Abstract
The packaging of viral genomic RNA into nucleocapsids and subsequently into virions is not completely understood. Phosphoprotein (P) and nucleoprotein (NP) interactions link NP-RNA complexes with P-L (polymerase) complexes to form viral nucleocapsids. The nucleocapsid then interacts with the viral matrix protein, leading to specific packaging of the nucleocapsid into the virion. A mammalian two-hybrid assay and confocal microscopy were used to demonstrate that Ebola virus VP35 and VP40 interact and colocalize in transfected cells. VP35 was packaged into budding virus-like particles (VLPs) as observed by protease protection assays. Moreover, VP40 and VP35 were sufficient for packaging an Ebola virus minignome RNA into VLPs. Results from immunoprecipitation-reverse transcriptase PCR experiments suggest that VP35 confers specificity of the nucleocapsid for viral genomic RNA by direct VP35-RNA interactions.
The Ebola (EBOV) and Marburg viruses are members of the family Filoviridae of the order Mononegavirales. Both viruses are associated with recurrent outbreaks of deadly hemorrhagic fevers with mortality rates as high as 90% (25, 26). Currently, there are no approved vaccines or treatments for Ebola virus infection. A better understanding of the molecular aspects of the Ebola virus life cycle is paramount for developing specific treatments and vaccines for Ebola virus infection.
The Ebola virus genome is 19 kb in length and includes seven genes coding for eight proteins: nucleoprotein (NP), VP40, phosphoprotein (VP35), VP24, VP30, glycoprotein (GP), and the large protein (polymerase [L]). The GP gene has an alternate transcription editing site leading to production of a soluble form of GP, sGP (29). Like those of other members of the Mononegavirales, the Ebola virus nucleocapsid is composed of NP, VP35, L, and the viral RNA. The Ebola virus nucleocapsid also contains an accessory protein, VP24, whose function remains uncertain (9, 30). The nucleocapsid is associated with viral replication and transcription and is likely packaged within virions by interactions with the VP40 matrix protein (20, 21).
Since Ebola virus is a biosafety level 4 agent, direct studies of viral processes is cumbersome. The use of expression plasmids to study individual viral proteins and their biological effects has provided a wealth of information about the virus life cycle. For example, Muhlberger et al. developed a system to study replication and transcription using a 3E-5E minigenome (20, 21). The 3E-5E minigenome is composed of the complete leader and trailer sequences of the EBOV genome, as well as a chloramphenicol acetyltransferase (CAT) reporter gene in the genome sense orientation. Muhlberger et al. have demonstrated that the 3E-5E minigenome is transcribed by viral proteins to yield CAT mRNAs. Packaging of an Ebola virus-derived RNA has been accomplished by using a derivative of the 3E-5E minigenome. For example, Watanabe et al. were able to package a green fluorescent protein minigenome following coexpression of virus nucleocapsid proteins (VP35, NP, and L), VP40, and GP. They were able to infect recipient cells that had been transfected previously with the viral proteins VP35, NP, and L, which are necessary for transcription and replication (30). Similar experiments by Groseth et al. using an RNA polymerase I-driven replicon also demonstrated that the 3E-5E minigenome could be packaged (8). These studies confirmed that the 3E-5E minigenome contained the cis-acting signals necessary for transcription, replication, and packaging.
VP40 is the major virion protein and plays a fundamental role in virus assembly and budding. VP40 buds from the cell surface in the form of virus-like particles (VLPs). Budding is mediated by L domains present at the N terminus of VP40, which interact with specific host factors (11, 14, 23, 24). VP40 has been shown to form octameric ringlike structures, and these octameric structures have RNA binding properties (7, 28). Ablation of the octameric rings did not disrupt VLP budding but did impair replication (12). Recent evidence from our laboratory and subsequent evidence from Kallstrom et al. and Licata et al. suggest that GP and NP enhance VP40 VLP budding (15, 16). Although interactions between VP40 and the nucleocapsid are poorly understood, it is likely that NP-VP40 interactions are important for incorporation of the nucleocapsid into the assembling virion, as has been reported for human parainfluenza virus (6).
Less is known about VP35's role in the virus life cycle, although VP35 has been implicated in regulating the interferon response to Ebola virus infection (2, 3, 10). In addition to its role in modulating the cell immune response, VP35 plays a role in the formation of the viral nucleocapsid (13). VP35 is necessary for viral replication and transcription (20, 21); however, the mechanism by which VP35 functions in nucleocapsid assembly, replication, and transcription is currently unknown. Presumably, VP35 confers specificity of the nucleocapsid protein for the viral RNA, as has been predicted for other negative-sense RNA viruses (4, 19, 32).
We used a transfection-based system to identify the minimal set of viral proteins required for packaging the 3E-5E minigenome. Our results indicate that VP35 and VP40 interact and colocalize in transfected cells and that these two proteins are sufficient to package the 3E-5E minigenome RNA into VLPs.
MATERIALS AND METHODS
Cells.
Vero, Cos-1, and 293T cells were maintained at 37°C and 5% CO2 in Dulbecco minimal essential medium (Mediatech) supplemented with 10% fetal calf serum (Invitrogen) and 1× penicillin-streptomycin (Invitrogen).
Plasmids and antibodies.
Plasmids pCAGGs VP40 and pCAGGs NP have been described previously (9, 16, 17). The N-terminal VP35 pCAGGs construct and VP35 monoclonal antibody were kindly provided by Chris Basler (3). The pCAGGSVP35 construct contains an influenza virus hemagglutinin (HA) epitope tag at the amino terminus of VP35. VP40 antibodies (kindly provided by Roland Grunow) and HA antibodies (Roche) were used as previously described (9, 17).
Mammalian two-hybrid assays.
VP40 and VP35 were inserted into activation and binding domain vectors supplied with the Mammalian 2-Hybrid Assay kit (Stratagene) using standard procedures. The resultant plasmids were sequenced, and the chimeric viral proteins were immunoprecipitated with anti-VP35 and anti-VP40 antibodies. 293T cells were transfected with 200 ng of ADVP40, BDVP40, ADVP35, BDVP35, BDp53 (negative control plasmid), ADTRAF (negative control plasmid), and pLUC (reporter plasmid) using Lipofectamine (Invitrogen) in triplicate in 24-well tissue culture dishes. Forty-eight hours posttransfection, the cells were washed twice with phosphate-buffered saline (PBS) and lysed in the RLB buffer supplied with the Luciferase Assay System (Promega). The lysates were centrifuged briefly, and the supernatants were loaded into a 96-well plate. An equal volume of freshly prepared luciferase assay substrate (Promega) was added to each lysate. The relative intensity of each sample was measured using a luminometer (Marathon 3200R; Fisher Scientific).
VLP budding assays.
