Abstract
MicroRNAs (miRNAs) are key regulators of gene expression in higher eukaryotes. Recently, miRNAs have been identified from viruses with double-stranded DNA genomes. To attempt to identify miRNAs encoded by herpes simplex virus 1 (HSV-1), we applied a computational method to screen the complete genome of HSV-1 for sequences that adopt an extended stem-loop structure and display a pattern of nucleotide divergence characteristic of known miRNAs. Using this method, we identified 11 HSV-1 genomic loci predicted to encode 13 miRNA precursors and 24 miRNA candidates. Eight of the HSV-1 miRNA candidates were predicted to be conserved in HSV-2. The precursor and the mature form of one HSV-1 miRNA candidate, which is encoded ∼450 bp upstream of the transcription start site of the latency-associated transcript (LAT), were detected during infection of Vero cells by Northern blot hybridization. These RNAs, which behave as late gene products, are not predicted to be conserved in HSV-2. Additionally, small RNAs, including some that are roughly the expected size of precursor miRNAs, were detected using probes for miRNA candidates derived from sequences encoding the 8.3-kilobase LAT, from sequences complementary to UL15 mRNA, and from the region between ICP4 and US1. However, no species the size of typical mature miRNAs were detected using these probes. Three of these latter miRNA candidates were predicted to be conserved in HSV-2. Thus, HSV-1 encodes at least one miRNA. We hypothesize that HSV-1 miRNAs regulate viral and host gene expression.
MicroRNAs (miRNAs) are noncoding small RNA molecules with important regulatory functions in higher eukaryotic development and gene expression (reviewed in references 2 and 5). The vast majority of known mature miRNAs are about 21 to 23 nucleotides (nt) long. They are derived from longer pol II (19) or pol III (25) primary transcripts (pri-miRNAs) that are processed in the nucleus by the RNase III enzyme Drosha. The excised fold-back precursor miRNA (pre-miRNA) is typically 60 to 80 nt long and assumes a stem-loop structure with an imperfectly duplexed stem. Pre-miRNA is then exported to the cytoplasm by the export factor Exportin 5 (39). The pre-miRNA is later cleaved by the RNase III enzyme Dicer to excise the miRNA in the form of a small interfering RNA (siRNA)-like duplex (16) which then unwinds, leaving one 21- to 23-nt strand energetically favored to enter the multiprotein RNA-induced silencing complex (RISC). The other strand is usually degraded. Mature miRNAs in RISC regulate protein-coding gene expression via the RNA silencing machinery, typically by forming imperfect duplexes with target messenger RNAs (mRNAs) (reviewed in references 2 and 5). Depending on the extent of complementarity and the location of the binding site, miRNAs can direct cleavage and/or regulate translation of their target mRNAs. Perfect complementarity is generally thought to result in cleavage and imperfect complementarity usually in impaired translation, most often when the target sequences are located in the 3′ untranslated region of the mRNA.
Thousands of miRNAs have been identified in different organisms to date (13). The discovery of miRNAs encoded by DNA viruses suggests that viruses have evolved to exploit RNA silencing for regulation of host and viral genes (reviewed in reference 32). Of the viral miRNAs identified so far, most are encoded by viruses in the herpesvirus family (6, 25, 26, 29). Specifically, three members of the γ (lymphotropic) subfamily, Epstein-Barr virus (EBV) (26), Kaposi's sarcoma-associated herpesvirus (6, 25, 29), and mouse gammaherpesvirus 68 (25), and one member of the β subfamily, human cytomegalovirus (25), have been shown to encode miRNAs. To date, none of the α subfamily members has been experimentally proven to encode miRNAs, although two of these viruses, herpes simplex viruses 1 and 2 (HSV-1 and -2), have been predicted to do so (25).
HSV-1 and -2 cause a spectrum of diseases, from debilitating genital infections and sight-threatening ocular infections in immunocompetent adults to more severe infections in newborns and immunosuppressed individuals, such as patients with AIDS (37). The HSV genome is approximately 150 kbp and encodes at least 80 gene products (28). The genome is composed of two covalently linked segments, called Long (L) and Short (S), based upon their relative length. These segments each contain unique regions (UL or US) flanked by inverted repeat regions (35). The genes found in the unique regions are present in the genome as a single copy, while the genes that are encoded in the repeat regions are present in the genome in two copies.
Most viral miRNAs have been identified by cDNA cloning of small RNAs from virus-infected cells (6, 25, 26, 29), whereas others have been identified following computational prediction and hybridization analysis (25, 33). Experimental screening of viral miRNAs via high-throughput sequencing of large numbers of cDNA clones from infected cells is technically challenging and could be incomplete, given that viral gene expression can have highly constrained tissue-, time-, and replication state-specific patterns. Additionally, cellular miRNAs would be expected to dominate in small RNA molecule samples extracted from infected cells. When we started this project, there was no software reported for predicting viral miRNAs; all of the existing miRNA gene prediction software was designed for prediction of eukaryotic miRNAs and heavily relied on cross-species conservation (17, 20). As the viral miRNAs that had been identified lacked homologs in other viruses (26), new software to predict viral miRNAs needed to be developed and validated.
Hypothesizing that HSV-1 encodes miRNAs, we applied a bioinformatic method to the HSV-1 genome and predicted 24 miRNA candidates from 13 precursors. The sequences of some predicted HSV-1 miRNAs are conserved in HSV-2. We investigated whether any of these predicted miRNAs or ones predicted by others (25) were expressed in HSV-1-infected cells.
MATERIALS AND METHODS
Computational prediction of viral miRNAs.
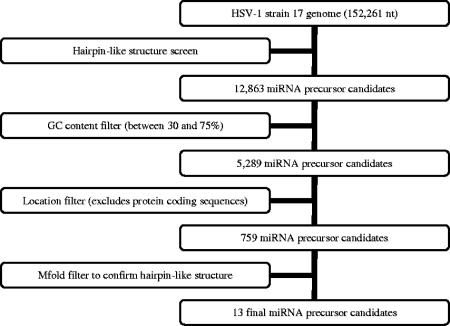
The EBV miRNA prediction was carried out using the complete genome sequence of EBV (NC_007605) obtained from the National Center for Biotechnology Information (NCBI). The HSV-1 miRNA prediction was performed using the complete genome sequence of HSV-1 strain 17 (NC_001806) obtained from NCBI. Figure 1 shows a flowchart of the computational prediction process. Briefly, both strands of the viral genome were scanned for hairpin-structured miRNA precursors using a 21-nt query window with 1-nt increments at a time in a way similar to that previously reported for Arabidopsis thaliana (36). Sequences with potential hairpin-like structures were extracted as candidate miRNA precursors. A GC content requirement of 30% to 75% for the 21-nt query sequences was applied. Additionally, low-complexity sequences, such as those with dinucleotides repeated ≥4 times (for example, ATATATAT), trinucleotides repeated ≥3 times (for example, ATGATGATG), or any single nucleotide repeated >6 times (for example, AAAAAAA), were removed using a repeat filter. Such sequences have not been observed in known miRNAs. Candidates within or antisense to protein-coding regions were subsequently removed according to the NCBI genome annotations. The resulting candidate miRNA precursors were analyzed with the program mfold (42) for secondary structure prediction. Sequences with a hairpin-like secondary structure as their lowest energy form were selected as potential miRNA precursors. The corresponding 21-nt query sequences and their pairing sequences on the other arm of the hairpin precursor (if both passed the GC content and the repeat filters) were selected as miRNA candidates.
