Abstract
Cytolytic T cells play a major role in controlling herpes simplex virus type 2 (HSV-2) infections in humans. In an effort to more thoroughly evaluate the response to HSV-2 directly, ex vivo, we developed an enzyme-linked immunospot (ELISPOT) assay that utilized pools of overlapping synthetic peptides presented by autologous dendritic cells to purified CD8+ T cells. Donor response rates to individual open reading frames (ORFs) ranged from fewer than 5% responding to as many as 70% responding, with the greatest frequency of responses (by ORF) being directed against UL39, UL25, UL27, ICP0, UL46, and UL47 in descending order of frequency. HSV-2-seropositive subjects responded to as few as 3 or as many as 46 of the 48 ORFs tested, with a median of 11 ORFs recognized. HLA-B*07 expression correlated with stronger responses overall that were directed primarily against UL49 and UL46. Cumulative precursor frequencies in the blood ranged from 500 to almost 6,000 HSV-2 spot-forming units/106 CD8+ T cells. The magnitude and breadth of the response in the infected population were greater than previously appreciated. Whether this variability in the CD8+ T-cell response within individuals is associated with the frequency of viral reactivation warrants further study.
Several lines of evidence support the importance of T-cell immunity in the resolution of herpes simplex virus (HSV) infections in humans. Patients with T-cell immune deficiencies, whether genetic or acquired, suffer more frequent reactivation and greater disease severity than immunocompetent persons (8). In animal models, deficiencies in CD8+ T cells reduce viral clearance in neural tissue (16, 18). In previous studies, we have shown that the reduction of viral shedding in lesional biopsies coincides with the appearance of HSV-2-specific cytolytic activity in lesions (13). These observations have largely been based upon qualitative assays of T-cell immunity. The last 5 years has seen the development of standardized assays to quantify T-cell responses to viral proteins. While a multitude of these studies have used whole viral antigens to evaluate responses to HSV-1 and HSV-2, little is known about the breadth and magnitude of the response to specific HSV proteins or about how these responses differ between individuals. To explore these issues, we initiated a study to define the predominant responses to specific HSV proteins among HSV-2-seropositive persons.
(This work was presented in part at the 28th International Herpesvirus Workshop, Madison, Wis., 2003 and the 27th International Herpesvirus Workshop, Cairns, Australia, 2002.)
MATERIALS AND METHODS
Human subjects.
Forty donors were recruited for this trial: 33 were attendees of the Virology Research Clinic at the University of Washington, Seattle, and 7 were recruited at Corixa Corp. Subjects electing to participate in the study gave informed consent. Thirty-seven of the donors were HSV-2 seropositive. Infection with HSV-1 and HSV-2 was diagnosed by type-specific immunoblot (1) or glycoprotein G-based (15) immunoassays. HLA typing was performed by low-resolution PCR-based methods at Puget Sound Blood Center, Seattle, WA. High resolution HLA typing was performed for certain HLA alleles (A*02, A*24, and B*07) on selected subjects.
Preparation of PBMC from human subjects.
Subjects were leukapheresed at the Puget Sound Blood Center. Peripheral blood mononuclear cells (PBMC) recovered from the leukapheresis product by Ficoll-Hypaque density gradient separation were washed thoroughly, suspended in 10% dimethyl sulfoxide (DMSO)/50% human serum/40% RPMI 1640, frozen at approximately 2.5 × 107 cells per cryovial, and stored in liquid nitrogen.
Preparation of DC.
PBMC were suspended in RPMI plus 2% human serum (HS) and allowed to adhere to wells of six-well plates for 2 h at 37°C. Nonadherent PBMC were removed and transferred to a second six-well plate, and IL-4 (10 ng/ml) (gift from Immunex, Seattle, WA) and granulocyte-macrophage colony-stimulating factor (GM-CSF; 10 ng/ml) (gift from Immunex, Seattle, WA) were added. Dendritic cell (DC) culture medium (RPMI plus 5% fetal calf serum [FCS], 10 ng/ml IL-4, 10 ng/ml GM-CSF) was added to the first plate. Both plates were cultured at 37°C. The following day, nonadherent cells were removed from the second plate, DC medium was added, and the cells were returned to 37°C. DC were harvested from both plates after a total of 5 to 7 days and used as antigen-presenting cells (APC) in enzyme-linked immunospot (ELISPOT) assays.
Reagents.
Human type AB serum (HS) was prepared by pooling and heat-inactivating serum from groups of 30 to 50 healthy donors. Cell culture medium consisted of RPMI 1640 (Gibco/BRL), 10% HS, 1 × 10−5 M 2-mercaptoethanol (Sigma, St. Louis, MO), 1 × 10−2 M HEPES (Invitrogen, Carlsbad, CA), 1 × 10−4 M sodium pyruvate (Invitrogen, Carlsbad, CA), 2 × 10−3 M glutamine (Invitrogen, Carlsbad, CA), and 50 μg/ml gentamicin (Invitrogen, Carlsbad, CA). Dithiothreitol (DTT; 2 × 10−4 M; Sigma) was added to the ELISPOT medium because sulfhydryl modification after storage in DMSO can significantly reduce the antigenicity of synthetic peptides (4). In preliminary experiments, DTT did not alter any aspect of ELISPOT assays other than significantly improving CD8 T-cell recognition of cysteine-containing epitopes (e.g., UL47/550; see below).