A functional budding assay for VP40 has been described previously (16, 17). Briefly, 293T cells were transfected with 500 ng of the indicated plasmids using Lipofectamine (Invitrogen). For experiments where VP40 budding efficiency was measured in the presence of the 3E-5E minigenome, cells were transfected with 1 μg 3E-5E minigenome 18 h posttransfection of plasmid DNA. Following the RNA transfection, the cells were metabolically labeled with [35S]Met-Cys (Perkin-Elmer; 100 μCi/ml) for 6 h. The culture medium was harvested and clarified in a microcentrifuge at 3,000 rpm for 10 min at 4°C. The supernatant was then layered onto a 20% sucrose cushion in STE buffer (0.01 M Tris-HCl [pH 7.5], 0.01 M NaCl, 0.001 M EDTA [pH 8.0]) and centrifuged for 2 h at 36,000 rpm to isolate VLPs. The VLPs were resuspended in STE buffer. RIPA buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1.0% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) was used to lyse the cells and purified VLPs. Proteins associated with cell extracts and VLPs were immunoprecipitated with appropriate antibodies and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were visualized by autoradiography and quantified by phosphorimager analysis.
Protease protection assay.
VLP budding assays were performed as described previously (14, 17, 31) with minor modifications as follows. 293T cells on 100-mm tissue culture dishes (Corning) were transfected with 10 μg of pCAGGs, VP35, VP40, or VP35 plus VP40 plasmids using Lipofectamine (Invitrogen). [35S]Met-Cys (1,600 μCi; Perkin-Elmer) was added to each 100-mm dish 24 h posttransfection. At 30 hours posttransfection, the culture medium was clarified at 1,500 rpm for 10 min and layered over 20% sucrose in an STE cushion, and the VLPs were purified at 36,000 rpm for 2 h at 4°C. The VLPs were suspended in 400 μl of STE buffer and divided into six 60-μl fractions. The fractions were treated with either 6 μl of STE buffer, 3 mg/ml soybean trypsin inhibitor (Roche Biochemicals), 1% Triton X-100 (Fisher), 0.1 mg/ml trypsin (Promega), 1% Triton X-100, and 0.1 mg/ml trypsin or 0.1 mg/ml trypsin and 3 mg/ml trypsin inhibitor. All fractions were incubated at room temperature for 30 min, followed by the addition of 25 μl of soybean trypsin inhibitor at a concentration of 100 mg/ml to quench the reactions. Each fraction was lysed in RIPA buffer in a final volume of 500 μl. Proteins were immunoprecipitated with appropriate antibodies and resolved by SDS-PAGE. As a control for protein expression, the transfected cells were lysed in RIPA buffer and split into equal fractions. Immunoprecipitated proteins were resolved by SDS-PAGE and visualized by autoradiography.
IP-RT-PCR.
Immunoprecipitation (IP)-reverse transcriptase (RT) PCR was carried out essentially as described by Nanda et al. (22). 293T cells were transfected with 500 ng of pCAGGs, pCAGGsVP40, pCAGGsVP35, pCAGGsNP, pCAGGSVP40 plus pCAGGsVP35, or pCAGGsVP35 plus pCAGGsNP, followed by 1 microgram of 3E-5E minigenome RNA transfection 18 h posttransfection of DNA plasmids. The cells were incubated in hypotonic buffer (10 mM HEPES [pH 7.6], 1.5 mM MgCl2, 10 mM KCl, 0.2 mM phenylmethylsulfonyl fluoride) and incubated on ice for 5 min. The cells were lysed with NP-40 at a final concentration of 0.5%, vortexed, incubated on ice for an additional 10 min, vortexed, and centrifuged at 4,000 × g at 4°C for 15 min, and glycerol was added to a final concentration of 5% (33). Two hundred micrograms of each cell lysate was incubated with 50 μl protein G-agarose (Invitrogen) for 30 min at room temperature. The cell lysates were removed from the protein G-agarose, VP40, and HA; NP antibodies were added to a dilution of 1:500; and samples were incubated for an additional 1 h at room temperature. Fifty microliters of protein G-agarose (Invitrogen) per sample was washed three times in 0.1% bovine serum albumin in PBS, pH 7.6; prepared in diethyl pyrocarbonate (DEPC)-treated water; and added to the cell lysate. After 1 hour of incubation with protein G-agarose beads, the beads were washed three times with wash buffer (10 mM Tris [pH 7.6], 100 mM KCl, 5 mM MgCl2, and 1 mM dithiothreitol) and eluted in 100 μl 50 mM Tris (pH 8.0), 1% SDS, and 10 mM EDTA at 65°C for 10 min. Immunocomplexes were treated with 100 μg proteinase K at 37°C for 30 min, followed by two rounds each of phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform extraction, and precipitated by ethanol with 10 μg glycogen. RNA was purified by centrifugation for 30 min at 13,000 × g, washed with 70% ethanol, and suspended in 5 mM Tris (pH 8.0). RT-PCR was carried out using primer 3E 3′ leader (5′GGACACAAAAACAAG3′) and Superscript II reverse transcriptase (Invitrogen), followed by PCR with Taq DNA polymerase (Invitrogen). Thirty-five cycles with primers 3E 3′ leader and 3E 5′ trailer (5′GTGGACACACAAAAAAG3′) were carried out under the following conditions: 95°C for 30 s, 45°C for 30s, and 68°C for 1 min, followed by 10 min at 68°C final extension, and the PCR products were resolved on 1% agarose gels stained with ethidium bromide. As a control, the purified RNA samples were used in RT-PCRs with primers specific for the cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) RNA. Primer GAPDH 3′ (5′GTAGAGGCAGGGATGATGTTC3′) was used as the RT primer and 3′ PCR primer, while primer GAPDH 5′ (5′GCCAAAAGGGTCATCATCTC3′) was used as the 5′ primer in PCRs. Western blot analysis was performed on a small aliquot of each cell lysate to ensure that viral proteins were produced.
Confocal microscopy.
Cos-1 cells were grown on coverslips and transfected with pCAGGs, VP35, VP40, or VP35 plus VP40 plasmids as described above. The cells were washed twice with PBS and fixed with 1:1 acetone-methanol at 30 h posttransfection. The coverslips were incubated with rat anti-HA and mouse anti-VP40 antibodies for 30 min at room temperature, washed twice with PBS, incubated with Alexafluor anti-rat 488 (Invitrogen/Molecular Probes) and Alexafluor anti-mouse 594 (Invitrogen/Molecular Probes), washed twice, and affixed to slides with Prolong antifade (Invitrogen/Molecular Probes). Confocal images were obtained using a Zeiss LSM-510 Meta confocal microscope.
