Abstract
Upon entering a cell, alphaherpesvirus capsids are transported toward the minus ends of microtubules and ultimately deposit virus DNA within the host nucleus. The virus proteins that mediate this centripetal transport are unknown but are expected to be either viral tegument proteins, which are a group of capsid-associated proteins, or a surface component of the capsid itself. Starting with derivatives of pseudorabies virus that encode a fluorescent protein fused to a structural component of the virus, we have made a collection of 12 mutant viruses that lack either the VP26 capsid protein or an individual tegument protein. Using live-cell fluorescence microscopy, we tracked individual virus particles in axons following infection of primary sensory neurons. Quantitative analysis of the VP26-null virus indicates that this protein plays no observable role in capsid transport. Furthermore, viruses lacking tegument proteins that are nonessential for virus propagation in cell culture were also competent for axonal transport. These results indicate that a protein essential for viral propagation mediates transport of the capsid to the nucleus.
Alphaherpesviruses are neurotropic pathogens that are the causative agents of both mild (herpes labialis) and severe (herpes keratitis and encephalitis) disease. Manifestation of each of these disease forms is dependent upon the transport of virus particles in axons of peripheral nerves. The initial infection of neurons is mediated by retrograde axonal transport of viral particles from peripheral nerve endings in the skin or mucosa to neuronal cell bodies present in sensory or sympathetic ganglia. The transported virus particle consists of a capsid, which contains the virus genome, and a subset of associated virus “tegument” proteins (10, 14). Virus membrane proteins and additional tegument proteins are shed when the virus first enters the cytosol of a cell. Retrograde axonal transport is dependent upon intact microtubules, and the dynein motor complex is the only known cellular motor that can account for the directed transport of capsids toward the nucleus (13, 21). Support for the involvement of dynein in retrograde capsid transport has been forthcoming in transformed cell lines (8, 22).
How herpesvirus capsids co-opt dynein has yet to be resolved. Several herpesvirus proteins (UL9, UL34, VP11/12, and VP26) bind to components of the dynein motor complex in yeast two-hybrid or in vitro assays (9, 16, 24). The relevance of these interactions is not immediately clear. The UL34 protein is not incorporated into extracellular virions and therefore cannot be associated with capsids during transport to the nucleus (12, 17). The UL9 protein plays an essential role in virus replication but, similar to UL34, has not been reported as a structural component of virions. The VP11/12 protein is not present on capsids undergoing retrograde transport toward the nucleus and is therefore unlikely to participate in this process (10). In contrast, the VP26 protein is a surface component of the capsid and is thus a candidate to recruit dynein (3, 11).
VP26 was further implicated in the process of capsid transport by assembling capsids with a baculovirus expression system. Capsids assembled in the absence of VP26 show a reduced propensity to cluster around the nucleus after microinjection into cells, compared to capsids assembled in the presence of VP26 (9). However, a mutant of herpes simplex virus type 1 (HSV-1) lacking VP26 is transported to sensory ganglia following inoculation into the mouse cornea (4). This indicates either that VP26 does not participate in retrograde capsid translocation during infection or that VP26 is only partly responsible for this process. To clarify the role of the VP26-dynein interaction in retrograde capsid transport, we provide the first examination of the dynamics of VP26-null capsids in living cells and find that VP26 plays no detectable role in capsid transport. Because this finding strengthens a possible role for the tegument proteins in capsid transport, we made a collection of viruses, each with a deletion of one of the tegument genes. We find that, of the 11 tegument proteins that are not essential for virus propagation in cell culture and could therefore be included in this study, none were required for retrograde axonal transport. These findings are consistent with an essential virus protein, such as the VP1/2 tegument protein, being the mediator of intracellular capsid transport to the nucleus.
MATERIALS AND METHODS
Virus and cells.
All viruses were derived from the infectious clone of pseudorabies virus (PRV) strain Becker, pBecker3 (19). Each mutant virus was isolated following transfection of the corresponding infectious clone into pig kidney epithelial (PK15) cells, as previously described (14). Single-step growth curves were used to determine rates of virus propagation, and viral titers were measured by plaque assay as previously described (18).
