes. J. Virol. 76:525-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haruna, Y., N. Hayashi, N. Hiramatsu, T. Takehara, H. Hagiwara, Y. Sasaki, A. Kasahara, H. Fusamoto, and T. Kamada. 1993. Detection of hepatitis C virus RNA in liver tissues by an in situ hybridization technique. J. Hepatol. 18:96-100. [DOI] [PubMed] [Google Scholar]
- 20.Her, L. S., E. Lund, and J. E. Dahlberg. 1997. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science 276:1845-1848. [DOI] [PubMed] [Google Scholar]
- 21.Hiscox, J. A. 2003. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res. 95:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holtzman, E., I. Smith, and S. Penman. 1966. Electron microscopic studies of detergent-treated HeLa cell nuclei. J. Mol. Biol. 17:131-135. [DOI] [PubMed] [Google Scholar]
- 23.Humbert, J.-P., N. Matter, J.-C. Artault, P. Köppler, and A. N. Malviya. 1996. Inositol 1,4,5-trisphosphate receptor is located to the inner Nuclear membrane vindicating regulation of nuclear calcium signaling by inositol 1,4,5-trisphosphate. J. Biol. Chem. 271:478-485. [DOI] [PubMed] [Google Scholar]
- 24.Hunsperger, E., and J. T. Roehrig. 2005. Characterization of West Nile viral replication and maturation in peripheral neurons in culture. J. Neurovirol. 11:11-22. [DOI] [PubMed] [Google Scholar]
- 25.Jans, D. A., L. J. Briggs, S. E. Gustin, P. Jans, S. Ford, and I. G. Young. 1997. A functional bipartite nuclear localisation signal in the cytokine interleukin-5. FEBS Lett. 406:315-320. [DOI] [PubMed] [Google Scholar]
- 26.Jans, D. A., and P. Jans. 1994. Negative charge at the casein kinase II site flanking the nuclear localization signal of the SV40 large T-antigen is mechanistically important for enhanced nuclear import. Oncogene 9:2961-2968. [PubMed] [Google Scholar]
- 27.Jans, D. A., T. Moll, K. Nasmyth, and P. Jans. 1995. Cyclin-dependent kinase site-regulated signal-dependent nuclear localization of the SW15 yeast transcription factor in mammalian cells. J. Biol. Chem. 270:17064-17067. [DOI] [PubMed] [Google Scholar]
- 28.Kapoor, M., L. Zhang, M. Ramachandra, J. Kusukawa, K. E. Ebner, and R. Padmanabhan. 1995. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 270:19100-19106. [DOI] [PubMed] [Google Scholar]
- 29.Kos, K. A., B. A. Osborne, and R. A. Goldsby. 1975. Inhibition of group B arbovirus antigen production and replication in cells enucleated with cytochalasin B. J. Virol. 15:913-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, P., P. D. Uchil, P. Sulochana, G. Nirmala, R. Chandrashekar, M. Haridattatreya, and V. Satchidanandam. 2003. Screening for T cell-eliciting proteins of Japanese encephalitis virus in a healthy JE-endemic human cohort using recombinant baculovirus-infected insect cell preparations. Arch. Virol. 148:1569-1591. [DOI] [PubMed] [Google Scholar]
- 31.Lad, V. J., A. K. Gupta, S. N. Ghosh, and K. Banerjee. 1993. Immunofluorescence studies on the replication of some arboviruses in nucleated and enucleated cells. Acta Virol. 37:79-83. [PubMed] [Google Scholar]
- 32.Lanzetta, P. A., L. J. Alvarez, P. S. Reinach, and O. A. Candia. 1979. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100:95-97. [DOI] [PubMed] [Google Scholar]
- 33.Leibowitz, J. L., and J. R. DeVries. 1988. Synthesis of virus-specific RNA in permeabilized murine coronavirus-infected cells. Virology 166:66-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindenbach, B. D., and C. M. Rice. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchison, T. J., and M. W. Kirschner. 1986. Isolation of mammalian centrosomes. Methods Enzymol. 134:261-268. [DOI] [PubMed] [Google Scholar]