Three-channel confocal microscopy was used to visualize RNA-VP35-VP40 complexes. Vero cells grown on coverslips were transfected with VP35, VP40, or VP35 plus VP40. As a negative control, the antisense CAT gene was excised from the 3E-5E minigenome using enzymes NotI and NdeI, inserted into the pGEMT vector, and used as a transcription template. One microgram (each) of total 293T, 3E-5E, antisense CAT (derived from the 3E-5E minigenome), and pTRI-Xef RNAs was labeled using the ULYSIS Alexafluor 488 nucleic acid labeling kit (Molecular Probes/Invitrogen), and 1 μg of RNA per sample was used in all double-transfection experiments. The Alexafluor 488-labeled RNA was transfected 18 h after the transfection of DNA plasmids. Vero cells were washed twice with PBS and fixed with acetone-methanol (1:1). Samples were incubated with anti-rat HA and anti-mouse VP40 antibodies for 30 min at room temperature, washed twice with PBS, and incubated with Alexafluor anti-rat 647 (Invitrogen/Molecular Probes) and Alexafluor anti-mouse 568 (Invitrogen/Molecular Probes). Samples were analyzed using a Bio-Rad 1024 ES standard confocal microscope.
3E-5E minigenome packaging.
Elke Muhlberger kindly provided the 3E-5E Ebola virus minigenome construct (20, 21). The 3E-5E minigenome plasmid was linearized with RsrII (New England Biolabs), phenol-chloroform (Sigma) extracted, and used as a template in transcription reactions using the Megascript T7 Transcription kit (Ambion). RNA was purified from transcription reactions with the MEGAclear kit (Ambion) and quantified by spectrophotometry. One microgram RNA per sample was used in all double-transfection experiments.
In experiments investigating the packaging of the 3E-5E minigenome, the transfection procedures were identical to those used for quantification of VP35 and 3E-5E effects on VP40 VLP budding, except that the pellets from the 20% sucrose cushion were resuspended in buffer RLT (QIAGEN) and purified using QIAGEN′s RNAeasy kit. Cell culture monolayers were washed twice with PBS, resuspended in 1 ml of PBS, divided into two equal portions, centrifuged, and stored at −70°C for future analysis by Western blotting and RT-PCR. RT-PCR was carried out using the primers Ebola 3′leader (5′GGACACAAAAACAAG3′) and Ebola 5′trailer (5′GTGGACACACAAAAAAG3′). These primers correspond to the extreme 3′ and 5′ ends of the 3E-5E minigenome. Reverse transcription was accomplished with the Ebola 3′leader primer using Superscript II RT (Invitrogen) following the manufacturer's recommended protocol. cDNAs were amplified through 35 cycles of PCR under the following conditions: 95°C for 30 s, 45°C for 30 s, and 68°C for 1 min, followed by 10 min at 68°C final extension. Controls excluding RT were performed to ensure that only 3E-5E minigenome-derived cDNAs were amplified. GAPDH was used as a control for the specificity of 3E-5E packaging. Primer GAPDH 3′ (5′GTAGAGGCAGGGATGATGTTC) was used as the RT primer and 3′ PCR primer, while primer GAPDH 5′ (5′GCCAAAAGGGTCATCATCTC) was used as the 5′ primer in PCRs. Controls excluding RT were performed to ensure that only GAPDH cDNAs were amplified. PCR products were electrophoresed through 1.0% agarose and visualized by staining them with ethidium bromide.
VLP RNase protection assay.
Seventy-five percent confluent 293T cells in 10-cm dishes (Corning) were transfected with 10 micrograms of pCAGGsVP40 plus pCAGGSVP35, followed by transfection of 5 micrograms of 3E-5E minigenome RNA 18 hours later. VLPs were collected 6 h posttransfection of the 3E-5E minigenome RNA and purified over 20% sucrose for 2 h at 36,000 rpm. The VLPs were resuspended in 180 μl DEPC-treated water and divided into four 40-μl fractions. Samples were prepared as follows: untreated or RNase treated with 1 μl RNase cocktail (Ambion), 0.5 μl of Triton X-100, and 1 μl of RNase cocktail and Triton X-100. Samples were brought to a final volume of 50 μl by the addition of DEPC water, followed by incubation at 37°C for 30 min. RNA was purified using the RNAeasy kit (QIAGEN), and RT-PCR was performed as described above for 3E-5E minigenome packaging.
RESULTS
VP35 and VP40 interact.
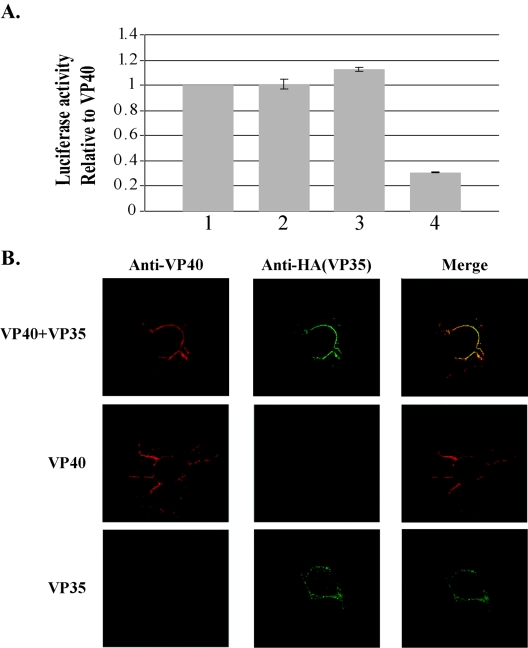
Investigations of VP35 and VP40 interactions were carried out in mammalian two-hybrid assays. Activation domain (AD) and binding domain (BD) fusion proteins were constructed with VP40 and VP35, resulting in the plasmids ADVP40, BDVP40, ADVP35, and BDVP35. Constructs were sequenced and tested for expression and recognition by immunoprecipitation with VP40 and VP35 antibodies (data not shown). 293T cells were transfected with combinations of the fusion protein constructs and a luciferase reporter construct that contained Gal4 binding domains. ADVP40 and BDVP40 were used as positive controls for EBOV protein-protein interactions, as VP40-VP40 interactions are well documented (24, 28) (Fig. 1A). Both ADVP40-BDVP35 and ADVP35-BDVP40 interacted as well as the VP40-VP40 positive control. As expected, the negative control resulted in a fivefold-weaker signal. These data indicate that VP40 interacts with VP35 at a level virtually identical to that of VP40-VP40 interactions.
FIG. 1.