FIG. 1.
Flowchart of the HSV-1 miRNA prediction procedure. See the text for details.
Conservation of predicted HSV-1 miRNAs in HSV-2 genome.
The predicted 21-nt sequences of HSV-1 miRNA candidates were compared with the HSV-2 strain HG52 genome (NC_001798) using the NCBI blastn program (1). For each HSV-2 sequence that is orthologous to a candidate HSV-1 miRNA, sequences upstream and downstream in the HSV-2 genome were extracted and searched for a hairpin-like secondary structure using mfold (42). A candidate HSV-1 miRNA was predicted to be conserved in HSV-2 if the putative HSV-2 pre-miRNA also possessed hairpin-like secondary structure, and the putative orthologous miRNA was located in the stem region of the hairpin.
Cells and virus.
Vero (African green monkey) cells were cultured at 37°C in Dulbecco's modified Eagle's medium (Mediatech, Herndon, VA) supplemented with 5% newborn calf serum. Wild-type HSV-1 strain KOS was prepared and titrated as previously described (8).
RNA preparation and Northern blot analyses.
Vero cells were mock infected or infected with strain KOS at a multiplicity of infection of 10 as previously described (27) and were harvested at various times postinfection. Small RNA (<200 nt) and large RNA fractions were isolated using the mirVana miRNA Isolation kit according to the manufacturer's protocol (Ambion, Austin, TX). Large RNA samples were treated with TURBO DNase (Ambion) to remove contaminating DNA, followed by RNeasy MinElute column purification to remove the DNase according to the manufacturer's protocol (QIAGEN, Valencia, CA).
For Northern blot analysis of small RNAs, 1-μg aliquots of each small RNA fraction, as well as radiolabeled Decade Markers (Ambion), were separated in 15% denaturing polyacrylamide gel electrophoresis (PAGE) gels (acrylamide:bis ratio, 19:1) containing 8 M urea in 0.089 M Tris/0.089 M borate/0.002 M EDTA buffer (TBE). The gels were soaked briefly in 0.5 μg/ml ethidium bromide in TBE to allow visualization of the RNA using a UV transilluminator (Bio-Rad, Hercules, CA). RNAs were transferred by electroblotting to a positively charged nylon membrane (Pierce, Rockford, IL) using a semidry apparatus (Bio-Rad) and UV cross-linked to the membrane. PAGE-purified DNA oligonucleotides (Integrated DNA Technologies, Coralville, IA) with the reverse complementary sequence to candidate miRNAs (boldfaced sequences in Table 1; see supplemental Table 1 posted at http://coen.med.harvard.edu) or to let-7, a cellular miRNA (16), were end labeled with [γ-32P]ATP (MP Biomedicals, Irvine, CA) to high specific activity (at least 5.3 × 106 dpm/pmol, measured using a liquid scintillation counter [PerkinElmer, Wellesley, MA] at 95% efficiency for 32P). The labeled probes were purified using the mirVana Probe & Marker kit (Ambion) or Micro Bio-Spin Columns (Bio-Rad) according to the manufacturers' protocols. Hybridizations and washes were carried out using the ULTRAhyb-Oligo hybridization buffer according to the manufacturer's directions (Ambion), and signals were detected by phosphor storage technology (Bio-Rad). Blots were stripped by pouring boiling 0.1% sodium dodecyl sulfate (SDS) solution over the membrane, allowing to cool to room temperature, and repeating at least once.
TABLE 1.
Genomic coordinates and sequences of predicted HSV-1 pre-miRNAs and miRNAs
| Precursor no. | Predicted pre-miRNA sequence, 5′ to 3′ (predicted miRNA sequence highlighted in boldface) | Position, orientationd |
|---|---|---|
| 1 | GCCGGACCCCCUCGCGAUGGGAAUGGACGGGAGCGACGGGGCCGGCGCAAAAAAACGCAGUAUCUCCCGCGAAGGCUACCCGCCGCCCCAGCCCCCGGCCAAAUGCGGAAACGGUCCCGCGCUCUCGCCUUUAUACGCGGGCCGCCCUGC | 103206-103355, + |
| 2a | CGAGGGGAACGGGGGAUGGAAGGACGGGAAGUGGAAGUCCUGAUACCCAUCC UACACCCCCCUGCCUUCCACCCUCCGGCCCCCCG | 118316-118401, + |
| 3 (T-3b) | GCGUGCCGGGGUGGUAGAGUUUGACAGGCAAGCAUGUGCGUGCAGAGGCGAGUAGUGCUUGCCUGUCUAACUCGCUAGUCUCGGCCGC | 125870-125957, + |
| 4 | GCGCGUCCCGCGCUCCCUCGGGGGGGUUCGGGCAUCUCUACCUCAGUGCCGCCAAUCUCAGGUCAGAGAUCCAAACCCUCCGGGGGCGCCCGCGC | 126708-126802, + |
| 4-RCc | GCGCGGGCGCCCCCGGAGGGUUUGGAUCUCUGACCUGAGAUUGGCGGCACUGAGGUAGAGAUGCCCGAACCCCCCCGAGGGAGCGCGGGACGCGC | 126708-126802, − |
| 5 | CGCAUGGCAUCUCAUUACCGCCCGAUCCGGCGGUUUCCGCUUCCGUUCCGCAUGCUAACGAGGAACGGGCAGGGGGCGGGGCCCGGGCCCCGACUUCCCGGUUCGGCGGUAAUGAGAUACGAGCCCCGCG | 131677-131806, + |
| 6 | AGUGAGAACGCGAAGCGUUCGCACUUCGUCCCAAUAUAUAUAUAUUAUUAGGGCGAAGUGCGAGCACUGGCGCCGUGCCCGACU | 131959-132042, + |
| 6-RCc | AGUCGGGCACGGCGCCAGUGCUCGCACUUCGCCCUAAUAAUAUAUAUAUAUUGGGACGAAGUGCGAACGCUUCGCGUUCUCACU | 131959-132042, − |
| 7 | GGACGUCAGUGUUGGGGGAGACGGUUUCGGUGAGGUUAUUUAGCUGCUCGAGCAGAUACUCGACCGGGUCGGUCUGCAGGCGCACCGUCGCGGAAACGAUCUCCUGGGAGGCCAUGACGCC | 34568-34688, − |
| 8 | GGCGCAACGGUCGGGGGGCGGGCGGAAAGGCGAGAGCGAAUGCUAAACUAAACGCUAACCCAGCUCCCGCCGUUGCGUUCACGCCAACCGCCGGGCC | 50672-50768, − |
| 9 | GGGUGGGGAACAACCCAUACCGGACAGAUGCCGAUGAGCCACCGCACCCUUGGGUGCGGGUGGUACGGGGUGGUUUGUUCAUCCUAUGGUUCCGACCC | 143938-144035, − |
| 10 | GGAUUGGCUGGUGUAGUGGGCGCGGCCAGAGACCACCCAGCGCCCGACCCCCCCCUCCCCACAAACACGGGGGGCGUCCCUUAUUGUUUUCCCUCGUCCCGGGUCGACGCCCCCUGCUCCCCGGACC | 120802-120928, + |
| 11 | GCUUCGUCCGCGUAUCGGCGUCCCGGCGCGGCGAGCGUCUGACGGUCUGUCUCUGGCGGUCCCGCGUCGGGUCGUGGAUCCGUGUCGGCAGC | 131324-131415, + |
| T-1b | GGCGACCCCGGUCCCUGUAUAUAUAGGGUCAGGGGGUUCCGCACCCCCUAACAUGGCGCCCCCGGUCCCUGUAUAUAUAGUGUCACGGGGUUCCACGCC | 2482-2580, − |
| T-4b | CGCUAUUAUAAAAAAAGUGAGAACGCGAAGCGUUCGCACUUUGUCCUAAUAAUAUAUAUAUUAUUAGGACAAAGUGCGAACGCUUCGCGUUCUCACUUUUUUUAUAAUAGCG | 62420-62531, + |
| T-5b | GCGUUCGGCCAUGUUGUGGGCCAGCACCUGCAGCGUGAGCAUGGCGGGCCCGUCCACUACCACGCGCCCGUUGUGAAACAUGGCGUUGACCGUGUUGGCCACCAGAUUGGCCGGGUGC | 37817-37934, + |
| T-7b | GCCGUGUGCCCCAGUCGCACUCGUCCCUGGCUCAGGCCGCGAACCAAGAACAGAGUCUGUGCCGGGCGCGUGCGACGGUGGCGCGCGGC | 4465-4553, + |
The sequence of HSV-1 miR-H1 (see the text) is underlined.