Dorsal root ganglia explants were isolated from E8-E10 chicken embryos and cultured on poly-dl-ornithine and laminin as previously described (20). Neurons were cultured for 2 to 3 days before being infected.
Construction of recombinant virus strains.
PRV-GS443 and PRV-GS847 encode green fluorescent protein (GFP) and monomeric red-fluorescent protein (mRFP1) fused to the UL35 gene product (VP26 capsid protein), respectively, and were previously described (2, 20, 21). PRV-GS962 encodes mRFP1 fused to the N terminus of the UL36 gene product (VP1/2 tegument protein) and is identical to the previously described PRV-GS935, with the coding sequence for mRFP1 in place of that for the latter's GFP (14). PRV-GS962 was made by RecA-dependent homologous recombination by previously described methods (18).
Disruption of UL4, UL14, UL21, UL35, UL41, UL46, UL47, UL48, UL49, and UL51 was accomplished by first inserting a kanamycin marker flanked with Flp recombination target (FRT) sites into the infectious clone by RED-GAM mutagenesis, followed by FLP recombinase-mediated excision of the marker, as previously described (14). The mutations are summarized in Table S1 in the supplemental material. In each case, primers were designed to encode 40 bp of homology flanking the gene targeted for deletion. Each primer also encoded an FRT sequence (underlined sequence in Table S1 in the supplemental material) with one FRT site preceded by a stop codon (shown in bold in Table S1 in the supplemental material). The linear PCR product was recombined into either pGS962 (for deletion of UL35) or pGS443 (for deletion of tegument-encoding genes) by RecA-independent homologous recombination in the Escherichia coli strain EL250. The kanamycin marker was subsequently removed by Flp-mediated recombination at the two FRT sites. The resulting infectious clones carry a stop codon and a single 34-bp FRT site in the target gene and, in all but one case (UL14), a deletion within the target coding sequence. A deletion was not introduced into the UL14 coding sequence, due to the proximity of the UL13 and UL15 genes, which overlap the UL14 coding sequence. Deletions removed the entire coding sequence of the target gene when possible but in some instances were designed to avoid polar effects on neighboring genes (UL4, UL47, UL49, and UL51).
Deletion of either the US3 or UL13 gene was achieved by RecA-dependent homologous recombination with the pGS847 infectious clone. The deletion alleles were made with the following primers: for deletion of UL13, 5′-GGAGGAGGCGTGAGCTAACTGCCGTACGAGGTGG-3′ and, for deletion of US3, 5′-CCAACTCGCGCACCATGTAATTGACGTTTGATCCCGTCC-3′. Each primer was paired with a reverse-complement primer and used in a sequential overlap extension PCR to produce the mutant allele, which was subsequently cloned into the pGS284 allelic exchange vector and recombined with pGS847, resulting in pGS950 and pGS1015 (18). The UL13 deletion replaces codons 12 to 278 (of 398 codons total) with a single TAA stop codon, thereby leaving the overlapping UL12 and UL14 genes intact. The US3 deletion replaces codons 2 to 368 (of 390 codons total and relative to the minor US3-coding sequence) with a TAA stop codon.
Fluorescence microscopy.
All images were captured with an inverted wide-field Nikon Eclipse TE2000-U (Sutter Instruments, Novato, Calif.) and a Cascade:650 camera (Photometrics; Roper Scientific). The microscope was housed in a box, with the environment kept at 37°C (Life Imaging Services, Reinach, Switzerland). Images were acquired and processed using the Metamorph software package (Molecular Devices, Downington, Pa.).
Living primary neurons from chicken dorsal root ganglia were imaged in sealed chambers with a 60 × 1.4 numeric aperture oil objective as previously described (21). In axons, capsid transport toward the cell body was recorded by time-lapse fluorescence microscopy of mRFP1 or GFP emissions, and the emissions were tracked and analyzed as previously described (21).
RESULTS
Construction of a fluorescently tagged capsid virus lacking the VP26 capsid protein.
Using time-lapse fluorescence microscopy, we have previously imaged and tracked individual capsids undergoing retrograde axonal transport to the nuclei of sensory neurons in culture (21). Because a recombinant virus encoding the GFP-coding sequence fused to the VP26 gene was used for these studies, this approach is not amenable to the study of a VP26-null virus. However, we have recently determined that two tegument proteins, VP1/2 and UL37, remain associated with capsids during retrograde transport toward the nucleus and, importantly, can be detected in axons only as part of the capsid transport complex (14). Using VP1/2 as an alternative target for tagging capsids with a fluorescent protein, two derivates of the alphaherpesvirus, pseudorabies virus, were made.