(A) Mammalian two-hybrid assays. 293T cells were transfected and assayed for luciferase production in the following combinations: (1) ADVP40 plus BDVP40 (positive control), (2) ADVP40 plus BDVP35, (3) ADVP35 plus BDVP40, and (4) ADTRAF plus BDP53 (negative control). Three independent experiments were performed in triplicate. The error bars indicate ±1 standard error. (B) VP40 and VP35 colocalize in Cos-1 cells. (Top row) Cells were transfected with VP40 and VP35 and then probed with mouse anti-VP40 (red) and rat anti-HA (green). The merged image shows colocalization (yellow) of VP40 and VP35 at the plasma membrane and the cell projections. (Middle row) Cells were transfected with VP40 and probed with mouse anti-VP40 (red) and rat anti-HA (green). No cross-reactivity of the HA antibody was detected. (Bottom row) Cells were transfected with VP35 and probed with mouse anti-VP40 and rat anti-HA. No cross-reactivity of the VP40 antibody was detected.
Confocal microscopy was used to support mammalian two-hybrid data. Cos-1 cells grown on coverslips were transfected with pCAGGs alone, pCAGGsVP40, pCAGGsVP35, or both pCAGGSVP40 and pCAGGsVP35. As shown in Fig. 1B, top row, distinct areas of colocalization between VP40 and VP35 were readily detectable at the plasma membrane, as well as in the filamentous projections that formed at the surfaces of VP40- plus VP35-cotransfected cells. These findings support a strong colocalization of VP40 and VP35. Cells transfected with VP35 alone showed staining throughout the cell body (Fig. 1B, bottom row). In contrast, cells cotransfected with VP35 and VP40 exhibited localization of VP35 primarily at the cell surface and in the cell projections, where VP35 colocalized with VP40. This finding suggests that VP40 may recruit VP35 to regions of the cell where VLPs are assembled and released.
VP35 is packaged within VP40 VLPs.
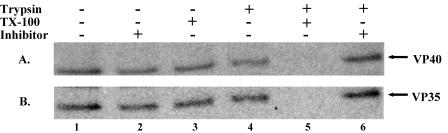
To assess whether VP40-VP35 interactions would lead to enclosure of VP35 within VP40 VLPs, we performed a VLP protease protection assay. 293T cells were transfected with pCAGGs, VP40, VP35, or VP40 plus VP35. Purified VLPs were divided into six equal fractions, as described in Materials and Methods. As reported previously, VP40 was digested only in the presence of both Triton X-100 and trypsin (Fig. 2A, lane 5) (17). Similarly, we found that VP35 was also degraded completely only in the presence of both Triton X-100 and trypsin (Fig. 2B, lane 5). Treatment with trypsin alone was insufficient to digest VP35 (Fig. 2B, lane, 4), indicating that VP35 is packaged within budding VP40 VLPs. It should be noted that VP35 was unable to bud from cells as a VLP when expressed alone in mammalian cells.
FIG. 2.
Protease protection assay of VLPs containing VP35 and VP40. VLP pellets were split into six fractions and treated with STE buffer (lane 1), soybean trypsin inhibitor (lane 2), Triton X-100 (lane 3), trypsin (lane 4), trypsin and Triton X-100 (lane 5), and trypsin and soybean trypsin inhibitor (lane 6). The fractions were divided equally, and viral proteins were immunoprecipitated with α-VP40 (A) or α-HA (B). Samples were resolved by SDS-PAGE and detected by autoradiography.
Effects of VP35 and the 3E-5E minigenome on release of VP40 VLPs.
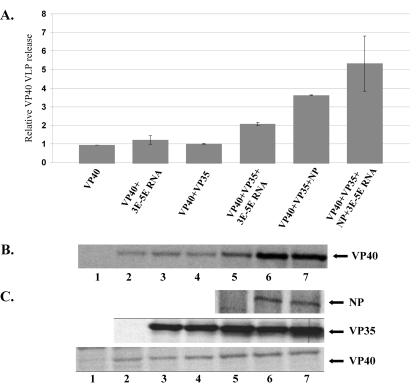
We next sought to determine whether VP35 or VP35-RNA complexes enhance the release of VP40 VLPs. 293T cells were transfected with vector alone, VP40, VP35, VP40 plus VP35, VP35 plus the 3E-5E minigenome, VP40 plus the 3E-5E minigenome, VP40 plus VP35 plus the 3E-5E minigenome, VP40 plus VP35 plus NP, or VP40 plus VP35 plus NP plus the 3E-5E minigenome. The 3E-5E minigenome RNA was transfected as described in Materials and Methods, and VLP budding assays were performed as described. VP40 release was quantified by phosphorimager analysis and averaged (Fig. 3A). Our results indicate that coexpression of VP35 did not enhance VP40 budding (Fig. 3A and B, compare lanes 2 and 4), in contrast to the enhancement observed following coexpression of NP and GP (15, 16). The addition of the 3E-5E minigenome to VP40-transfected cultures also failed to enhance budding of VP40 (Fig. 3A and B, compare lanes 2 and 3). However, when VP35 and the 3E-5E minigenome were present together, VP40 VLP budding was enhanced slightly, by 2.2-fold, compared to that of VP40 alone (Fig. 3A and B, compare lanes 2 and 5). VP35 did not further enhance the increase in budding of VP40 VLPs when VP40 and VP35 were coexpressed with NP (Fig. 3A and B, lane 6), demonstrating a 3.6-fold increase in budding compared to VP40 alone. Addition of the 3E-5E minigenome to VP40- plus VP35- plus NP-transfected cells demonstrated a slight increase in budding of VP40 VLPs over VP40 plus VP35 plus NP samples (Fig. 3A and B, lane 7) and a 5.4-fold increase over VP40 alone. Immunoprecipitation was carried out on all transfected cell monolayers to ensure that all proteins were expressed in transfected cells (Fig. 3C). These results suggest that although VP35 interacts and colocalizes with VP40, it does not significantly enhance VP40 budding.
FIG. 3.
Enhancement of VP40 VLP budding. (A) Summary of the results of three independent budding assays. Cells were transfected with the indicated viral protein constructs and RNA. VP40 release was quantified by immunoprecipitation and phosphorimager analysis. (B) A representative experiment of VP40-transfected 293T cells. Lane 1, pCAGGS (mock); lane 2, VP40 alone; lane 3, VP40 plus 3E-5E minigenome; lane 4, VP40 plus VP35 cotransfected; lane 5, VP40 plus VP35 plus 3E-5E minigenome cotransfected; lane 6, VP40 plus VP35 plus NP cotransfected; lane 7, VP40 plus sVP35 NP plus 3E-5E minigenome cotransfected. (C) Cell lysate control immunoprecipitation to ensure expression of the proteins of interest.
VP35 interacts with the 3E-5E minigenome RNA.