Selection of HSV-2 ORFs for study.
Open reading frames (ORFs) for peptide synthesis (n = 48) were selected from among the known ORFs for HSV-2 (5) at the time the study was initiated (Table 1). Resource limitations necessitated selection of a subset of HSV-2 ORFs. Selection of ORFs was primarily based on published data suggesting human CD8 T-cell responses tend to be specific for immediate-early (IE) and virion proteins (capsid, tegument, and glycoprotein) (6, 7, 10, 11, 14, 19). Five additional ORFs (UL23, UL29, UL39, UL50, and UL55) were selected because they had been discovered, using expression-cloning methods, to be novel antigens recognized by HSV-specific CD4 T cells (9, 17; Hosken et al., unpublished).
TABLE 1.
Overlapping 15-mer peptides synthesized for this study
| HSV-2 ORF | ORF class | Common name | ORF length (aa) | No. of 15-mer peptides | Peptide overlap (aa) | No. of library pools | No. of peptides/library pool |
|---|---|---|---|---|---|---|---|
| UL54 | IE | ICP27 | 512 | 126 | 11 | 2 | 63, 63 |
| RL2 | IE | ICP0 | 825 | 164 | 10 | 3 | 50, 50, 64 |
| US1 | IE | ICP22 | 413 | 81 | 10 | 2 | 41, 41 |
| US12 | IE | ICP47 | 86 | 16 | 10 | 1 | 16 |
| RS1 | IE | ICP4 | 1,318 | 161 | 10 | 5 | 50, 50, 50, 50, 62 |
| UL6 | Capsid | 678 | 134 | 10 | 2 | 67, 67 | |
| UL15 | Capsid | 735 | 145 | 10 | 2 | 73, 74 | |
| UL18 | Capsid | VP23 | 318 | 69 | 11 | 1 | 69 |
| UL19 | Capsid | VP5/ICP5 | 1,374 | 273 | 10 | 5 | 50, 50, 50, 50, 73 |
| UL25 | Capsid | 585 | 115 | 10 | 2 | 50, 65 | |
| UL26/UL26.5 | Capsid | VP21, VP24 | 625 | 123 | 10 | 2 | 61, 62 |
| UL35 | Capsid | VP26 | 113 | 21 | 10 | 1 | 21 |
| UL38 | Capsid | ICP32 | 467 | 92 | 10 | 1 | 92 |
| UL4 | Tegument | 202 | 39 | 10 | 1 | 39 | |
| UL11 | Tegument | 97 | 18 | 10 | 1 | 18 | |
| UL13 | Tegument | 519 | 102 | 10 | 2 | 51, 51 | |
| UL14 | Tegument | 220 | 42 | 10 | 1 | 42 | |
| UL21 | Tegument | 530 | 104 | 10 | 2 | 50, 52 | |
| UL36 | Tegument | ICP1-2 | 3,122 | 623 | 10 | 7 | 89, 89, 89, 89, 89, 89, 89 |
| UL37 | Tegument | 1,114 | 221 | 10 | 4 | 50, 50, 50, 71 | |
| UL41 | Tegument | VHS | 492 | 97 | 10 | 2 | 50, 47 |
| UL46 | Tegument | VP11/12 | 722 | 146 | 10 | 3 | 50, 50, 46 |
| UL47 | Tegument | VP13/14 | 696 | 172 | 11 | 3 | 50, 50, 72 |
| UL48 | Tegument | VP16 | 490 | 96 | 10 | 2 | 48, 48 |
| UL49 | Tegument | VP22 | 300 | 73 | 11 | 1 | 73 |
| UL51 | Tegument | 245 | 47 | 10 | 1 | 47 | |
| US3 | Tegument | 481 | 100 | 10 (11)a | 2 | 33, 67 | |
| US9 | Tegument | 90 | 16 | 10 | 1 | 16 | |
| US10 | Tegument | 303 | 59 | 10 | 1 | 59 | |
| US11 | Tegument | 151 | 29 | 10 | 1 | 29 | |
| UL1 | Glycoprotein | gL | 225 | 43 | 10 | 1 | 43 |
| UL10 | Glycoprotein | gM | 468 | 92 | 10 | 1 | 92 |
| UL20 | Membrane protein | 223 | 43 | 10 | 1 | 43 | |
| UL22 | Glycoprotein | gH | 839 | 166 | 10 | 2 | 83, 83 |
| UL27 | Glycoprotein | gB | 905 | 179 | 10 | 2 | 89, 90 |
| UL44 | Glycoprotein | gC | 481 | 94 | 10 | 1 | 94 |
| UL45 | Membrane protein | 173 | 33 | 10 | 1 | 33 | |
| UL53 | Glycoprotein | gK | 339 | 66 | 10 | 1 | 66 |
| US4 | Glycoprotein | gG | 700 | 138 | 10 | 2 | 69, 69 |
| US5 | Glycoprotein | gJ | 93 | 17 | 10 | 1 | 17 |
| US6 | Glycoprotein | gD | 394 | 77 | 10 | 1 | 77 |
| US7 | Glycoprotein | gI | 373 | 73 | 10 | 1 | 73 |
| US8 | Glycoprotein | gE | 545 | 107 | 10 | 2 | 50, 57 |
| UL23 | Other | 136 | 31 | 11 | 1 | 31 | |
| UL29 | Other | ICP8 | 1,196 | 238 | 10 | 5 | 50, 50, 50, 50, 38 |
| UL39 | Other | ICP6 | 1,142 | 227 | 10 | 4 | 50, 50, 50, 77 |
| UL50 | Other | dUTPase | 369 | 72 | 10 | 1 | 72 |
| UL55 | Other | 187 | 36 | 10 | 1 | 36 |
The N-terminal 33 peptides overlapped by 10 amino acids; the remainder overlapped by 11 amino acids.