The coding sequences for the monomeric red-fluorescent protein were inserted into the 5′ end of the VP1/2 gene (UL36) by RecA-dependent homologous recombination with the pBecker3 infectious clone, resulting in pGS962 (2, 19). Deletion of the entire VP26-coding sequence from pGS962 resulted in pGS1205. Transfection of pGS962 and pGS1205 into pig kidney epithelial (PK15) cells produced the viruses PRV-GS962 and PRV-GS1205, respectively.
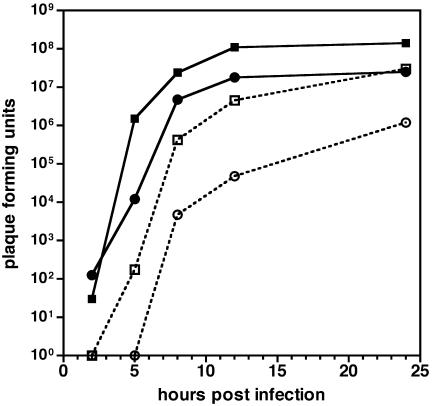
Although a VP26-null strain of HSV-1 has previously been described, the propagation kinetics of a VP26-null alphaherpesvirus have not previously been examined (4). Therefore, the kinetics of virus entry and propagation of PRV-GS962 and PRV-GS1205 were examined by single-step growth analysis in PK15 cells. PRV-GS962 propagation occurred at a rate equivalent to that of our previous reports for the wild-type PRV-Becker (14). Intracellular PFU were detected as early as 2 h postinfection at similar titers for both viruses, indicating that the absence of VP26 did not impact the ability of PRV-GS1205 to reach the nucleus and undergo replication (Fig. 1). We noted that PRV-GS1205 released fewer PFU in the culture media than PRV-962, indicating a role for VP26 in virus egress or capsid/virion stability. In a separate experiment, an approximate twofold decrease in overall burst size was observed with PRV lacking VP26 (PRV-GS962, 6 × 108 PFU/ml; PRV-GS1205, 3 × 108 PFU/ml), similar to that of a previous report for HSV-1 lacking VP26 (4).
FIG. 1.
Single-step growth kinetics of VP26- and VP26-null fluorescent viruses. Virions were harvested from media (dashed lines, open symbols) and PK15 cells (solid lines, filled symbols) at the indicated times. Squares, PRV-GS962; circles, PRV-GS1205.
The VP26 capsid protein does not actively participate in microtubule transport of capsids toward the nucleus.
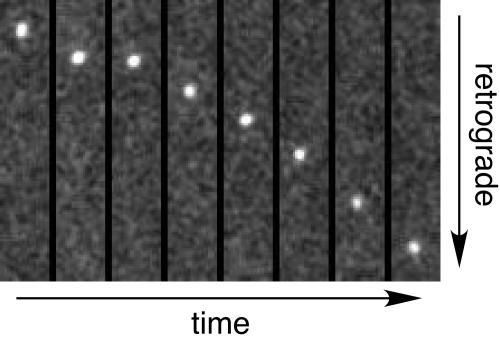
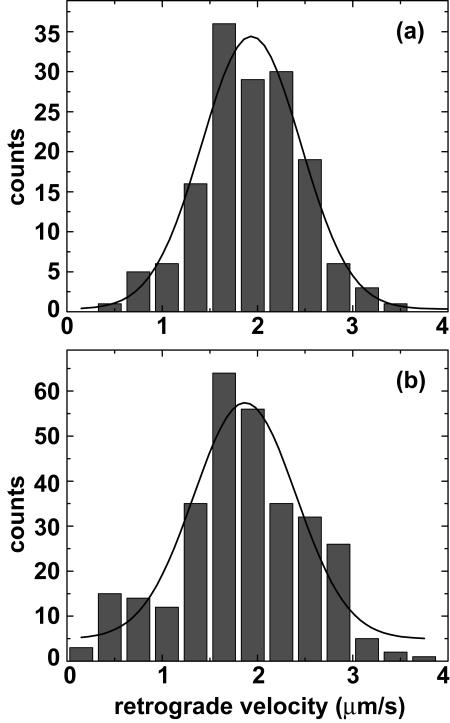
To observe the retrograde transport of capsids, axons of primary sensory neurons were imaged within the first hour postinfection by time-lapse fluorescence microscopy using 200-ms exposure times (5 frames/s), as previously described (21). Capsid-VP1/2 complexes of PRV-GS962 and PRV-GS1205 were both readily observed moving processively in the retrograde direction toward the neuronal cell body (Fig. 2, movie M1 in the supplemental material, and data not shown). To compare the transport dynamics of PRV-GS962 and PRV-GS1205 capsids, individual capsid-VP1/2 fluorescent punctae were tracked for each virus. A statistical analysis of average capsid velocities during continuous runs of uninterrupted retrograde