These loci and precursors were predicted by Pfeffer et al. (25) and are numbered T-1, T-3, T-4, T-5, and T-7. Mature miRNA candidates within these predicted precursor sequences were identified by us.
Because these two precursors do not represent unique genomic locations, they are not given a unique number. RC indicates that they are from the reverse complementary strand of precursors 4 and 6, respectively.
Numbers in this column correspond to genomic coordinates of HSV-1 strain 17 (NC_001806), while + and − indicate transcription from left to right and from right to left, respectively, in the prototype orientation of the HSV-1 genome.
For Northern blot analysis of large RNAs, 2-μg aliquots of each RNA fraction, as well as 10 μg of Century-Plus RNA Markers (Ambion) and 10 μg of Millennium RNA Markers (Ambion), were separated in 1% agarose gels. Electrophoresis, transfer of RNA to the membrane (Pierce), prehybridization, hybridization, and washes were performed using the NorthernMax-Gly kit according to the manufacturer's directions (Ambion). Probes to UL23 (tk) mRNA were generated from a PCR-amplified tk gene product (with the primers 5′-CCGAACCCCGCGTTTATGAACA-3′ and 5′-GTCCACTTCGCATATTAAGG-3′) using the North2South Biotin Random Prime Labeling kit according to the manufacturer's directions (Pierce). Detection of biotin-labeled blots was performed using the Chemiluminescent Nucleic Acid Detection Module according to the manufacturer's directions (Pierce).
RESULTS
Prediction of HSV-1 miRNAs.
In an effort to identify miRNAs encoded by HSV-1, we applied a computational method previously applied to identify plant miRNAs (36), which predicts possible locations of miRNA precursors in the HSV-1 genome using information about the local sequence composition and the predicted secondary structure of miRNA precursors. As a test, we first applied the algorithm to the EBV genome, from which expression of miRNAs had been experimentally validated (26). Our method successfully identified all five validated EBV miRNAs (26) out of a total of seven predicted candidates. The two EBV miRNA precursor candidates predicted by us and not yet validated correspond to EBV genomic coordinates 10411 to 10299 and 139534 to 139443. Both precursors were predicted to be transcribed from right to left on the viral genome.
We then applied the algorithm to the HSV-1 genome (Fig. 1). By searching for 21-nt sequences with hairpin-structured precursors, we identified 11 genomic loci and 13 precursor sequences encoding 24 miRNA candidates (in boldface in Table 1). We numbered the loci 1 to 11 (Fig. 2), with two loci potentially encoding predicted precursors on both strands (4 and 4-RC as well as 6 and 6-RC, respectively) (Table 1). For each of two of the predicted precursors (8 and 10), only one miRNA was predicted, as only one side of the predicted pre-miRNA passed our filters. Predicted precursor 7 was later found to be encoded by the sequence antisense to the UL15 and UL15.5 open reading frames. It passed through our filters, because UL15 encodes spliced mRNA and there is no annotation for UL15.5 in the NCBI sequence. Nevertheless, we decided to investigate 7 along with the other predictions.
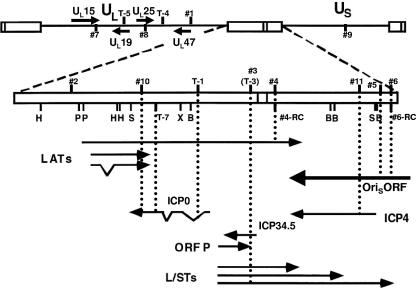
FIG. 2.
Genomic positions of predicted HSV-1 miRNA precursors. The HSV-1 genome in the prototype orientation is shown at the top. UL and US denote the unique sequences of the long (L) and short (S) components of the genome, respectively, which are labeled with solid lines, and the open boxes denote repeat sequences. The L-S junction region is expanded below, with restriction endonuclease cleavage sites abbreviated as follows: B, BamHI; H, HpaI; P, PstI; S, SalI; and X, XhoI. Locations and orientations of transcripts in this region are denoted by solid arrows, with angled lines denoting portions removed by splicing. Numbers indicate locations of predicted miRNA precursors in the viral genome, with those in UL and US shown on the top and those in the repeat sequences below. Those above the line or box are predicted to be transcribed from left to right, while those below indicate the opposite direction of transcription. Dotted lines indicate positions of the predicted miRNA precursors relative to different transcripts. In the unique sequences, only transcripts antisense to predicted miRNA precursors are shown. See Table 1 for exact genomic coordinates of the predicted HSV-1 miRNA precursors.
While our studies were in progress, another group predicted eight HSV-1-encoded miRNA precursors (25). Due to the existence of repeat regions in the HSV-1 genome, these eight actually represent five unique precursors, which we numbered T-1, T-3, T-4, T-5, and T-7, corresponding to 1, 3, 4, 5, and 7 in reference 25 (Fig. 2; Table 1). Of these five, only one (T-3) corresponds to one of our predicted locations (coincidentally, precursor 3).
Some of the predicted HSV-1 miRNAs are predicted to be conserved in HSV-2.
Viral miRNAs have generally been reported to lack sequence conservation across different viral species (25), with the exception of the primate polyomaviruses (33). Because of the high homology between the HSV-1 and HSV-2 genomes, we wanted to investigate whether the HSV-1 miRNAs predicted by us and by others (25) are conserved in HSV-2. Each of the 24 mature HSV-1 miRNA candidates that we predicted was compared with the HSV-2 genome independently. Of these 24, 8 (from predicted precursors 4, 4-RC, 5, 6, 6-RC, and 7) were predicted to be conserved in the HSV-2 genome in terms of both sequence and location (Table 2). (For each of four of the precursors, only one of the predicted miRNAs was predicted to be conserved in HSV-2.) However, the sequences of the predicted precursors are not well conserved in HSV-2, even though the corresponding HSV-2 sequences assume hairpin-like secondary structures that passed our criteria (compare Tables 1 and 2). Of the five HSV-1 miRNA precursors predicted by others (25), two (T-4 and -5) were predicted to be conserved in the HSV-2 genome. The predicted mature miRNAs within these two predicted precursors were also conserved in HSV-2 (Table 2).