Synthetic peptides.
Table 1 summarizes the peptides that were synthesized. Amino acid sequences were derived from the HSV-2 strain HG52 genome (GenBank accession no. NC-001798). Peptides (15 amino acids long, overlapping by 10 or 11 amino acids) were synthesized in crude form by either Multiple Peptide Systems (San Diego, CA) (ORFs UL47, UL54, and ICP0) or Mimotopes (Raleigh, NC) (all other peptides) and lyophilized. Preliminary experiments using UL47 and ICP0 and using epitopes from non-HSV antigens demonstrated that identical results could be obtained with peptides overlapping by either 10 or 11 amino acids; therefore, an overlap of 10 amino acids was selected for the evaluation portion of the study. Each peptide was dissolved at 10 mg/ml in sterile, endotoxin-free DMSO (Sigma), transferred to a 0.5-ml sterile cryovial (Sarstedt, Newton, NC), and stored at 4°C. The mass of each HSV-2 peptide made by Mimotopes was approximately 1 mg. To improve solubilization in DMSO, peptide pellets were initially dissolved in 5% acetonitrile/water and relyophilized to powder form prior to addition of DMSO.
Preparation of peptide pools.
Peptide pools were prepared for each HSV-2 ORF from the individual peptide stocks by combining individual peptides so that each was present at 100 μg/ml in a final volume of 100 μl of DMSO. Peptide pools were stored in cryovials at 4°C. Two types of peptide pools were prepared: library pools and array pools. Library pools were prepared by grouping peptides linearly across an ORF (Table 1). The average library pool contained 50 peptides, a number which was shown to be optimal for detection of most CD8+ T-cell responses. Array pools were prepared by arranging the peptides within a single library pool in a row-and-column format and pooling the peptides in each column or row.
ELISPOT assay for detection of CD8+ T-cell responses.
Because the purpose of the study was to define CD8 responses to HSV-2, purified CD8+ T cells were used as responders in ELISPOT assays. Preliminary experiments also showed that T-cell responses to previously known (6) CD8 epitopes (e.g., UL47/289 and UL47/551) were often at or below the limit of detection when whole PBMC were used, but were significantly above the limit of detection with purified CD8+ responder cells. The use of autologous dendritic cells and highly enriched CD8+ T cells reduces the ELISPOT assay background as compared to when whole PBMC are used (see below).
CD8+ T cells were prepared by positive selection using CD8+ Microbeads (Miltenyi, Auburn, CA) using MACS columns per the manufacturer's directions. T cells enriched by either positive or negative selection using magnetic beads gave identical responses to known CD8 T-cell epitopes in preliminary experiments. Positive selection was used because it yielded higher-purity CD8+ T cells, estimated at >95% in preliminary analyses as assessed by flow cytometry.
Peptides were screened by 24-h coculture of CD8+ T cells (5 × 105/well), autologous dendritic cells (5 × 104/well), and peptides in 96-well ELISPOT plates that had been precoated with anti-human gamma interferon (IFN-γ) antibody 1D1K (mAbTech, Mariemont, OH). Each library pool was screened once. Each peptide in the pool was present at a final concentration of 0.5 μg/ml, which was determined in preliminary experiments to be optimal for detection of responses while minimizing false positives. ELISPOT plates were subsequently developed by sequential incubation with biotinylated 7-B6-1 monoclonal antibody (MAb) (mAbTech), Avidin-peroxidase (Vectastain ABC kit; Vector Labs, Burlingame CA), and AEC substrate (AEC kit; Vector Labs, Burlingame, CA). ELISPOTS were counted with an automated video-microscopy ELISPOT reader (Zeiss, Berlin, Germany). ELISPOTS from selected wells were also visually screened under a stereo microscope to verify the results.