movement was performed (Fig. 3). PRV-GS962 moved with an average velocity of 1.94 ± 0.04 μm/s, with uninterrupted motion proceeding across an average distance of 11.1 ± 0.5 μm (n = 153). PRV-GS1205 velocity was 1.87 ± 0.07 μm/s and occurred across an average distance of 10.1 ± 0.4 μm (n = 300). The differences between the two viruses were determined to be statistically insignificant (t test; alpha = 0.05).
FIG. 2.
VP26-null capsid retrograde axonal transport. Example of virus particle transport resulting from infection of dorsal root sensory neurons with PRV-GS1205 and imaged within the first hour postinfection. A montage of eight frames from a subregion of a time-lapse recording are shown (see movie M1 in the supplemental material for the entire time-lapse recording). Each frame is a 200-ms exposure representing every fourth frame of the original recording (the montage represents a 6.4-s time window). A single VP26-null capsid (mRFP1-VP1/2) complex is shown in the montage. The frames are each 2.7 μm × 15.2 μm.
FIG. 3.
Analysis of capsid transport velocities. Histogram of retrograde transport velocities of individual virus particles resulting from infections of dorsal root sensory neurons with PRV-GS962 (a) and PRV-GS1205 (b). The smooth curve in each panel represents the best-fit Gaussian curve for each sample.
Tegument proteins that are nonessential for virus propagation in cultured cells are dispensable for retrograde axonal transport of capsids.
The failure of the VP26-null mutation to impact retrograde capsid transport supports the possibility that capsid-associated tegument proteins may be required for this process. We therefore made a collection of 11 viruses that each lacked a single tegument protein (see Table S1 in the supplemental material). Each mutant virus genome was confirmed by restriction analysis and DNA sequencing (data not shown).
Infection of cultured dorsal root sensory neurons was performed as described above; however, imaging of axons was performed with 50-ms exposure times (20 frames/s), which was possible due to the brighter fluorescence achieved by fusion of a fluorescent protein to VP26 as opposed to VP1/2. Capsids moving progressively in the retrograde direction were readily observed for each mutant virus, although, as expected, mutant viruses that produced low titers produced fewer observable moving capsids (data not shown). An initial assessment of capsid velocities, which included tracking 30 or more uninterrupted runs of capsid motion from at least 12 recordings each, indicated that transport was not grossly affected by the absence of any of the 11 tegument proteins examined (Table 1).
TABLE 1.