TABLE 2.
Genomic coordinates, sequences, and locations of predicted HSV-1 miRNA orthologs in HSV-2
| Precursor no.a | Predicted pre-miRNA sequence, 5′ to 3′ (predicted miRNA ortholog sequence highlighted in boldface), in HSV-2 | Position, orientation | Location |
|---|---|---|---|
| 4 | CCUGCGGGGGGGCUCGGGCCACCUGACCUUCGUAACCUGCACUCAGGUCAGAGCCCCAGACCCCCCGCGGG | 127588-127658, + | Within minor LAT, ∼250 bp upstream of its poly (A) site |
| 4-RC | CCCGCGGGGGGUCUGGGGCUCUGACCUGAGUGCAGGUUACGAAGGUCAGGUGGCCCGAGCCCCCCCGCAGG | 127588-127658, − | Antisense to minor LAT, ∼250 bp upstream of its poly (A) site |
| 5 | CCGCGCGGCAUCUCAUUAGCGCCCGGCGCGGGCGGCUUCCGCUUCCGCCCGCGAUGCUAAUGAGACCCUCGUCGCGG | 132429-132505, + | Within intergenic region between ICP4 and US1 |
| 6 | AGUGAGAACGCGAAGCGUUCGCACUUCGUCCUAAUAGUAUAUAUAUUAUUAGGGCAAAGUGCGAGCGCUGGCGCCCU | 132661-132737, + | Within intergenic region between ICP4 and US1 |
| 6-RC | AGGGCGCCAGCGCUCGCACUUUGCCCUAAUAAUAUAUAUACUAUUAGGACGAAGUGCGAACGCUUCGCGUUCUCACU | 132661-132737, − | Within intergenic region between ICP4 and US1 |
| 7 | AGUACAUGCGGACGUCGGUGUUGGGAGAGACGGUUUCGAUGAGGUUGUUGAGCUGCUCGGACAGAUACUCGACCGGGUCGGUCUGCAGGCGCACCGUCACGGAGACGAGCUCCUGGGACGCCAUGACGCCCCCGGAGUUG | 34517-34656, − | Antisense to UL15 and UL15.5 mRNAs, upstream of UL17 |
| T-4 | GUGAGAACGCGAAGCGUUCGCACUUUGUCCUAAUAAUAUAUAUACUAUUAGGACAAAGUGCGAACGCUUCGCGUUCUCAC | 62890-62969, + | Within intergenic region between UL29 and UL30 |
| T-5 | UCGGCCAUGUUGUGCGCCAGCACCUGCAGCGUGAGCAUGGCGGGCCCGUCGACGACGACGCGCCCGUUGUGGAACAUGCGCUUGACCGUGUUGGCCACCAGAUUGGC | 37865-37971, + | Antisense to ICP5 open reading frame |
These numbers correspond to the locations of predicted pre-miRNA and miRNA sequences in HSV-1. Refer to Table 1 for their sequence, position, and orientation in the HSV-1 genome.
Detection of an miRNA encoded upstream of the transcription start site of the latency-associated transcript (LAT).
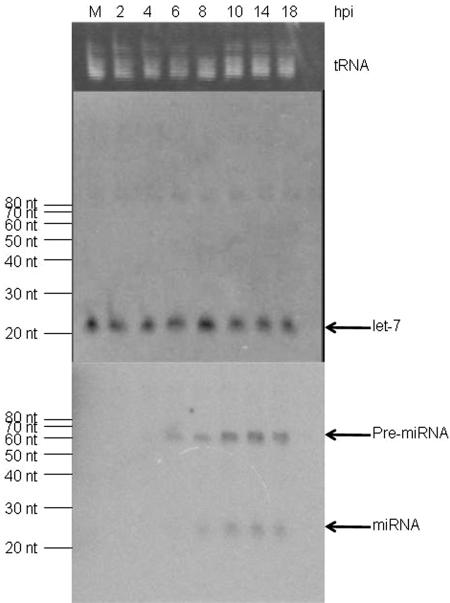
To test whether any predicted HSV-1 miRNAs are expressed during productive infection, Northern blot hybridizations were performed using samples containing small RNAs harvested from HSV-infected Vero cells at different time points postinfection. As a loading control, we stained the gel with ethidium bromide prior to membrane transfer, which confirmed similar amounts of tRNA in each lane (Fig. 3, upper panel). We first investigated whether HSV-1 infection has any effect on the level of a cellular miRNA by Northern blot hybridization, using a probe antisense to let-7. We observed little change, if any, in the level of mature let-7 during infection (Fig. 3, middle panel). Thus, the level of let-7 was useful as a control in our experiments. We sometimes observed a species of small RNA of a size of ∼85 nt. However, this RNA was larger than the size of the let-7 precursor, which is 72 nt (16). Thus, it could be a result of nonspecific hybridization. Nonetheless, it also served as an internal control.
FIG. 3.
Analysis of HSV-1 miRNAs. Small RNAs harvested from mock-infected (M) or from HSV-1-infected cells at the times indicated at the top of the figure were separated by PAGE, stained for tRNA with ethidium bromide (top panel), and blotted to a membrane for hybridization with a probe for let-7 miRNA (middle panel) or a pool of probes for predicted HSV-1 miRNAs (bottom panel). This pool contained probes 1-5′, 2-5′, 3-5′, 4-5′, 5-5′, 6-5′, 7-5′, 8, 9-5′, and 11-5′. The position of tRNA is shown to the right of the top panel. The sizes of RNA markers are indicated to the left of the middle and the bottom panels. The positions of let-7 miRNA, HSV-1 miRNAs, and their precursors are indicated to the right of the middle and the bottom panels, respectively.
Our computational prediction was based on the genome sequence of strain 17, the only complete HSV-1 genome sequence available at the time we started the project. We used this sequence to design probes for the predicted miRNAs. However, our experiment validation used strain KOS. Fortunately, regions containing the predicted precursor sequences in KOS have all been sequenced at least once in an ongoing KOS genome project (A. Griffiths, R. C. Colgrove, Jr., C. Cui, P. A. Schaffer, D. M. Knipe, and D. M. Coen, unpublished results). Except for predicted precursor 9, in which each predicted mature miRNA may have a single base difference in sequence between 17 and KOS, all the other predicted mature miRNA sequences are the same in these two viral strains. Thus, the probes we designed were applicable for detection of HSV-1 strain 17 and KOS miRNAs.
Strand-specific oligonucleotides antisense to each predicted HSV-1 miRNA were end labeled to high specific activity with [γ-32P]ATP. As a first test of our hypothesis, we pooled 10 labeled probes that would not hybridize with each other and performed Northern blot hybridization. No hybridizing species were detected in RNA harvested from mock-infected cells. However, positive signals were detected in RNA harvested from cells infected for 6 to 18 h at positions corresponding to both miRNAs (∼23 nt) and their precursors (∼60 nt; Fig. 3, lower panel). In contrast, when a similar blot was hybridized with a pool of probes complementary to sequences not predicted to be miRNAs, no positive signals specific to virus-infected cells were detected (see supplemental Fig. 1 posted at http://coen.med.harvard.edu).