Statistical methods.
Statistical analyses used SAS for Windows 9.1 (SAS Institute Inc., Cary, NC.). CD8 T-cell responses to HSV-2 proteins were analyzed both collectively and separately by ORFs. Each assay run utilized a single medium control. Each medium control (n = 40) had ≤8 spot-forming units (SFU)/106 CD8+ T cells. Their value was not utilized in analyses. The criterion for scoring an ORF as positive, chosen as an absolute number of SFU, was selected to minimize the number of samples from HSV-seronegative persons that produced a response above the cutoff. ORF-level CD8 T-cell responses were examined individually for ORFs found to be positive in at least 25% of HSV-2-seropositive participants. ORFs found to be positive in fewer than 25% of subjects were not evaluated individually. As assays were sometimes performed by dividing the complete ORF into two to seven segments, responses to each ORF were scored as positive when any of the segments were found to be positive. ELISPOT results were analyzed both as frequencies of subjects with positive responses, and as amino acid-adjusted frequency, where the proportion positive was divided by ORF length. Associations with participant-level characteristics were again tested using the chi-square test for frequencies and the Wilcoxon test for length-adjusted frequencies. Multiple-comparison techniques were used to adjust for the potential risk of false significance when testing many ORFs against each characteristic; specifically, the method of Benjamini and Hochberg was applied to control the false discovery rate of 5% per characteristic (2).
The collection of ORF-specific CD8 responses for each participant was graphically summarized using a magnitude-breadth curve. The magnitude-breadth curve for each participant plots the proportion of ORF-specific ELISPOT responses (net SFU/106 CD8 cells) larger than a threshold value, x, as a function of x; therefore, at each point the y axis indicates the average proportion of responses exceeding the response level designated on the x axis. A simple quantitative summary of the overall magnitude of each individual's CD8 response is computed as the area under the magnitude-breadth curve (AUC). The AUC value is equivalent to the average of the ORF-specific responses. To construct the curve, individuals are grouped by common characteristics and the proportions of each individual's responses that exceed a threshold are averaged within the group for each threshold. These average proportions are then plotted against the threshold and, for each group, connected by a line. Differences in this averaged response by other participant-level characteristics (HLA type, gender, etc.) were tested using the Wilcoxon rank sum test. Only HLA types present in at least 25% of subjects were considered.
RESULTS
Frequency of CD8+ T-cell responses to HSV proteins among HSV-2-seropositive persons.
We studied 40 subjects, 37 of whom were HSV-2 seropositive, 2 of whom were HSV seronegative, and 1 of whom was HSV-1 seropositive and HSV-2 seronegative (Table 2). Of the 37 HSV-2 positives, 16 were also HSV-1 seropositive. Sixty-three percent were women, and 84% were Caucasian. Their median age was 34 years (range, 24 to 63 years). Approximately 70% of the subjects who had HSV cultures obtained were HSV-2 culture positive on at least one occasion (Table 2). We compared the individual ELISPOT results for each well containing HSV peptides (library pools) between the 37 HSV-2-seropositive persons and 2 HSV-seronegative persons. Of the 194 separate ELISPOT wells used to screen the two HSV-seronegative subjects, only 1 well exceeded 20 SFU/106 CD8+ T cells. Of the 2,694 separate ELISPOT assays performed from the screening peptide pools, 478 (17%) had readings above 20 SFU/106 CD8+ T cells. Overall, 36 of 37 HSV-2-seropositive subjects responded to at least 1 of the HSV-2 ORFs. Thus, a cutoff of 20 spots/106 CD8+ T cells had 99.5% specificity for defining a positive response to an HSV ORF in our assay.
TABLE 2.