- 37.Mori, Y., T. Okabayashi, T. Yamashita, Z. Zhao, T. Wakita, K. Yasui, F. Hasebe, M. Tadano, E. Konishi, K. Moriishi, and Y. Matsuura. 2005. Nuclear localization of Japanese encephalitis virus core protein enhances viral replication. J. Virol. 79:3448-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muralikrishna, B., S. Thanumalayan, G. Jagatheesan, N. Rangaraj, A. A. Karande, and V. K. Parnaik. 2004. Immunolocalization of detergent-susceptible nucleoplasmic lamin A/C foci by a novel monoclonal antibody. J. Cell Biochem. 91:730-739. [DOI] [PubMed] [Google Scholar]
- 39.Ng, M. L., J. S. Pedersen, B. H. Toh, and E. G. Westaway. 1983. Immunofluorescent sites in Vero cells infected with the flavivirus Kunjin. Arch. Virol. 78:177-190. [DOI] [PubMed] [Google Scholar]
- 40.Nickerson, J. A., G. Krockmalnic, K. M. Wan, C. D. Turner, and S. Penman. 1992. A normally masked nuclear matrix antigen that appears at mitosis on cytoskeleton filaments adjoining chromosomes, centrioles, and midbodies. J. Cell Biol. 116:977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nouri Aria, K. T., R. Sallie, D. Sangar, G. J. Alexander, H. Smith, J. Byrne, B. Portmann, A. L. Eddleston, and R. Williams. 1993. Detection of genomic and intermediate replicative strands of hepatitis C virus in liver tissue by in situ hybridization. J. Clin. Investig. 91:2226-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penman, S. 1966. RNA metabolism in the HeLa cell nucleus. J. Mol. Biol. 17:117-130. [DOI] [PubMed] [Google Scholar]
- 43.Peranen J., M. Rikkonen, P. Liljestrom, and L. Kaariainen. 1990. Nuclear localization of Semliki Forest virus-specific nonstructural protein nsP2. J. Virol. 64:1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 45.Takeda, H., A. Oya, K. Hashimoto, T. Yasuda, and M. A. Yamada. 1978. Association of virus specific replicative ribonucleic acid with nuclear membrane in chick embryo cells infected with Japanese encephalitis virus. J. Gen. Virol. 38:281-291. [DOI] [PubMed] [Google Scholar]
- 46.Takegami, T., and S. Hotta. 1990. Synthesis and localization of Japanese encephalitis virus RNAs in the infected cells. Microbiol. Immunol. 34:849-857. [DOI] [PubMed] [Google Scholar]
- 47.Tata, J. R., M. J. Hamilton, and R. D. Cole. 1972. Membrane phospholipids associated with nuclei and chromatin: melting profile, template activity and stability of chromatin. J. Mol. Biol. 67:231-246. [DOI] [PubMed] [Google Scholar]
- 48.Uchil, P. D., and V. Satchidanandam. 2003. Architecture of the flaviviral replication complex: protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 278:24388-24398. [DOI] [PubMed] [Google Scholar]
- 49.Uchil, P. D., and V. Satchidanandam. 2003. Characterization of RNA synthesis, replication mechanism, and in vitro RNA-dependent RNA polymerase activity of Japanese encephalitis virus. Virology 307:358-371. [DOI] [PubMed] [Google Scholar]
- 50.Weidman M. K., R. Sharma, S. Raychaudhuri, P. Kundu, W. Tsai, and A. Dasgupta 2003. The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res. 95:75-85. [DOI] [PubMed] [Google Scholar]
- 51.Westaway, E. G., A. A. Khromykh, M. T. Kenney, J. M. Mackenzie, and M. K. Jones. 1997. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology 234:31-41. [DOI] [PubMed] [Google Scholar]
- 52.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whittaker, G. R., M. Kann, and A. Helenius. 2000. Viral entry into the nucleus. Annu. Rev. Cell Dev. Biol. 16:627-651. [DOI] [PubMed] [Google Scholar]
- 54.Zebovitz, E., J. K. Leong, and S. C. Doughty. 1974. Involvement of the host cell nuclear envelope membranes in the replication of Japanese encephalitis virus. Infect. Immun. 10:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]