Retrograde transport of capsids following infection of dorsal root sensory neurons
| Virus | Knocked out tegument gene | Avg. capsid velocity (μm/s ± SEM) | No. of capsids tracked | Total no. of runs analyzed |
|---|---|---|---|---|
| PRV-GS443 | 1.91 ± 0.07 | 53 | 104 | |
| PRV-GS717 | UL4 | 1.52 ± 0.09 | 24 | 79 |
| PRV-GS950 | UL13 | 1.69 ± 0.08 | 25 | 55 |
| PRV-GS729 | UL14 | 1.63 ± 0.09 | 18 | 52 |
| PRV-GS762 | UL21 | 1.57 ± 0.09 | 17 | 56 |
| PRV-GS747 | UL41 (VHS) | 1.53 ± 0.08 | 28 | 73 |
| PRV-GS776 | UL46 (VP11/12) | 1.85 ± 0.07 | 37 | 79 |
| PRV-GS731 | UL47 (VP13/14) | 1.60 ± 0.10 | 17 | 45 |
| PRV-GS1081 | UL48 (VP16) | 1.90 ± 0.10 | 25 | 39 |
| PRV-GS764 | UL49 (VP22) | 1.60 ± 0.10 | 12 | 30 |
| PRV-GS735 | UL51 | 1.52 ± 0.08 | 24 | 80 |
| PRV-GS1015 | US3 | 1.64 ± 0.08 | 24 | 60 |
DISCUSSION
The delivery of viral DNA to the host cell nucleus is a critical step in the establishment of a herpesvirus infection. This process is made efficient by the recruitment of the dynein motor complex to capsids deposited in the cytosol following virus entry into cells (8). Association of dynein with capsids is presumably essential for retrograde axonal transport and alphaherpesvirus neurotropism. To better understand the mechanism by which herpesviruses recruit dynein and move intracellularly, we made a collection of fluorescent viruses carrying knockout mutations for the capsid surface protein, VP26, and for each of the viral tegument proteins.
VP26 binds the VP5 major capsid protein at a 1:1 ratio on hexons, resulting in 900 copies of VP26 on the capsid surface and making VP26 an attractive candidate as a recruiter of dynein (1, 23, 26). However, reports on the role of VP26 in capsid transport have yielded somewhat contradictory conclusions. Assembly of capsid structures with or without VP26 in insect cells and subsequent microinjection of the particles into mammalian cells support a role for VP26 in the retrograde transport of capsids along microtubules (9). However, a virus with a deletion of the gene encoding VP26, UL35, is competent for retrograde axonal transport in a mouse model of infection (4). We therefore set out to image and track individual VP26-null capsids during retrograde axonal transport in cultured sensory neurons. Although previous studies imaged capsids in living cells by virtue of a GFP-VP26 fusion, imaging VP26-null capsids was made possible by fusing mRFP1 to the capsid-associated tegument protein, VP1/2 (5-7, 14, 20, 21).
The absence of VP26 was found to have no impact on the kinetics of capsid retrograde transport in axons. This finding was unexpected, as the absence of the 900 copies of VP26 results in a significant alteration in capsid surface topology (26). Yet we find that transport of these aberrant capsids proceeds with kinetics that are indistinguishable from those of fully assembled capsids. The simplest explanation is that the VP26 protein plays no role in the retrograde transport of capsids toward the nucleus. This indicates that another capsid protein or capsid-associated protein (i.e., tegument protein) mediates the relevant interaction with dynein. On the capsid surface, the absence of VP26 at pentons exposes the VP5 major capsid protein. Although VP5 coprecipitates with dynein from infected cells, this interaction is likely indirect, as the capsid vertices are binding sites for the tegument, and capsid-tegument interactions persist after virus entry into cells (14, 24, 25). Our findings further demonstrate that VP1/2 association with capsids is independent of VP26, which is consistent with its binding via the vertices. Therefore, VP5 is probably not available for direct interactions with dynein.
We therefore examined the roles of the tegument proteins in retrograde capsid transport. Eleven mutant viruses were made, each lacking a single tegument protein. Two additional mutant viruses, with the gene encoding either the VP1/2 or UL37 tegument protein deleted, were previously made but could not be included in the current study (15). The VP1/2 tegument protein is required for viral propagation and therefore could not be used to infect cells in the absence of the VP1/2 protein. Similarly, propagation of the ΔUL37 isolate of PRV resulted in titers that were insufficient for the subsequent imaging of capsid transport in our neuronal cultures following infection. Of the remaining 11 tegument-knockout viruses examined, none were notably impaired for retrograde transport. Small variations in the rate of transport were observed among the mutant viruses, but these differences may result from the small number of infected cells from which capsid velocities were analyzed (data not shown). A thorough examination of capsid transport dynamics, similar to that conducted for the VP26-null virus, for all 11 mutant viruses was beyond the scope of this study. However, we expect that the observed variations in transport velocities may not be relevant, as at least one of the mutants with reduced velocity lacked a protein (VP13/14) that is not associated with intracellular capsids after entry into cells and is therefore unlikely to participate in the transport process (10, 14).