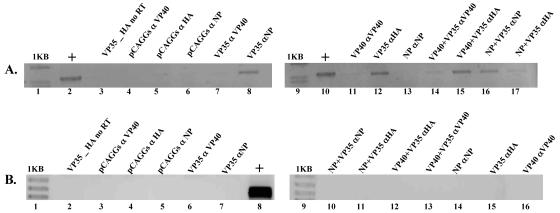
IP-RT-PCR was used to investigate EBOV protein interactions with the 3E-5E minigenome. Cell monolayers were lysed, immunoprecipitated, and assayed for the 3E-5E minigenome as described in Materials and Methods. 3E-5E minigenome RNA could be detected in the VP35- plus 3E-5E minigenome RNA-transfected cell lysates immunoprecipitated with anti-HA antibody, suggesting direct VP35-3E-5E minigenome RNA interactions (Fig. 4A, lane 12). 3E-5E minigenome RNA could also be detected in VP35- plus VP40- plus 3E-5E minigenome RNA-transfected samples immunoprecipitated with anti-HA and anti-VP40 (Fig. 4A, lanes 15 and 14, respectively), as well as in samples transfected with NP plus VP35 plus the 3E-5E minigenome RNA immunoprecipitated with anti-HA and anti-NP antibodies (Fig. 4A, lanes 16 and 17, respectively). To establish the selectivity of our IP-RT-PCR assay, pCAGGS- plus 3E-5E minigenome RNA-transfected cell lysates were immunoprecipitated with the anti-VP40, anti-HA, and anti-NP antibodies, and 3E-5E minigenome RNA was not detected in any sample (Fig. 4A, lanes 4, 5, and 6). As additional controls, VP35- plus 3E-5E minigenome RNA-transfected lysates were immunoprecipitated with anti-VP40 and anti-NP antibodies (Fig. 4A, lane 7). The 3E-5E minigenome RNA could be detected in anti-NP-immunoprecipitated samples (Fig. 4A, lane 8). This was likely due to cross-reactivity between anti-NP antibodies and VP35 (data not shown). To determine whether VP35 specifically bound to 3E-5E minigenome RNA, RT-PCR for the abundant cellular GAPDH RNA was carried out on the same RNA eluted during the immunoprecipitation step of our assay. As shown in Fig. 4B, GAPDH RNA could not be detected in any of the samples.
FIG. 4.
IP-RT-PCR of 3E-5E-transfected cell lysates. All samples were transfected with the 3E-5E minigenome and combinations of expression vectors, including empty pCAGGs vector controls, as indicated. (A) IP-RT-PCR to detect 3E-5E minigenome RNA. IP-RT-PCR was carried out as described in Materials and Methods. (B) RT-PCR of GAPDH RNA from immunoprecipitated complexes. RT-PCR was carried out as described in Materials and Methods. +, positive control.
VP40-VP35-3E-5E complexes were visualized by three-channel confocal microscopy.
VP40-VP35-3E-5E minigenome complexes were visualized using fluorescently tagged 3E-5E minigenome transcripts, which were cotransfected into VP40, VP35, or VP40 plus VP35 Vero cells. As a control for specificity of the VP40-VP35-3E-5E minigenome complexes, total RNA extracted from 293T cells, TRI-Xef mRNA transcripts (Xenopus elongation factor 1α cDNA provided as a control with the Ambion Megascript Transcription Kit), and antisense CAT RNAs were fluorescently labeled and transfected into parallel cultures. At 30 h posttransfection of plasmid DNA, the Vero cells were fixed, stained, and visualized as described in Materials and Methods. The emission spectra for the secondary antibodies and labeled RNA were chosen to minimize cross-excitation. When visualized by confocal microscopy, VP35 fluoresces blue, RNA fluoresces green, and VP40 fluoresces red. Therefore, the possible color combinations indicating colocalization are as follows: VP35 plus RNA, turquoise/aquamarine; VP40 plus RNA, yellow; VP35 plus VP40, violet/purple; and VP35 plus VP40 plus RNA, white.
Formation of VP40-VP35-3E-5E minigenome complexes is supported by colocalization of VP40, VP35, and the 3E-5E minigenome at the edges of the cell, presumably just under the plasma membrane (Fig. 5A and B). In some fields, colocalization is prominent in a region where a tendril emanates from the body of the cell (Fig. 5A, area of colocalization in lower right corner). To show the specificities and contributions of the Ebola virus-derived regions of the 3E-5E minigenome to the formation of EBOV RNP complexes, we excised the CAT reporter gene from the 3E-5E minigenome in the antisense orientation. As shown in Fig. 5C, the leaderless and trailerless control antisense CAT RNA failed to colocalize with either VP40 or VP35 and failed to form RNP complexes. These data support the specificity of 3E-5E minigenome RNP complex formation. In this control experiment, VP35-VP40 complexes were readily detectable (violet color), supporting our previous confocal data (Fig. 1B). In cells transfected with VP40 plus 3E-5E RNA or VP40 plus antisense CAT RNA, no colocalization was detected (data not shown). Experiments were also carried out with fluorescently labeled total 293T and pTRI-Xef mRNAs. Again, no colocalization was detected between these negative control RNAs and VP40 or VP35 (data not shown). In summary, the confocal-microscopy data support the formation of specific VP40-VP35-3E-5E minigenome RNA complexes. Moreover, these complexes most likely form due to direct interaction of VP35 with 3E-5E minigenome RNA, as viral RNA-VP40 complexes were not readily detected (data not shown).
FIG. 5.
Three-channel confocal microscopy. The possible color combinations indicating colocalization are as follows: VP35 plus RNA, turquoise/aquamarine; VP40 plus RNA, yellow; VP35 plus VP40, violet/purple; VP35 plus VP40 plus RNA, white. (A and B) Vero cells were transfected with VP40 plus VP35 plus the 3E-5E minigenome. (C) Vero cells were transfected with pCAGGs VP40 plus VP35 plus antisense CAT RNA. The cells were fixed and stained as described in Materials and Methods. All images were analyzed using the LSM 5 Image Browser software from Zeiss. The red boxes indicate the regions of magnification. Only the 3E-5E minigenome colocalized with VP40-VP35 complexes.
The 3E-5E minigenome can be packaged into VP40 plus VP35 VLPs.
Since VP40-VP35-3E-5E complexes were detected by confocal microscopy, we sought to determine whether the 3E-5E minigenome could be packaged into budding VLPs in the presence of VP40 and VP35. Briefly, VLPs were purified from cell culture medium supernatants and purified over 20% sucrose in STE, and 3E-5E minigenome RNA was purified using the RNAeasy kit (QIAGEN). RT-PCR was performed using primers specific to the trailer and leader sequences of the 3E-5E minigenome. To ensure that viral proteins were expressed and that minigenome RNA was present in all cultures, the cell monolayers were washed in PBS, split into equal portions, and pelleted. One-half of the cells were lysed in RIPA buffer, followed by Western blotting for Ebola virus proteins. RNA was extracted from the remaining cell fraction, and RT-PCR was performed to confirm the presence of the 3E-5E minigenome RNA. For RT-PCRs, controls excluding RT were included to ensure that no 3E-5E template was present (Fig. 6B).