Participant characteristics
| Subject-level characteristic | Result for:
|
||
|---|---|---|---|
| All subjects (n = 40) | HSV-2 positive only (n = 37) | HSV-2 positive with all ORFS completed (n = 21) | |
| Gender [no. (%)] | |||
| Female | 25 (63) | 24 (65) | 15 (71) |
| Male | 15 (38) | 13 (35) | 6 (29) |
| Race [no. (%)] | |||
| White | 32/38 (84) | 30/36 (83) | 18 (86) |
| Nonwhite | 6/38 (16) | 6/36 (17) | 3 (14) |
| Age [median (range) yr] | 34 (24-63) | 35 (24-63) | 37 (24-63) |
| HSV status [no. (%)] | |||
| HSV-2 only | 21 (53) | 21 (57) | 11 (52) |
| HSV-1 and -2 | 16 (40) | 16 (43) | 10 (48) |
| HSV-1 only | 1 (3) | 0 (0) | 0 (0) |
| Negative | 2 (5) | 0 (0) | 0 (0) |
| HSV-2 culture positive [no. (%)] | |||
| Yes | 15 (65) | 15 (68) | 7 (70) |
| No | 8 (35) | 7 (32) | 3 (30) |
| Culture unavailable | 17 | 15 | 11 |
| HLA [no. (%)] | |||
| A*01 | 13 (33) | 12 (32) | 6 (29) |
| A*02 | 23 (58) | 20 (54) | 7 (33) |
| A*03 | 9 (23) | 9 (24) | 7 (33) |
| B*07 | 10 (25) | 10 (27) | 6 (29) |
| B*44 | 11 (28) | 11 (30) | 6 (29) |
| C*01a | 9 (23) | 8 (22) | 3 (14) |
| C*03 | 9 (23) | 8 (22) | 4 (19) |
| C*077 | 20 (50) | 18 (49) | 10 (48) |
| HLA homozygous alleles [no. (%)] | |||
| Yes | 10 (25) | 9 (24) | 6 (29) |
| No | 30 (75) | 28 (76) | 15 (71) |
HLA allele B*27 was present in slightly fewer than 25% of participants, but was closely related to HLA allele C*01 (Fisher's test, P < 0.001). Of the nine participants with C*01, six also had B*27, and only one participant without C*01 had B*27.
Complete analyses of library and array pools were performed on 21 of the 37 HSV-2-seropositive subjects. Sixteen HSV-2 subjects were not fully evaluated due to the unavailability of sufficient PBMC. For the subjects that were evaluated with less than the complete available peptide set, we measured reactivity with peptides covering 11 ORFs in eight subjects and 22 ORFs in the remaining eight subjects. We utilized all of the available data from the 40 subjects for defining the parameters of the ELISPOT assay and for analyses involving percentages out of total ORFs measured (frequencies, amino acid-adjusted frequencies, and magnitude and breadth). We used data from the 21 patients with complete data when computing cumulative SFU and the number of positive responses out of all 97 peptide pools. The demographic and clinical characteristics of the 21 persons tested with the complete peptide set did not differ from those of the complete group of 37 HSV-2-infected subjects (Table 2).
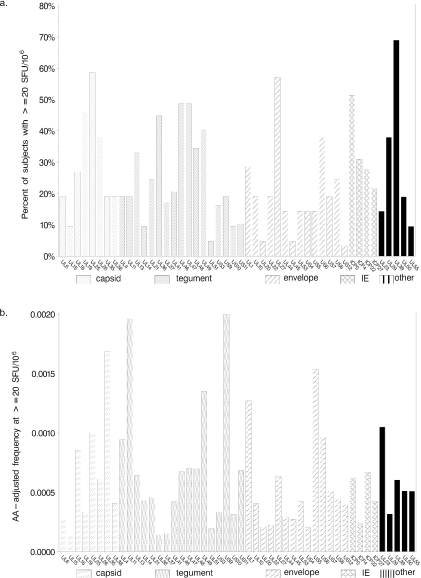
Figure 1a depicts the frequency of CD8+ T cell responses for all 37 HSV-2-seropositive persons. The number of subjects studied for each ORF varied as discussed above. The response rates varied from <5% to 70% of persons, with the highest frequency of responses (by ORF) to UL39, UL25, UL27, ICP0, UL46, UL47, UL19 U36, UL49, and UL26 in descending order of frequency. Responses to these ORFs ranged from 38% to 70% of the tested subjects. When categorized by functional group, 83% of the subjects developed a CD8+ T-cell response to a capsid protein, 83% to at least one tegument protein, 70% to an IE protein, and 45% to an envelope protein. The median number of ORFs to which an individual responded was 11 (range, 3 to 46; Fig. 2). The frequencies of the response were strongly correlated with the length of the ORF and hence the number of peptides utilized to cover an ORF (Spearman correlation statistic, ρ = 0.4, P < 0.001). As such, we also calculated an amino acid-adjusted response rate to each HSV-2 ORF. The amino acid-adjusted frequency of responses to the top ORFs were as follows in descending order: US9, UL11, UL35, US5, UL39, UL1, UL23, UL25, US6, and UL4 (Fig. 1b).
FIG. 1.
Frequency of CD8+ T-cell responses to HSV proteins among 37 subjects with HSV-2 infection. (a) Proportion of subjects with responses (defined as >20 SFU/106 CD8+ cells) to ORFs listed on the x axis. ORFs are organized by structural and functional categories. (b) CD8 responses adjusted for lengths of the predicted HSV-2 proteins. Amino acid (AA)-adjusted frequencies were computed by dividing the proportion of subjects with positive responses by the number of amino acids in the ORF.
FIG. 2.
Breadth of the CD8+ T-cell response to HSV-2. The number of ORFs to which positive (≥20 SFU/106 CD8+ T cells) responses were detected is plotted versus the number of responding subjects among the 21 HSV-2-seropositive subjects with genital herpes who were assayed against the complete set of peptides covering 48 HSV-2 ORFs.