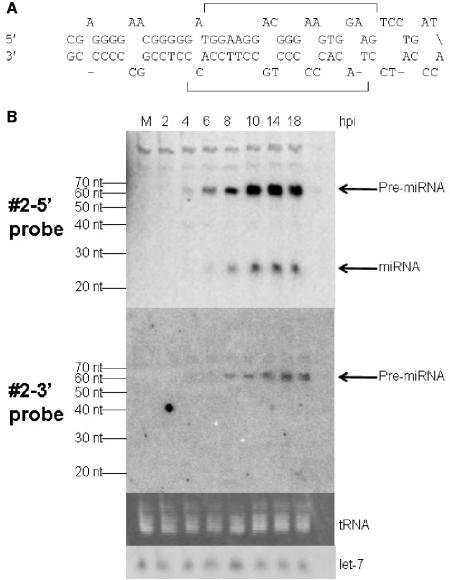
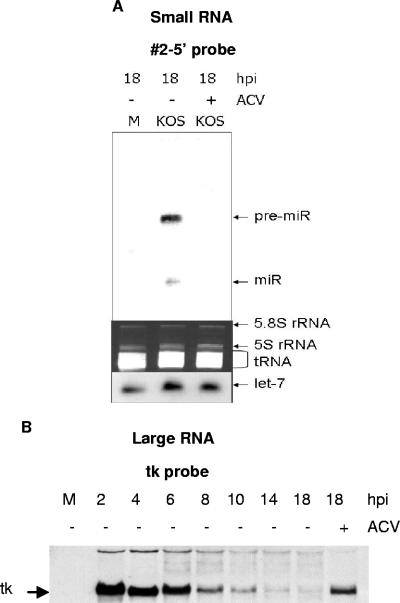
We then repeated the hybridization using each labeled probe complementary to a predicted miRNA individually. One probe, antisense to the predicted miRNA in the 5′ strand of predicted precursor 2 (Fig. 4A), which is encoded ∼450 bp upstream of the transcription start site of the latency-associated transcript (LAT), detected species with the sizes of precursor (∼60 nt) and mature miRNA (∼23 nt; Fig. 4B, upper panel). The signal for the precursor became detectable at 4 h postinfection (hpi), and the signal for the mature miRNA became detectable at 6 hpi. Both signals became strong at 8 hpi and remained at high levels through 18 hpi. The probe antisense to the 3′ strand of predicted precursor 2 (Fig. 4A) detected a species from infected cells the same size as the precursor detected using the 5′ probe but did not detect a mature miRNA (Fig. 4B, second panel). This is exactly the pattern of hybridization expected for a mature miRNA that is encoded in the 5′ strand of precursor 2. This RNA meets expression criterion A (detection of a distinct ∼22 nt RNA) and biogenesis criterion C (prediction of a potential fold-back precursor) established for miRNAs by Ambros et al. (3). Moreover, we detected the predicted precursor. We designated this miRNA as HSV-1 miR-H1 (hsv1-miR-H1) according to the microRNA annotation system (3), and it has been deposited in the microRNA registry (13) with accession no. MI004730.
FIG. 4.
Identification of an HSV-1 miRNA. (A) The sequence of predicted HSV-1 miRNA precursor 2 is shown. The upper strand is the 5′ strand, and the lower strand is the 3′ strand. The predicted mature miRNAs, which were used to design probes, are indicated with brackets. (B) Northern blot analysis of predicted HSV-1 miRNA precursor 2 using small RNAs harvested from mock-infected (M) or HSV-1-infected cells at the times indicated at the top of the figure. The top panel shows a phosphorimage of the pattern of hybridization using the probe to the 5′ strand of predicted precursor 2, and the next panel shows the pattern using the probe to the 3′ strand. The third panel shows a photograph under UV light of a portion of the ethidium bromide-stained gel prior to membrane transfer, and the bottom panel shows a phosphorimage of the pattern of hybridization using a let-7 probe. The sizes of RNA markers are indicated to the left of the top and the second panels. The positions of HSV-1 pre-miRNA, miRNA, tRNA, and let-7 are indicated to the right of the figure.
HSV-1 miR-H1 and its precursor are late gene products.
The time course of expression of HSV-1 miR-H1 and its precursor resembled that of a late gene product, being detectable starting at 4 hpi and remaining at high levels after 8 hpi (Fig. 4B). In contrast, an early transcript, tk, was detected by 2 hpi but declined in abundance after 4 hpi (Fig. 5B). We therefore investigated whether, like late gene products, the accumulation of HSV-1 miR-H1 was affected by an inhibitor of viral DNA synthesis, acyclovir (ACV). At 18 hpi, neither the precursor nor the mature form could be detected in RNAs extracted from Vero cells infected with HSV-1 in the presence of 100 μM ACV (Fig. 5A). In contrast, tk mRNA was detected in the presence of ACV, as expected (Fig. 5B). These data and the time course of expression (Fig. 4B) indicate that HSV-1 miR-H1 and its precursor are expressed with late kinetics.
FIG. 5.
Effect of acyclovir (ACV) on HSV-1 miR-H1 expression. (A) Small RNAs were harvested from mock-infected (M) or from HSV-1-infected cells in the absence (−) or presence (+) of ACV at 18 h. The top panel shows a phosphorimage of the pattern of hybridization with probe 2-5′. The middle panel shows a photograph under UV of a portion of ethidium bromide-stained gel prior to membrane transfer. The bottom panel shows a phosphorimage of the pattern of hybridization using a let-7 probe. The positions of pre-miR-H1 (pre-miR), miR-H1 (miR), 5.8S rRNA, 5S rRNA, tRNA, and let-7 are indicated to the right of the figure. (B) Large RNAs were harvested from mock-infected (M) or from HSV-1-infected cells in the absence (−) or presence (+) of ACV at the times indicated at the top of the figure, separated by agarose gel electrophoresis, and blotted to membrane for hybridization with a tk probe. The position of tk mRNA is indicated to the left of the phosphorimage.
Detection of small RNA species using other HSV-1 miRNA candidate probes.
We then investigated whether we could detect small RNAs corresponding to other predicted HSV-1 miRNAs. Results from such experiments are presented in Fig. 6. Probes antisense to both the 5′ and the 3′ strands of predicted precursor 4 detected a species of ∼70 nt, starting at 10 hpi and accumulating through 18 hpi (Fig. 6A). The 5′ probe also detected a larger species of RNA in both mock-infected and infected cells, evidently due to nonspecific hybridization (Fig. 6A). However, we did not definitively detect a species with the size of mature miRNA using either the 5′ or the 3′ probe. A probe antisense to the 5′ strand of predicted precursor 6-RC detected virus-specific species, one with a size of ∼38 nt, starting at 8 hpi and remaining at a constant level through 18 hpi, the other of ∼110 nt, starting at 10 hpi and accumulating through 18 hpi (Fig. 6B). Additionally, a “smear” of RNAs migrating between these two was detected at late times (Fig. 6B). Using probes to the 5′ (see supplemental Fig. 2 posted at http://coen.med.harvard.edu) and to the 3′ strands of predicted precursor 7, we detected a small RNA species ∼100 nt in size (Fig. 6C). This small RNA species was detected as early as 4 hpi and continued to accumulate through 18 hpi.
FIG. 6.