FIG. 6.
RT-PCR of 3E-5E minigenome RNA in VLPs. (A) RT-PCR of VLP pellets. 293T cells were transfected with expression vectors as indicated, followed by transfection of the 3E-5E minigenome. Inclusion of reverse transcriptase is indicated by +. (B) RT-PCR of cell monolayers transfected with the 3E-5E minigenome to ensure that 3E-5E was transfected into each culture. The lane numbers correspond to lane identifications and lane numbers in panel A. (C) GAPDH controls for VLP fractions to demonstrate specificity of 3E-5E minigenome packaging by VP40 plus VP35. The VLP total-RNA samples were used as a template for GAPDH reverse transcription.
Further investigation into VP35's role in packaging the 3E-5E minigenome was accomplished by transfecting parallel cultures with the 3E-5E minigenome (pCAGGs and 3E-5E minigenome transfected), VP40 plus 3E-5E, VP35 plus 3E-5E, VP40 plus NP plus 3E-5E, and VP40 plus NP plus VP35 plus 3E-5E, followed by isolation of VLPs through 20% sucrose. 3E-5E minigenome RNA was detected only in VP40 plus VP35 plus 3E-5E and VP40 plus NP plus VP35 plus 3E-5E VLP fractions from parallel cultures (Fig. 6A, lanes 7, 15, and 17). These data, in combination with the VP40-VP35 interaction data described above, suggest that VP40, VP35, and the 3E-5E minigenome are the minimal viral components required for packaging of RNA into VLPs. RT-PCR failed to detect the 3E-5E minigenome RNA in VLP fractions transfected with only the 3E-5E minigenome-, VP40- plus 3E-5E-, VP35- plus 3E-5E-, NP- plus 3E-5E-, and VP40- plus NP- plus 3E-5E-transfected cultures (Fig. 6A, lanes 3 to 6, 8 to 10, 13, and 14). Surprisingly, VP40 plus NP cultures were not able to package 3E-5E minigenome RNA in this assay (Fig. 6A, lane 9), although NP is a major component of the Ebola virus nucleocapsid (13) and is predicted to bind RNA and interact with VP40 (16, 19; R. F. Johnson, S. E. McCarthy, and R. N. Harty, unpublished data).
VP40, VP35, or NP alone was insufficient for packaging of the minigenome RNA (Fig. 6A, lanes 4, 5, and 6). The 3E-5E RNA was not detected in VLPs from 3E-5E minigenome-transfected cultures (Fig. 6A, lanes 3 and 13), confirming a role for viral proteins in packaging of the viral RNA into VLPs. These data suggest that packaging was specific in the presence of VP35 and VP40 and not a result of the copurification of 3E-5E minigenome-liposome complexes. As a further control, the 3E-5E minigenome could be detected by RT-PCR in all cell culture monolayers transfected with the 3E-5E minigenome (Fig. 6B). RT-PCR was carried out using primers for GAPDH, as described in Materials and Methods, on total RNA extracted from the VLP fraction to demonstrate that the 3E-5E minigenome RNA was specifically packaged into VP40 plus VP35 VLPs (Fig. 6C, lane 5). RT-PCR using the same primers was performed on total RNA isolated from the transfected cells (Fig. 6C, lanes 9 and 11). Western blot analysis of the cell fractions indicated that VP35, VP40, and NP were expressed appropriately (data not shown).
RNase protection assay.
Packaging of the 3E-5E minigenome RNA into VLPs was further confirmed by an RNase protection assay. 293T cells were transfected with VP40 and VP35, followed by transfection of 3E-5E minigenome RNA. VLPs were purified over 20% sucrose and resuspended in 5 mM Tris, pH 8.0. As shown in Fig. 7, the resuspended VLPs were divided into four equal fractions. One portion was not treated, while the others were treated with RNase cocktail, RNase cocktail plus Triton X-100, and Triton X-100 followed by RNA purification. RT-PCR analysis indicated that the 3E-5E RNA was digested by RNase in the presence of detergent (Fig. 7, lane 3), whereas the 3E-5E minigenome RNA was not digested in the fraction treated with RNase cocktail alone (Fig. 7, lane 2) or the fraction treated with Triton X-100 alone (Fig. 7, lane 4). These data further indicate that the 3E-5E minigenome RNA is packaged within a VP40 plus VP35 VLP.
FIG. 7.
RNase protection assay of VP40 plus VP35 plus 3E-5E minigenome VLPs. VLPs were purified, divided into equal fractions, and treated as indicated. Lane 1, untreated; lane 2, RNase treated and no Triton X-100; lane 3, RNase treated and Triton X-100; lane 4, Triton X-100 treated and no RNase treatement.
DISCUSSION
Here, we have described a transfection-based system to study the requirements for packaging of the Ebola virus-derived 3E-5E minigenome. We demonstrated that VP35 and VP40 associate (Fig. 1) and that this interaction is sufficient for packaging of the 3E-5E minigenome (Fig. 6). Although VP35 is packaged within VP40 VLPs (Fig. 2), VP35 did not enhance VP40 VLP budding (Fig. 3), as has been shown for NP and GP (16). Addition of the 3E-5E minigenome to VP40- plus VP35-cotransfected cultures enhances VP40 VLP budding by a modest 2.2-fold (Fig. 3). We have also demonstrated that VP35, VP40, and the 3E-5E minigenome form RNP complexes, resulting in recovery of the 3E-5E minigenome from VP40 plus VP35 VLP pellets (Fig. 4, 5, and 6, respectively). We have also demonstrated that the 3E-5E minigenome RNA is packaged within membrane-bound particles, further supporting our hypothesis that VP40 plus VP35 VLPs can package the 3E-5E minigenome RNA. These data suggest that VP35 binds 3E-5E minigenome RNA and is packaged into VLPs through VP35-VP40 interactions. This methodology also provides a means to study the requirements for VP40, VP24, VP35, NP, and the 3E-5E minigenome in the packaging and assembly of nucleocapsids into VLPs.
The role of VP40 in filovirus budding has been clearly established (17, 23, 24). Recent work has demonstrated roles for NP and GP in enhancing VP40 VLP budding (15, 16). However, little is known about the role of VP35 in the assembly of nucleocapsids or the specific encapsulation of viral RNA by the nucleocapsid and the nucleocapsid's subsequent assembly into a mature virion. The current hypothesis is that phosphoprotein and nucleoprotein interactions link nucleoprotein-RNA complexes with phosphoprotein-polymerase complexes to form viral nucleocapsids (18, 27, 32). The nucleocapsid then interacts with the viral matrix protein, leading to specific packaging into virions. Disrupting phosphoprotein-nucleoprotein interactions leads to defects in viral-RNA replication and/or transcription, presumably through the loss of proper nucleocapsid formation (5). However, the current hypothesis does not explain why nonspecific RNAs are not packaged into virions, as NP associates with nonviral RNA (18, 32). Our data suggest that VP35 specifically interacts with RNA. It is possible that specificity of VP35 for viral RNA, in conjunction with VP35-NP, VP40-VP35, and NP-VP40 interactions, may confirm specificity for packaging complete nucleocapsids in EBOV-infected cells.