Quantitative responses to HSV proteins.
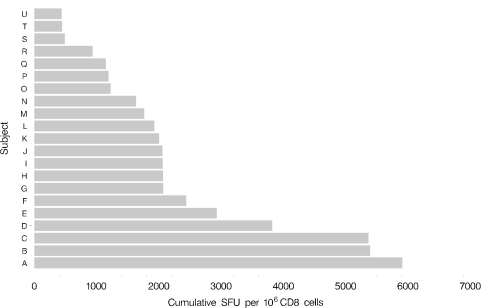
Figure 3 demonstrates the cumulative quantitative response of CD8 T-cell responses in each of the 21 individuals who were screened using all 97 peptide pools. Cumulative responses varied from about 500 SFU/106 CD8 T cells to almost 6,000 SFU/106 cells.
FIG. 3.
Magnitude of the CD8+ T-cell response to HSV-2. Shown are cumulative CD8+ T-cell responses for 21 subjects to peptide pools covering 48 HSV-2 ORFs.
HLA alleles and response.
HLA alleles influenced the breadth and magnitude of the response to HSV antigens. Among HSV-2-seropositive persons, those with the HLA B*07 allele had a stronger overall response to the entire set of tested peptides than did subjects who did not have the HLA B*07 allele (P = 0.008, Fig. 4a). In contrast, the presence of the HLA A*02 allele was associated with a weaker response to the HSV-2 peptide set (P = 0.034, Fig. 4b). We found other prevalent HLA class I alleles that did not correlate with CD8 IFN-γ responses to HSV peptides. These included A*01 (P = 0.66), A*03 (P = 0.27), B*44 (P = 0.40), C*01 (P = 0.37), C*03 (P = 0.57), and C*07 (P = 0.19). Nine persons with homozygous HLA class I A or B alleles had similar CD8+ T-cell responses to those heterozygous at both the A and B loci (P = 0.64). The 16 subjects with HSV-1 coinfection in addition to HSV-2 infection had similar overall magnitude of CD8 responses to those with HSV-2 infection alone (P = 0.92).
FIG. 4.
Magnitude-breadth curve representations of CD8 T-cell reactivity to HSV-2 peptide pools for subsets of HLA-defined subjects. (a) Comparison of HLA-A02 versus non-A02 subjects. (b) Comparison of HLA-B07 versus non-B07 subjects. (c) Comparison of HLA-A02 persons versus non-A02 subjects by class of HSV-2 proteins. (d) Comparison of HLA-B07 versus non-B07 by class of HSV-2 protein.
Subjects with the HLA-B*07 allele tended to have large responses to tegument and IE proteins (Fig. 4C). Specifically, subjects with HLA-B*07 were found to have significantly higher responses to UL49, UL46, and ICP22 compared to persons without HLA B*07 (P < 0.001, 0.003, and 0.033, respectively). Persons with the A*02 allele did not show the same preferential response to tegument or IE proteins (Fig. 4D) or to the specific proteins mentioned above, when compared to persons without HLA A*02. As expected (2), HLA alleles B*07 and C*07 were highly coexpressed (P < 0.001 by Fisher's exact test). No other significant associations were found between HLA alleles A*02, B*07, C*01, and C*07. Persons with the HLA A*02 allele exhibited a significantly higher response to UL47 than those who did not have the A*02 allele, and persons with HLA C*01 exhibited a significantly lower response to UL18 than those without the C*01 allele.
DISCUSSION
This study is the first to evaluate CD8+ T-cell responses to a substantial proportion of the predicted HSV-2 proteome. We used CD8+ cells isolated directly ex vivo to minimize artifacts that could arise from preliminary in vitro restimulation, and peptide sets based on the complete genome of a laboratory strain of HSV-2. Our subjects had well-defined HSV-2 and HSV-1 infection status, and all HSV-2-infected subjects had a clinical history consistent with genital herpes.
We describe several novel findings about the human CD8 T-cell effector response to HSV-2. The diversity and breadth of this response are far greater than previously appreciated. The predominant CD8+ T-cell responses to HSV-2 are directed at IE, tegument, and capsid proteins within the set of 48 proteins we evaluated. Significant variations exist in the breadth of responses and reactivity to individual HSV-2 proteins between individuals. Some individuals demonstrate a low to moderate level of response (20 to 50 SFU/106) per ORF to many HSV-2 proteins, while others exhibit relatively high levels of response to few proteins and others modest responses to only one or two HSV-2 proteins. In addition, the magnitude and breadth of the CD8+ T-cell responses were influenced by HLA type in that persons with HLA A*02 appear to have a relatively lower response to HSV-2 than do people who do not have the HLA A*02 allele, while those with B*07 exhibited a higher frequency of responses to the peptides evaluated than persons who did not have HLA B*07.