Identification of small RNA species specific to HSV-1 infection. Small RNAs harvested from mock-infected (M) or from HSV-1-infected cells at the times indicated at the top of the figure were separated by PAGE, stained with ethidium bromide (UV visualization of a portion of the gel is shown in the third gel in panel A and in the second gels in panels B, C, and D), and blotted to membrane for hybridization with 4-5′ probe (A, top gel), 4-3′ probe (A, second gel), 6-RC probe (B, top gel), 7-3′ probe (C, top gel), and T-1 probe (D, top gel). The sizes of RNA markers are indicated to the left of the phosphorimages. The position of tRNA and the putative pre-miRNAs are indicated to the right of the figure. (A to C) The membrane was stripped and hybridized with a probe to let-7, the position of which is also shown to the right of the bottom gels.
To investigate whether T-1, T-4, T-5, and T-7, predicted by Pfeffer et al. (25), are expressed during infection of cultured cells, we analyzed the secondary structure of these miRNA precursors using mfold (42), identified the best scored 21-nt sequences as miRNA candidates from both arms of the hairpin-like precursor, designed 31-nt probes antisense to the 21-nt sequences plus 5 nt on each end, and performed Northern blot hybridizations. Using a probe for T-1, we detected small RNA species in RNA extracted from infected cells. One species was ∼110 nt in size, became detectable at 8 hpi, and accumulated through 18 hpi (Fig. 6D). After prolonged exposure of the same blot, smears of smaller RNA species of ∼40 to 100 nt could be seen from 8 to 18 hpi (Fig. 6D and data not shown).
Probes antisense to other predicted miRNA precursors did not detect any small RNA species specific to KOS-infected Vero cells (see supplemental Fig. 2 posted at http://coen.med.harvard.edu).
DISCUSSION
When we started this project, the only virus known to encode miRNAs was EBV, a gammaherpesvirus, and there was no computational method designed to predict viral miRNAs. In this study, we applied a computational method to predict miRNAs encoded by HSV-1 and experimentally identified a bona fide HSV-1 miRNA, HSV-1 miR-H1. This miRNA is encoded upstream of the LAT transcription start site and is expressed as a late gene product. Thus, miRNAs are expressed by all three herpesvirus subfamilies (6, 25, 26, 29, and this report). We also detected several other small RNAs encoded by HSV, some of which may be pre-miRNAs. We discuss our computational and experimental findings below.
Computational prediction of herpesvirus miRNAs.
The computational method we used was very similar to one applied previously to predict plant miRNAs (36). However, due to the GC-rich nature of the HSV genome, the upper limit of the GC content filter was extended from 60% to 75%. To limit the number of candidates to a reasonable number to investigate using Northern blotting, we focused our search only within non-protein-coding regions. Even with the limitations of the GC content filter and genomic location requirement, the computational method successfully identified all five previously identified EBV miRNAs out of a total of seven predicted candidates. The remaining two candidates could be false positives in the prediction, but it is also possible that they are expressed, either at low levels or in specific cells under specific conditions that have not yet been investigated.
Even though we decided to focus our attention on non-protein-coding regions of the HSV-1 genome in this study, we cannot eliminate the possibility that HSV miRNAs might be expressed from protein-coding regions, too. In fact, we detected a possible pre-miRNA using probes to predicted precursor 7, which slipped through our filter even though it is antisense to UL15. We are currently investigating other potential miRNA candidates from protein-coding regions of HSV-1.
It is interesting to compare our computational method to predict HSV-1 miRNAs with that applied by Pfeffer et al. (25). Pfeffer et al. used a set of properties ascribed to known miRNAs to train a support vector machine to identify miRNAs, while we predicted miRNAs solely based on the local sequence composition and secondary structure. The two methods identified only one pre-miRNA in common, from location 3. There are several specific differences between the two methods which may account for the different miRNA candidates identified. The method of Pfeffer et al. did not eliminate candidates with extended “UA” repeats or with perfect pairing in hairpin structures. We eliminated candidates with these features because they have not been observed in experimentally verified miRNAs. The method of Pfeffer et al. also did not eliminate sequences within or antisense to protein-coding regions. Presumably our predicted miRNAs were not included in those predicted by Pfeffer et al. because they did not pass one of their criteria, including, but not limited to, the free energy of folding; the length of the stem-loop; the length of the longest symmetrical stem; the count of A, C, G, and U nucleotides in the symmetrical stem; and the number of A-U, G-C, and G-U pairs in the predicted minimal energy structure (25).
Conservation of predicted HSV-1 miRNAs in HSV-2.
It is interesting that 8 out of the 24 HSV-1 candidate miRNAs identified by our computational method are, by our criteria, conserved in the HSV-2 genome (Table 2). Even though the precursor sequences of these orthologous miRNAs in HSV-2 might not be highly homologous to those in HSV-1, they passed our criteria for hairpin-like secondary structures. This contrasts with the candidate HSV-1 miRNAs predicted with the other method (25), two of which were predicted to be conserved at both the precursor- and the mature-form levels (Table 2). The ortholog of T-4 in the HSV-2 genome (Table 2) predicted by us was the highest scored HSV-2 miRNA candidate predicted by Pfeffer et al. (25), while the ortholog of T-5 was not included in their HSV-2 list (25). This discrepancy may be explained by the different algorithms used in predicting HSV miRNAs. We used cross-species conservation analysis to predict HSV-2 miRNAs based on the prediction of HSV-1 miRNAs, while Pfeffer et al. predicted miRNAs encoded by these two viruses in parallel, without consideration of possible conservation between the two viruses (25). Given the precedent from highly homologous primate polyomaviruses (33), conservation analysis may be helpful in identifying authentic viral miRNAs with conserved functions. This conservation lends support to the hypothesis that the small RNAs detected with probes for predicted precursors 4, 6-RC, and 7 (Fig. 6) are functional pre-miRNAs, as they are predicted to be conserved between HSV-1 and -2 (Table 2).
Does HSV infection affect the cellular miRNA machinery?
We observed a constant level of let-7, a cellular miRNA, during the course of HSV-1 infection (Fig. 3, middle panel). This is in contrast to most cellular mRNAs, which are destabilized and degraded by the virion host shutoff protein (vhs; reviewed in reference 31). Our stocks of strain KOS exhibit vhs activity (C. Cui and D. M. Coen, unpublished results). One possibility is that let-7 and other miRNAs might be too small to be recognized by vhs. Another possibility is based on the recent report of the interaction between vhs and cellular translation initiation factors (11), which suggests that vhs might specifically target translating mRNAs. In either case, miRNAs would remain unscathed. A third possibility is that let-7 does turn over during HSV infection but its biosynthesis is not affected by HSV, so that the amounts of let-7 are replenished.
On the other hand, it is striking that pre-miR-H1 is much more abundant at every time point than its mature form (Fig. 4B, upper panel) (although this was also observed for a simian virus 40 miRNA [33]). Moreover, probes for other candidate miRNAs detected species that could be pre-miRNAs but did not detect mature miRNAs (Fig. 6). If, in fact, miRNAs are stable during HSV infection, these observations raise the possibility that HSV interferes with miRNA biogenesis, particularly cleavage by Dicer. Alternatively, Dicer itself may be unaffected by HSV infection, and the viral pre-miRNAs may simply be relatively poor substrates for Dicer (34).
Biogenesis of HSV-1 miR-H1.