VP40-VP35 interactions were investigated with mammalian two-hybrid assays, which indicated an in vivo interaction roughly equal to that of VP40-VP40 (Fig. 1A). Confocal microscopy was used to confirm VP40-VP35 interactions. As shown in Fig. 1B, distinct areas of colocalization between VP40 and VP35 were evident. The merged image clearly shows colocalization at regions presumably just beneath the plasma membrane. Negative controls indicated that the VP40 and HA antibodies are specific.
VP35 colocalized with VP40 and was packaged into VP40 VLPs (Fig. 1 and 2), indicating that VP35 may aid in nucleocapsid packaging, in addition to its roles in viral replication and transcription. The step(s) in which VP35 acts in packaging is unclear. We demonstrated that coexpression of VP35 with VP40 did not enhance VP40 VLP budding, and cotransfection of VP35 and the 3E-5E minigenome led to only a slight, 2.2-fold, increase in budding (Fig. 3A). By comparison, NP enhanced VP40 VLP budding 3.5-fold (16). However, addition of VP35 to NP- and VP40-transfected samples enhanced budding 3.6-fold, which indicates that VP35 failed to further enhance the effect of NP on VP40 VLP budding. Compilation of both data sets implies that NP plays a predominant role in the packaging of the nucleocapsid but does not exclude a role for VP35.
These data also indicate a specific interaction between the 3E-5E minigenome and VP35. Similar findings were reported recently for Chandipura virus phosphoprotein and viral RNA (1). IP-RT-PCR was performed to investigate specific interactions between the 3E-5E minigenome RNA and EBOV proteins VP35, VP40, and NP. Our data suggest that VP35 specifically interacts with the 3E-5E minigenome RNA (Fig. 4A) and not with other cellular RNAs (Fig. 4B, lanes 12 and 15). 3E-5E minigenome RNA could be captured only by NP in the presence of VP35 (Fig. 4A, lane 17), supporting the hypothesis that VP35 confers specificity for NP on viral RNA, as has been demonstrated in other Mononegavirales members (4, 18, 32). 3E-5E minigenome RNA was not detected in VP40- plus 3E-5E minigenome RNA-transfected samples, but 3E-5E minigenome RNA could be detected in samples transfected with both VP40 and VP35. This could be due to coimmunoprecipitation of VP35 by VP40, as VP40 antibody could not detect 3E-5E minigenome RNA when incubated with VP35- plus 3E-5E-transfected samples. In cells transfected with NP alone, no 3E-5E minigenome RNA or GAPDH RNA (Fig. 4A and C) could be detected when immunoprecipitated with anti-NP antibody. This could be due to NP interacting with many host RNAs, and the level of GAPDH and 3E-5E minigenome RNA that was bound to NP fell below the levels of detection in our assay.
Three-channel confocal microscopy was used to further support the formation of VP40-VP35-3E-5E minigenome RNA-RNP complex formation. For this assay, Vero cells were used instead of Cos-1 cells because Cos-1 cells proved refractory to a second transfection step (data not shown). As shown in Fig. 1B, colocalization between VP40 and VP35 was evident in Cos-1 cells. Similarly, in Vero cells, VP40 and VP35 colocalized, as can be seen in Fig. 5. Packaging assays with Vero cells yielded results identical to those of experiments using 293T cells. These results suggest that the interactions of the viral proteins and unidentified host factors are common to both cell lines (data not shown). Regions of colocalization between RNA and VP35 could be detected readily, and VP40-3E-5E minigenome RNA signal overlap (identified by a yellow color) could only be detected at the fringes of VP40-VP35-3E-5E minigenome complexes (identified by a white color). These data suggest that VP35 interacts with the viral RNA, and VP40-RNA interactions could be observed only when VP35 was part of the RNP (Fig. 5A). To test the specificity of the RNP complex, nonviral RNAs were fluorescently tagged and transfected into parallel cultures. No colocalization of the nonviral RNA with VP40, VP35, or VP40 plus VP35 could be detected, implying that formation of RNP complexes is specific to the 3E-5E minigenome.
A modified budding assay was used to determine whether the 3E-5E minigenome could be recovered from VP40 VLP pellets. The 3E-5E minigenome could be recovered only when VP40 and VP35 were coexpressed (Fig. 6A), suggesting that VP40 and VP35 are the minimal viral proteins needed for Ebola virus genome packaging. When VP40, NP, and VP35 were coexpressed, the 3E-5E minigenome could also be detected. In repeated experiments, NP and VP40 were insufficient for recovery of the 3E-5E minigenome, possibly due to nonspecific interactions of NP with cellular RNAs (18, 19, 32). As a control for specificity of packaging, RT-PCR was carried out on RNA samples recovered from VLPs using primers specific for GAPDH (Fig. 6C). GAPDH cDNA could not be detected in any VLP fraction sample, suggesting that the 3E-5E minigenome is specifically packaged by VP40 and VP35. These data indicate that the selective packaging of genomic RNA is due to specific interactions between the RNA and VP35.
Inferences from the data presented suggest a mechanism for nucleocapsid assembly and tethering of the nucleocapsid to VP40 as virions form. We speculate that NP-RNA complexes are formed as NP and viral genomic RNAs are produced. As VP35 is produced and associates with L protein, NP and viral genomic RNA form RNPs. VP35 then makes two contacts, one with NP and one with the RNA associated with NP. If VP35 contacts a nonviral RNA, then an unstable nucleocapsid forms, which quickly dissociates and is not packaged. If VP35 contacts viral genomic RNA, then the RNP stabilizes and a suitable viral nucleocapsid forms. The nucleocapsid then contacts VP40 through interactions with both VP35 and NP. Contact between VP40 and both NP and VP35 likely protects against packaging of nonspecific NP-RNA complexes into developing virions. Using this working model, we will continue to explore VP35, VP40, and NP requirements for packaging as requirements of cis-acting structures within the RNA required for VP35-3E-5E minigenome interactions.
Acknowledgments
We thank members of the Harty laboratory and J. Paragas for critical review of the paper. We also thank Roland Grunow, Elke Muhlberger, and Chris Basler for materials they supplied.
R.F.J. was supported in part by NIH Training Grant T32AI07324-13. This work was supported by NIH grants to R.N.H.