Previous studies have reported a narrow diversity in the CD8 T-cell response to HSV. Previously described targets of the CD8 T-cell response include gB2, gD2, gE2, UL46, UL47, UL49, ICP0, ICP4, ICP22, ICP27, UL7, and UL25 (6, 10, 11, 14, 19, 20). Our study documented CD8+ T-cell responses to each of the 48 ORFs studied, expanding by threefold the ORFs to which CD8+ T-cell responses are directed. We have gained an initial insight into what might be the immunodominant and subdominant responses in human populations. It should be noted, however, that we have to date intensively studied only 21 persons and a much more extensive evaluation of a diverse group of persons with HSV-2 is needed before one can adequately characterize the response in human populations. In addition, the population we studied was overrepresented (3) in the frequency of those with HLA A*01, A*03, B*07, B*44, and C*01. Moreover, although our survey of 48 ORFs is more complete than previous studies, there are an additional approximately 30 ORFs that were not evaluated by us and require evaluation before a complete human CD8 T-cell response to HSV-2 can be measured.
As with all assays using peptides to define T-cell responses, the results are a function of the reagents used. The use of shorter peptides or different concentrations could influence the sensitivity and specificity of our assays. When ranking immunodominance between ORFs, we demonstrated a positive correlation between the length of the protein and the proportion of HSV-2-infected persons with positive responses. Hence, immunodominance may be influenced by the availability of peptides for binding to diverse HLA, as well as kinetic and cell biology considerations (23). Moreover, we based our peptides on the gene sequence of the HSV-2 HG52 strain, which is the only fully sequenced HSV-2 isolate in the public domain. While this isolate is derived from a patient with HSV-2 (5), we have recently undertaken sequencing of several of the ORFs from clinical isolates of the more frequently recognized proteins described herein. We clearly find mutations among isolates which can affect T-cell responses (12). Thus, it is likely that while our current study detects a large number of previously unknown responses, our data are likely an underestimation of the entire T-cell response to HSV-2. More complete responses can be defined with further refinement of peptides to the proteins we measured as well as the addition of the approximately 35 HSV-2 proteins we did not include in our study.
We used a novel ELISPOT strategy combining purified CD8+ responder cells and autologous dendritic cells as antigen-presenting cells. This provided clear differentiation between HSV-seronegative and HSV-2-seropositive persons, as shown by the low frequency of CD8+ responses in seronegative persons. As our initial goal was to pick up predominant responses, this was a useful strategy. However, this technique requires more blood volume than does the use of unfractionated PBMC as responder cells. Once we define immunologic responses, comparison with standard PBMC-based assays can be performed. Such an approach might be enhanced by optimizing the sensitivity of the immunogenic peptide pools.
One of the interesting features of our work is the diversity in the immune responses between individuals. This is especially seen in the magnitude-breadth curves. As the diversity of HSV-2 reactivation rates is also great (21, 22), our data suggest that studies defining the association between the magnitude and breadth of the CD8+ T-cell response to HSV-2 and viral reactivation can be performed. Studies to define the association between such responses and viral reactivation are approachable and appear warranted.
Acknowledgments
This study was supported in part by grants 1-R43AI48315-01 and 2-R44AI048315-02 to Nancy Hosken, grants AI30731 and R37 AI42528 to Lawrence Corey, grant AI50132 to David M. Koelle, and grant AI-49394 to Christine Posavad.
We thank Mark Alderson and Peter Probst (Corixa Corporation) and Louis Picker (University of Oregon) for technical advice and scientific discussion, Melody Smith for coordinating leukapheresis and HLA typing, and Karen Kinch for administrative oversight of grants.