An obvious issue regarding any miRNA is how it is expressed. For HSV-1 miR-H1, we have defined its precursor and categorized these RNAs as late gene products. HSV-1 miR-H1 is encoded ∼450 bp upstream of the LAT transcription start site (9). There are two reports of transcripts that could serve as pri-miRNAs from which pre-miR-H1 could be derived (30, 41). One possible pri-miR-H1 is a 1.8-kb polyadenylated transcript (30). In line with this idea, both the 1.8-kb transcript (30) and HSV-1 miR-H1 are expressed with late kinetics. However, this transcript is not abundant (30, 41). Another pri-miR-H1 candidate is a more abundant 0.7-kb transcript (41), which has been reported to encode a protein called UOL (24). However, this transcript was reported to be absent in cells infected with KOS (41), the strain used in this study. The biogenesis of HSV-1 miR-H1 requires more investigation.
Other small RNA species detected using probes antisense to HSV-1 miRNA candidates.
Aside from HSV-1 miR-H1, we detected several small RNA species with interesting properties (Fig. 6). For predictions 4, 6-RC, 7, and T-1, we detected RNAs ranging in size from ∼70 to 110 nt. In the case of precursors 4 (∼70 nt) and 7 (∼100 nt), the size is somewhat smaller than what we predicted (Table 1). However, we note that a number of other experimentally identified pre-miRNAs, including HSV-1 pre-miR-H1, Kaposi's sarcoma-associated herpesvirus pre-miR-K5 (6), and simian virus 40 pre-miRNA (33), are smaller than their computationally predicted counterparts. This may be due to Drosha cleaving within rather than at the ends of hairpin structures, as has been suggested for human let-7 pre-miRNA (16). In the cases of 6-RC and T-1, the sizes (∼110 nt) are larger than those predicted by us and others (Table 1). Although this may lessen the likelihood that these two RNAs are bona fide pre-miRNAs, the sizes do fall within the range of experimentally identified pre-miRNAs (3). Perhaps the primary transcripts for these species fold into longer hairpins than are predicted. Thus, it is certainly possible that one or more of these small RNAs are, indeed, pre-miRNAs, a possibility that is enhanced by the predicted conservation of 4, 6-RC, and 7 in HSV-2 (Table 2).
For prediction 6-RC, we also detected a virus-specific RNA of ∼38 nt, which is larger than any known miRNA. Moreover, this RNA was detected before the larger RNA species, suggesting that they are not related in a precursor-product relationship. For T-1, we observed a smear of species at late times that may reflect processing events.
Interestingly, the small RNAs corresponding to 4 and T-1 could be derived from the minor 8.3-kb LAT RNAs (9, 23, 43). Precursor 4 could also be derived from L/S junction spanning transcripts (L/STs) (18, 38). The small RNAs corresponding to 6-RC might be derived from a primary transcript called OriSORF (15). We are unaware of a possible primary transcript for precursor 7.
Failure to detect potential mature miRNAs derived from these putative pre-miRNAs and other predicted miRNAs may be due to our assay being insufficiently sensitive. For example, the actual mature miRNA might be positioned a few nucleotides shifted from our prediction, so that the number of nucleotides complementary between the probe and the miRNA was too low. Also, all of our experiments entailed wild-type virus infection of cultured Vero cells. It is possible that certain miRNAs are expressed by the virus only in certain cell types under certain conditions. For example, the candidate miRNA from location 3 may arise from minor LATs (9, 23, 43) or from the L/STs (18, 38), neither of which is expressed abundantly in cells productively infected with wild-type virus (18, 38). It is also possible that certain predicted miRNAs, particularly those that could be derived from LATs, are expressed during latent infection. We are investigating these possibilities.
Possible biological roles of HSV-1 miRNAs.
A second obvious issue regarding any miRNA is its function. Of all the viral miRNAs identified to date, only one has been shown to have a target, which is a viral mRNA with perfect complementarity to the miRNA (33). Others have been proposed to target a viral mRNA (26) or predicted to target cellular mRNAs (6). Upon inspection of the HSV-1 genome, we did not find any transcripts with exact complementarity to our predicted miRNAs other than those encoded from the strand opposite the predicted miRNAs (Fig. 2). Of the small RNAs we detected, the possible pre-miRNAs of 7 and T-1 could target UL15 and UL15.5 (4) and intron-containing ICP0 transcripts (7), respectively. Although no mature miRNA has been detected for these predictions, recent studies suggest that pre-miRNAs may be able to be incorporated into RISC as the source of siRNA (12, 22). A computational analysis indicates that our predicted HSV-1 miRNAs do not have perfect complementarity to human mRNAs (G. Li, C. Cui, D. M. Coen, and X.-J. Wang, unpublished results). We have used computational methods to search for human and HSV-1 mRNAs that could form imperfect duplexes with our predicted HSV-1 miRNAs (G. Li, C. Cui, D. M. Coen, and X.-J. Wang, unpublished). These are currently under investigation. Identification of the mRNA targets of HSV-1 miR-1 and the other candidate HSV-1 miRNAs is clearly a critical goal.
Even though we could not find an ortholog of HSV-1 miR-H1 in the HSV-2 genome, it is still possible that an miRNA with similar function but low homology to HSV-1 miR-H1 is expressed by HSV-2. However, sequence alignment (data not shown) of the HSV-1 and -2 genomes reveals that the 4-kb region encompassing HSV-1 miR-H1 has, on average, <50% sequence homology, much lower than the overall 75% sequence homology between these two viruses. This suggests that HSV-1 miR-H1 might be unique to HSV-1. If so, this miRNA might possibly contribute to biological differences between HSV-1 and -2. It is interesting that substitution of a 2.8-kb region from HSV-1 immediately downstream of pre-miR-H1 for native HSV-2 sequences resulted in an otherwise HSV-2 virus that exhibited an HSV-1 phenotype for reactivation from latency in animal models of recurrent ocular and genital herpes (40).
At present, there are few genetic clues for biological activities of HSV miRNAs. The DNA sequence encoding HSV-1 pre-miR-H1 spans the 5′ untranslated region and the open reading frame of UOL (24). It is also part of a segment of the LAT promoter that has been found to enhance expression in productively infected neuronal cells and acutely infected ganglia (10). There is a deletion mutant that removes a 437-bp fragment that includes the pre-miR-H1 sequence (21). This mutant, 17ΔS/N, expresses wild-type levels of the 2.0-kb LAT in latently infected mice (21) and rabbits (14) and reactivates with normal kinetics during explant cocultivation (14, 21). However, it exhibits significantly reduced ocular reactivation following adrenergic stimulation of latently infected rabbits (14). Precise genetic experiment will be required to dissect whether this phenotype results from effects on the LAT promoter, UOL, or HSV-1 miR-H1. Regardless, we hypothesize that HSV-1 miRNAs are likely to regulate viral and host gene expression and play important roles in HSV-1 replication and pathogenesis.
Acknowledgments
We thank Qing Xu for help in forming the collaboration and Robert C. Colgrove, Jr., Keith Ketterer, Yang Mao, and Lichen Chen for providing strain KOS genome sequencing results and for technical assistance in accessing and analyzing the KOS genome sequences.