REFERENCES
- 1.Basak, X., S. Polley, M. Basu, D. Chattopadhyay, and S. Roy. 2004. Monomer and dimer of Chandipura virus unphosphorylated P-protein binds leader RNA differently: implications for viral RNA synthesis. J. Mol. Biol. 339:1089-1101. [DOI] [PubMed] [Google Scholar]
- 2.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Z., T. J. Green, M. Luo, and H. Li. 2004. Visualizing the RNA molecule in the bacterially expressed vesicular stomatitis virus nucleoprotein-RNA complex. Structure 12:227-235. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary, S. K., A. G. Malur, Y. Huo, B. P. De, and A. K. Banerjee. 2002. Characterization of the oligomerization domain of the phosphoprotein of human parainfluenza virus type 3. Virology 303:373-382. [DOI] [PubMed] [Google Scholar]
- 6.Coronel, E. C., T. Takimoto, K. G. Murti, N. Varich, and A. Portner. 2001. Nucleocapsid incorporation into parinfluenza virus is regulated by specific interaction with matrix protein. J. Virol. 75:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomis-Ruth, F. X., A. Dessen, J. Timmins, A. Bracher, L. Kolesnikowa, S. Becker, H. D. Klenk, and W. Weissenhorn. 2003. The matrix protein VP40 from Ebola virus octamerizes into pore-like structures with specific RNA binding properties. Structure 11:423-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groseth, A., H. Feldmann, S. Theriault, G. Mmehmetoglu, and R. Flick. 2005. RNA polymerase I-driven minigenome system for Ebola viruses. J. Virol. 79:4425-4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han, Z., H. Boshra, J. O. Sunyer, S. H. Zwiers, J. Paragas, and R. N. Harty. 2003. Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding. J. Virol. 77:1793-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartman, A. L., J. S. Towner, and S. T. Nichol. 2004. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology 328:177-184. [DOI] [PubMed] [Google Scholar]
- 11.Harty, R. N., M. E. Brown, G. Wang, J. Hibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and funtionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoenen, T., V. Volchkov, L. Kolesnikova, E. Mittler, J. Timmins, M. Ottmann, O. Reynard, S. Becker, and W. Weissenhorn. 2005. VP40 octamers are essential for Ebola virus replication. J. Virol. 79:1898-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, Y., L. Xu, Y. Sun, and G. J. Nabel. 2002. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posstranslational modification of nucleoprotein. Mol. Cell 10:307-316. [DOI] [PubMed] [Google Scholar]
- 14.Jasenosky, L. D., G. Neumann, I. Lukashevich, and Y. Kawaoka. 2001. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 75:5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kallstrom, G., K. L. Warfield, D. L. Swenson, S. Mort, R. G. Panchal, G. Ruthel, S. Bavari, and M. J. Aman. 2005. Analysis of Ebola virus and VLP release using an immunocapture assay. J. Virol. Methods 127:1-9. [DOI] [PubMed] [Google Scholar]
- 16.Licata, J. M., R. F. Johnson, Z. Han, and R. N. Harty. 2004. Contribution of Ebola virus glycoprotein, nucleoprotein and VP24 to budding of VP40 virus-like particles. J. Virol. 78:7344-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS4. J. Virol. 77:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, P., J. Yang, X. Wu, and Z. F. Fu. 2004. Interactions amongst rabies virus nucleoprotein, phosphoprotein and genomic RNA in virus-infected and transfected cells. J. Gen. Virol. 85:3725-3734. [DOI] [PubMed] [Google Scholar]
- 19.Mavrakis, M., L. Kolesnikova, G. Schoehn, S. Becker, and R. W. H. Ruigrok. 2002. Morphology of Marburg virus NP-RNA. Virology 296:300-307. [DOI] [PubMed] [Google Scholar]
- 20.Muhlberger, E., B. Lotfering, H.-D. Klenk, and S. Becker. 1998. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J. Virol. 72:8756-8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muhlberger, E., M. Weik, V. E. Volchkov, H. D. Klenk, and S. Becker. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J. Virol. 73:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nanda, S. K., R. F. Johnson, Q. Liu, and J. L. Leibowitz. 2004. Mitochondrial HSP70, HSP40, and HSP60 bind to the 3′ untranslated region of the murine hepatitis virus genome. 149:93-111. [DOI] [PMC free article] [PubMed]
- 23.Noda, T., H. Sagara, E. Suzuki, A. Takada, H. Kida, and Y. Kawaoka. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 76:4855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panchal, R. G., G. Ruthel, T. A. Kenny, G. H. Kallstrom, D. Lane, S. S. Badie, L. Li, S. Bavari, and M. J. Aman. 2003. In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc. Natl. Acad. Sci. USA 100:15936-15941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters, C. J., P. Jahrling, T. G. Ksiazek, E. Johnson, and H. Lupton. 1992. Filovirus contamination of cell cultures. Dev. Biol. Stand. 76:267-274. [PubMed] [Google Scholar]
- 26.Sullivan, N., Z.-Y. Yang, and G. J. Nabel. 2003. Ebola virus pathogenesis: implications for vaccines and therapies. J. Virol. 77:9733-9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarbouriech, N., J. Curran, C. Ebel, R. W. Ruigrok, and W. P. Beurmeister. 2000. On the domain structure and the polymerization state of the Sendai virus P protein. Virology 266:99-109. [DOI] [PubMed] [Google Scholar]
- 28.Timmins, J., G. Schoehn, C. Kohlhaas, H. D. Klenk, R. W. Ruigrok, and W. Weissenhorn. 2003. Oligomerization and polymerization of the filovirus matrix protein VP40. Virology 312:359-368. [DOI] [PubMed] [Google Scholar]
- 29.Volchkov, V. E., V. Volchkova, A. A. Slenczka, H.-D. Klenk, and H. Feldmann. 1998. Release of viral glycoproteins during Ebola virus infection. Virology 245:110-119. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe, S., T. Watanabe, T. Noda, A. Takada, H. Feldmann, L. D. Jasenosky, and Y. Kawaoka. 2004. Production of novel Ebola virus-like particles from cDNAs: an alternative to Ebola virus generation by reverse genetics. J. Virol. 78:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wills, J. W., R. C. Craven, and J. A. Achacoso. 1989. Creation and expression of myristylated forms of Rous sarcoma virus gag protein in mammalian cells. J. Virol. 63:4331-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, J., D. C. Hooper, W. H. Wunner, H. Koprowski, B. Dietzschold, and Z. Fu. 1998. The specificity of rabies virus RNA encapsidation by nucleoprotein. Virology 242:107-115. [DOI] [PubMed] [Google Scholar]
- 33.Yu, W., and J. L. Leibowitz. 1995. Specific binding of host cellular proteins to multiple sites within the 3′ end of the mouse hepatitis virus genomic RNA. J. Virol. 69:2016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]