REFERENCES
- 1.Ashley, R. A., J. Militoni, F. Lee, A. Nahmias, and L. Corey. 1988. Comparison of Western blot (immunoblot) and glycoprotein G-specific immunodot assay for detecting antibodies to herpes simplex types 1 and 2 in human sera. J. Clin. Microbiol. 26:662-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 3.Cao, K., J. Hollenbach, X. Shi, W. Shi, M. Chopek, and M. A. Fernandez-Vina. 2001. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum. Immunol. 62:1009-1030. [DOI] [PubMed] [Google Scholar]
- 4.Chen, W., J. W. Yewdell, R. L. Levine, and J. R. Bennink. 1999. Modification of cysteine residues in vitro and in vivo affects the immunogenicity and antigenicity of major histocompatibility complex class I-restricted viral determinants. J. Exp. Med. 189:1757-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koelle, D. M., H. Chen, M. A. Gavin, A. Wald, W. W. Kwok, and L. Corey. 2001. CD8 CTL from genital herpes simplex lesions: recognition of viral tegument and immediate early proteins and lysis of infected cutaneous cells. J. Immunol. 166:4049-4058. [DOI] [PubMed] [Google Scholar]
- 7.Koelle, D. M., H. B. Chen, C. M. McClurkan, and E. W. Petersdorf. 2002. Herpes simplex virus type 2-specific CD8 cytotoxic T lymphocyte cross-reactivity against prevalent HLA class I alleles. Blood 99:3844-3847. [DOI] [PubMed] [Google Scholar]
- 8.Koelle, D. M., and L. Corey. 2003. Recent progress in herpes simplex virus immunobiology and vaccine research. Clin. Microbiol. Rev. 16:96-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koelle, D. M., J. M. Frank, M. L. Johnson, and W. W. Kwok. 1998. Recognition of herpes simplex virus type 2 tegument proteins by CD4 T cells infiltrating human genital herpes lesions. J. Virol. 72:7476-7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koelle, D. M., Z. Liu, C. M. McClurkan, M. Topp, S. R. Riddell, E. G. Pamer, A. S. Johnson, A. Wald, and L. Corey. 2002. Expression of cutaneous lymphocyte-associated antigen by CD8+ T-cells specific for a skin-tropic virus. J. Clin. Investig. 110:537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koelle, D. M., Z. Liu, C. L. McClurkan, R. C. Cevallos, J. Vieira, N. A. Hosken, C. A. Meseda, D. C. Snow, A. Wald, and L. Corey. 2003. Immunodominance among herpes simplex virus-specific CD8 T-cells expressing a tissue-specific homing receptor. Proc. Natl. Acad. Sci. USA 100:12899-12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koelle, D. M., C. Liu, B. Byrd, A. Sette, J. Sidney, and A. Wald. 2005. Presented at the 30th International Herpesvirus Workshop, Turku, Finland.
- 13.Koelle, D. M., C. M. Posavad, G. R. Barnum, M. L. Johnson, J. M. Frank, and L. Corey. 1998. Clearance of HSV-2 from recurrent genital lesions correlates with infiltration of HSV-specific cytotoxic T lymphocytes. J. Clin. Investig. 101:1500-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikloska, Z., M. Ruckholdt, I. Ghadiminejad, H. Dunckley, M. Denis, and A. L. Cunningham. 2000. Monophosphosphoryl lipid A and QS21 increase CD8 T lymphocyte cytotoxicity to herpes simplex virus-2 infected cell proteins 4 and 27 through IFN-gamma and IL-12 production. J. Immunol. 164:5167-5176. [DOI] [PubMed] [Google Scholar]
- 15.Morrow, R. A., D. Friedrich, and E. Krantz. 2003. Performance of the Focus and Kalon enzyme-linked immunosorbent assays for antibodies to herpes simplex virus type 2 glycoprotein G in culture-documented cases of genital herpes. J. Clin. Microbiol. 41:5212-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira, R. A., D. C. Tscharke, and A. Simmons. 1994. Upregulation of class I major histocompatibility complex gene expression in primary sensory neurons, satellite cells, and Schwann cells in mice in response to acute but not latent herpes simplex virus infection in vivo. J. Exp. Med. 180:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Posavad, C. M., A. Wald, N. Hosken, M.-L. Huang, D. M. Koelle, and L. Corey. 2003. T cell immunity to herpes simplex virus in seronegative persons: silent infection or acquired immunity. J. Immunol. 170:4380-4388. [DOI] [PubMed] [Google Scholar]
- 18.Simmons, A., and D. C. Tscharke. 1992. Anti-CD8 impairs clearance of herpes simplex virus from the nervous system: implications for the fate of virally infected neurons. J. Exp. Med. 175:1337-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tigges, M. A., D. M. Koelle, K. Hartog, R. E. Sekulovich, L. Corey, and R. L. Burke. 1992. Human CD8+ herpes simplex virus-specific cytotoxic T-lymphocyte clones recognize diverse virion protein antigens. J. Virol. 66:1622-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tigges, M. A., S. Leng, D. C. Johnson, and R. L. Burke. 1996. Human herpes simplex (HSV)-specific CD8+ CTL clones recognize HSV-2-infected fibroblasts after treatment with IFN-gamma or when virion host shutoff functions are disabled. J. Immunol. 156:3901-3910. [PubMed] [Google Scholar]
- 21.Wald, A., J. Zeh, S. Selke, R. Ashley, and L. Corey. 1995. Virologic characteristics of subclinical and symptomatic genital herpes infections. N. Engl. J. Med. 333:770-775. [DOI] [PubMed] [Google Scholar]
- 22.Wald, A., J. Zeh, S. Selke, T. Warren, A. Ryncarz, R. Ashley, J. N. Krieger, and L. Corey. 2000. Reactivation of genital herpes type 2 infection in asymptomatic seropositive persons. N. Engl. J. Med. 342:844-850. [DOI] [PubMed] [Google Scholar]
- 23.Yewdell, J. W., and J. R. Bennink. 1999. Immunodominance in major histocompatibility class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17:51-88. [DOI] [PubMed] [Google Scholar]