This work was supported by NIH grants to D.M.C. (RO1 AI26126 and PO1 NS35138) and by a research grant from the Chinese Academy of Sciences to X.-J.W.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros, V. 2004. The functions of animal microRNAs. Nature 431:350-355. [DOI] [PubMed] [Google Scholar]
- 3.Ambros, V., B. Bartel, D. P. Bartel, C. B. Burge, J. C. Carrington, X. Chen, G. Dreyfuss, S. R. Eddy, S. Griffiths-Jones, M. Marshall, M. Matzke, G. Ruvkun, and T. Tuschl. 2003. A uniform system for microRNA annotation. RNA 9:277-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines, J. D., C. Cunningham, D. Nalwanga, and A. Davison. 1997. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J. Virol. 71:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 6.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 102:5570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, K. L., and B. Roizman. 1996. Alternatively spliced mRNAs predicted to yield frame-shift proteins and stable intron 1 RNAs of the herpes simplex virus 1 regulatory gene alpha 0 accumulate in the cytoplasm of infected cells. Proc. Natl. Acad. Sci. USA 93:12535-12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coen, D. M., H. E. Fleming, Jr., L. K. Leslie, and M. J. Retondo. 1985. Sensitivity of arabinosyladenine-resistant mutants of herpes simplex virus to other antiviral drugs and mapping of drug hypersensitivity mutations to the DNA polymerase locus. J. Virol. 53:477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobson, A. T., F. Sederati, G. Devi-Rao, W. M. Flanagan, M. J. Farrell, J. G. Stevens, E. K. Wagner, and L. T. Feldman. 1989. Identification of the latency-associated transcript promoter by expression of rabbit beta-globin mRNA in mouse sensory nerve ganglia latently infected with a recombinant herpes simplex virus. J. Virol. 63:3844-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobson, A. T., T. P. Margolis, W. A. Gomes, and L. T. Feldman. 1995. In vivo deletion analysis of the herpes simplex virus type 1 latency-associated transcript promoter. J. Virol. 69:2264-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, P., D. N. Everly, Jr., and G. S. Read. 2005. mRNA decay during herpes simplex virus (HSV) infections: protein-protein interactions involving the HSV virion host shutoff protein and translation factors eIF4H and eIF4A. J. Virol. 79:9651-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory, R. I., T. P. Chendrimada, N. Cooch, and R. Shiekhattar. 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123:631-640. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths-Jones, S. 2004. The microRNA registry. Nucleic Acids Res. 32:D109-D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill, J. M., H. H. Garza, Jr., Y.-H. Su, R. Meegalla, L. A. Hanna, J. M. Loutsch, H. W. Thompson, E. D. Varnell, D. C. Bloom, and T. M. Block. 1997. A 437-base-pair deletion at the beginning of the latency-associated transcript promoter significantly reduced adrenergically induced herpes simplex virus type 1 ocular reactivation in latently infected rabbits. J. Virol. 71:6555-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubenthal-Voss, J., L. Starr, and B. Roizman. 1987. The herpes simplex virus origins of DNA synthesis in the S component are each contained in a transcribed open reading frame. J. Virol. 61:3349-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutvágner, G., J. McLachlan, A. E. Pasquinelli, É. Bálint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834-838. [DOI] [PubMed] [Google Scholar]
- 17.Lai, E. C., P. Tomancak, R. W. Williams, and G. M. Rubin. 2003. Computational identification of Drosophila microRNA genes. Genome Biol. 4:R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, L. Y., and P. A. Schaffer. 1998. A virus with a mutation in the ICP4-binding site in the L/ST promoter of herpes simplex virus type 1, but not a virus with a mutation in open reading frame P, exhibits cell-type-specific expression of γ34.5 transcripts and latency-associated transcripts. J. Virol. 72:4250-4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, Y., M. Kim, J. Han, K.-H. Yeom, S. Lee, S. H. Baek, and V. N. Kim. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23:4051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim, L. P., M. E. Glasner, S. Yekta, C. B. Burge, and D. P. Bartel. 2003. Vertebrate microRNA genes. Science 299:1540. [DOI] [PubMed] [Google Scholar]
- 21.Maggioncalda, J., A. Mehta, O. Bagasra, N. W. Fraser, and T. M. Block. 1996. Herpes simplex virus type 1 mutant with a deletion immediately upstream of the LAT locus establishes latency and reactivates from latently infected mice with normal kinetics. J. Neurovirol. 2:268-278. [DOI] [PubMed] [Google Scholar]
- 22.Maniataki, E., and Z. Mourelatos. 2005. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 19:2979-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell, W. J., R. P. Lirette, and N. W. Fraser. 1990. Mapping of low abundance latency-associated RNA in the trigeminal ganglia of mice latently infected with herpes simplex virus type 1. J. Gen. Virol. 71:125-132. [DOI] [PubMed] [Google Scholar]
- 24.Naito, J., R. Mukerjee, K. R. Mott, W. Kang, N. Osorio, N. W. Fraser, and G.-C. Perng. 2005. Identification of a protein encoded in the herpes simplex virus type 1 latency associated transcript promoter region. Virus Res. 108:101-110. [DOI] [PubMed] [Google Scholar]
- 25.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grässer, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 26.Pfeffer, S., M. Zavolan, F. A. Grässer, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 27.Pilger, B. D., C. Cui, and D. M. Coen. 2004. Identification of a small molecule that inhibits herpes simplex virus DNA polymerase subunit interactions and viral replication. Chem. Biol. 11:647-654. [DOI] [PubMed] [Google Scholar]
- 28.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., Lippincott Williams & Wilkins, Philadelphia, Pa.
- 29.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:9301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh, J., and E. K. Wagner. 1993. Transcriptional analysis of the herpes simplex virus type 1 region containing the TRL/UL junction. Virology 196:220-231. [DOI] [PubMed] [Google Scholar]
- 31.Smiley, J. R., M. M. Elgadi, and H. A. Saffran. 2001. Herpes simplex virus vhs protein. Methods Enzymol. 342:440-451. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan, C. S., and D. Ganem. 2005. MicroRNAs and viral infection. Mol. Cell 20:3-7. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan, C. S., A. T. Grundhoff, S. Tevethia, J. M. Pipas, and D. Ganem. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435:682-686. [DOI] [PubMed] [Google Scholar]
- 34.Vermeulen, A., L. Behlen, A. Reynolds, A. Wolfson, W. S. Marshall, J. Karpilow, and A. Khvorova. 2005. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA 11:674-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wadsworth, S., R. J. Jacob, and B. Roizman. 1975. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J. Virol. 15:1487-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, X.-J., J. L. Reyes, N.-H. Chua, and T. Gaasterland. 2004. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 5:R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitley, R. J. 2001. Herpes simplex viruses, p. 2461-2509. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., Lippincott Williams & Wilkins, Philadelphia, Pa.
- 38.Yeh, L., and P. A. Schaffer. 1993. A novel class of transcripts expressed with late kinetics in the absence of ICP4 spans the junction between the long and short segments of the herpes simplex virus type 1 genome. J. Virol. 67:7373-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi, R., Y. Qin, I. G. Macara, and B. R. Cullen. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17:3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshikawa, T., J. M. Hill, L. R. Stanberry, N. Bourne, J. F. Kurawadwala, and P. R. Krause. 1996. The characteristic site-specific reactivation phenotypes of HSV-1 and HSV-2 depend upon the latency-associated transcript region. J. Exp. Med. 184:659-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, J., W. Kang, M. E. Marquart, J. M. Hill, X. Zheng, T. M. Block, and N. W. Fraser. 1999. Identification of a novel 0.7-kb polyadenylated transcript in the LAT promoter region of HSV-1 that is strain specific and may contribute to virulence. Virology 265:296-307. [DOI] [PubMed] [Google Scholar]
- 42.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zwaagstra, J. C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, S. C. Wheatley, K. Lillycrop, J. Wood, D. S. Latchman, K. Patel, and S. L. Wechsler. 1990. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. J. Virol. 64:5019-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]