Based on the exclusion of the proteins examined in this report from the transport process, a role for the VP1/2 or UL37 tegument proteins in dynein binding and capsid transport is supported. This model is consistent with the finding that both VP1/2 and UL37 remain associated with capsids as they traverse the cytosol to the nucleus (10, 14). We have recently determined that VP1/2, but not UL37, is required for the transport of capsids along microtubules during the egress phase of infection (15). Whether the microtubule-based transport of viral particles following entry and during the egress phase proceeds by a related mechanism is unknown but is of much interest. Although yeast two-hybrid assays have so far failed to identify interactions between dynein and VP1/2 or UL37, we expect that additional genetic manipulations of these viruses will result in the identification of the relevant participants in the microtubule-based transport of herpesvirus capsids (9).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant 1R01AI056346 (to G.A.S.) and NIGMS grant GM-64624-01 (to S.P.G.). S.E.A. was supported by Cell and Molecular Basis of Disease training grant NIHT32GM08061. K.E.C. was supported by training program in Immunology and Molecular Pathogenesis grant NIHT32AI07476. J.I.L. was supported by Viral Replication training grant NIHT32AI060523. G.T.S. was supported by a fellowship from the Helen Hay Whitney Foundation.
We thank Dmitri Petrov for help in data analysis and Bruce Banfield for helpful discussions of the data.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Booy, F. P., B. L. Trus, W. W. Newcomb, J. C. Brown, J. F. Conway, and A. C. Steven. 1994. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus). Proc. Natl. Acad. Sci. USA 91:5652-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen, G. H., M. Ponce de Leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. J. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai, P., N. A. DeLuca, and S. Person. 1998. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247:115-124. [DOI] [PubMed] [Google Scholar]
- 5.Desai, P., and S. Person. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 72:7563-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai, P., G. L. Sexton, J. M. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dohner, K., A. Wolfstein, U. Prank, C. Echeverri, D. Dujardin, R. Vallee, and B. Sodeik. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13:2795-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas, M. W., R. J. Diefenbach, F. L. Homa, M. Miranda-Saksena, F. J. Rixon, V. Vittone, K. Byth, and A. L. Cunningham. 2004. Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J. Biol. Chem. 279:28522-28530. [DOI] [PubMed] [Google Scholar]
- 10.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heilman, C. J., Jr., M. Zweig, J. R. Stephenson, and B. Hampar. 1979. Isolation of a nucleocapsid polypeptide of herpes simplex virus types 1 and 2 possessing immunologically type-specific and cross-reactive determinants. J. Virol. 29:34-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristensson, K., E. Lycke, M. Röyttä, B. Svennerholm, and A. Vahlne. 1986. Neuritic transport of herpes simplex virus in rat sensory neurons in vitro. Effects of substances interacting with microtubular function and axonal flow [nocodazole, taxol and erythro-9-3-(2-hydroxynonyl)adenine]. J. Gen. Virol. 67:2023-2028. [DOI] [PubMed] [Google Scholar]
- 14.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luxton, G. W., J. I. Lee, S. Haverlock-Moyns, J. M. Schober, and G. A. Smith. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Moreno, M., I. Navarro-Lerida, F. Roncal, J. P. Albar, C. Alonso, F. Gavilanes, and I. Rodriguez-Crespo. 2003. Recognition of novel viral sequences that associate with the dynein light chain LC8 identified through a pepscan technique. FEBS Lett. 544:262-267. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, G. A., S. P. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. USA 98:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, G. A., L. Pomeranz, S. P. Gross, and L. W. Enquist. 2004. Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc. Natl. Acad. Sci. USA 101:16034-16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trus, B. L., F. L. Homa, F. P. Booy, W. W. Newcomb, D. R. Thomsen, N. Cheng, J. C. Brown, and A. C. Steven. 1995. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J. Virol. 69:7362-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye, G. J., K. T. Vaughan, R. B. Vallee, and B. Roizman. 2000. The herpes simplex virus 1 UL34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou, Z. H., J. He, J. Jakana, J. D. Tatman, F. J. Rixon, and W. Chiu. 1995. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol. 2:1026-1030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.