Abstract
Ebola virus (EBOV) infection blocks cellular production of alpha/beta interferon (IFN-α/β) and the ability of cells to respond to IFN-α/β or IFN-γ. The EBOV VP35 protein has previously been identified as an EBOV-encoded inhibitor of IFN-α/β production. However, the mechanism by which EBOV infection inhibits responses to IFNs has not previously been defined. Here we demonstrate that the EBOV VP24 protein functions as an inhibitor of IFN-α/β and IFN-γ signaling. Expression of VP24 results in an inhibition of IFN-induced gene expression and an inability of IFNs to induce an antiviral state. The VP24-mediated inhibition of cellular responses to IFNs correlates with the impaired nuclear accumulation of tyrosine-phosphorylated STAT1 (PY-STAT1), a key step in both IFN-α/β and IFN-γ signaling. Consistent with this proposed function for VP24, infection of cells with EBOV also confers a block to the IFN-induced nuclear accumulation of PY-STAT1. Further, VP24 is found to specifically interact with karyopherin α1, the nuclear localization signal receptor for PY-STAT1, but not with karyopherin α2, α3, or α4. Overexpression of VP24 results in a loss of karyopherin α1-PY-STAT1 interaction, indicating that the VP24-karyopherin α1 interaction contributes to the block to IFN signaling. These data suggest that VP24 is likely to be an important virulence determinant that allows EBOV to evade the antiviral effects of IFNs.
The filoviruses, Ebola virus (EBOV) and Marburg virus, cause periodic outbreaks of severe hemorrhagic fever in humans. In EBOV outbreaks consisting of more than 10 reported cases, mortality rates have ranged from 40 to 90% (41), and Marburg virus outbreaks have had reported case fatality rates ranging from 25 to 80% (13). This extreme virulence has made Ebola and Marburg viruses of concern both as naturally emerging pathogens and as potential bioweapons (41).
The molecular mechanisms contributing to the severe pathogenesis of filovirus infection are poorly understood. Several potential mechanisms contributing to EBOV virulence have been reviewed (41). These include cytotoxicity of the viral glycoprotein, the production of proinflammatory cytokines, and the dysregulation of the coagulation cascade due to the production of tissue factor (14, 20, 21, 62, 64). Infection also appears to induce a general immune suppression (11, 53). Possible mechanisms contributing to this suppression include inhibition of dendritic cell activation and an induction of lymphocyte apoptosis (2, 8, 18, 22, 43). Each of these pathogenic processes likely occurs as a result of the active replication of the virus. Thus, the ability of the virus to counteract early antiviral responses, including those of the host's interferon system, likely plays an important role in EBOV virulence (41).
EBOV encodes mechanisms to counteract the host interferon (IFN) response by blocking both production of IFN-α/β and cellular responses to IFN-α/β or -γ treatment (6, 24, 26, 27). We previously demonstrated that the EBOV VP35 protein suppresses IFN-α/β production by inhibiting the activation of interferon regulatory factor 3 (IRF-3) (5, 7, 51), and subsequent studies confirm that VP35 exerts this function (8, 28). However, the manner in which EBOV blocks signaling from the IFN-α/β or -γ receptor has remained incompletely defined.
IFN-α/β, a family of structurally related proteins, and IFN-γ bind to two distinct receptors but activate similar signaling pathways (reviewed in reference 38). For both pathways, ligand binding activates receptor-associated Jak family tyrosine kinases. These undergo auto- and transphosphorylation and phosphorylate the cytoplasmic domains of the receptor subunits. The receptor-associated phosphotyrosine residues then serve as docking sites for the SH2 domains of STAT proteins. The receptor-associated STATs then undergo tyrosine-phosphorylation and form homo- or heterodimers via reciprocal SH2 domain-phosphotyrosine interactions. Signaling from the IFN-α/β receptor results predominately in the formation of STAT1:STAT2 heterodimers which additionally interact with IRF-9. IFN-γ signaling results predominately in the formation of STAT1:STAT1 homodimers. Upon dimerization, the STAT1:STAT2 heterodimer or the STAT1:STAT1 homodimer interacts with a specific member of the karyopherin α (also known as importin α) family of nuclear localization signal (NLS) receptors, karyopherin α1 (importin α5) (45, 48, 55). This interaction with karyopherin α1 mediates the nuclear accumulation of these STAT1-containing complexes (45, 48, 55). The consequence of the activation and nuclear accumulation of these complexes is the specific transcriptional regulation of numerous genes, some of which have antiviral properties (39).
In the present study, we demonstrate that the Zaire EBOV VP24 protein, a minor viral structural protein previously implicated in viral nucleocapsid formation (31, 63), viral budding or assembly (3, 25), and host range determination (61), is capable of blocking both IFN-α/β and IFN-γ signaling. Mechanistically, we find that VP24 prevents the nuclear accumulation of tyrosine-phosphorylated STAT1 (PY-STAT1); moreover, we provide evidence that an interaction between VP24 and karyopherin α1 mediates this inhibition. The result is a subversion of the antiviral effects of IFNs. Consistent with the role of VP24 as an EBOV-encoded antagonist of IFN signaling pathways, we find that EBOV infection also imposes a block to the nuclear accumulation of PY-STAT1 following IFN treatment. Subversion of the IFN signaling pathways likely promotes EBOV propagation in vivo and contributes to viral pathogenesis. Strategies that target this function of VP24 may enhance the efficacy of IFNs toward EBOV.
MATERIALS AND METHODS
Cells and viruses.
293T cells, 293 cells, and Vero cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Sendai virus strain Cantell was grown in 10-day-old embryonated chicken eggs at 37°C for 48 h. Zaire EBOV was grown and EBOV infections were performed in Vero E6 cells at the Centers for Disease Control and Prevention under biosafety level 4 (BSL-4) containment. For analysis by immunofluorescence microscopy, Vero cells were plated onto 12-mm-diameter glass coverslips and infected at a multiplicity of infection (MOI) of 0.5. For analysis of tyrosine-phosphorylated STAT1 by Western blotting, cells were cultured in a 24-well plate and infected at an MOI of 1.0.
Reporter gene assays.
Vero cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as outlined in the manufacturer's protocol. Cell monoloayers, 90 to 95% confluent in six-well plates, were transfected with 1.5 μg of an ISG54 promoter chloramphenicol acetyltransferase (CAT) reporter construct (pHISG54-CAT) or of an IRF-1 gamma-activated sequence (GAS) element-driven luciferase reporter construct (35), 1 μg of a constitutively expressing luciferase reporter construct (pCAGGS-luc), and 2.5 μg of the relevant expression plasmid. Twenty-four hours posttransfection, cells were washed and maintained in Dulbecco's modified Eagle medium containing 0.3% bovine serum albumin, with or without (mock-treated control) 1,000 U/ml of human IFN-β or 1,000 U/ml of human IFN-γ (as indicated) (Calbiochem, San Diego, CA). Twenty-four hours post-IFN treatment, cells were harvested and analyzed for CAT and luciferase activities. The CAT activity was quantified by using a PhosphorImager and normalized to the luciferase activity.
Western blots and coimmunoprecipitations.
To detect STAT1 levels, 293T cells were transfected as described above with 10 ng of a human IRF-9 expression plasmid (pCAGGS-hIRF-9) and 2.5 μg of the expression plasmids indicated in the text. We included pCAGGS-hIRF-9 because these 293T cells do not efficiently respond to IFN-β without overexpression of IRF-9 (data not shown). Twenty-four hours posttransfection, the medium was replaced with DMEM, 0.3% bovine serum albumin (BSA), 1,000 U/ml human IFN-β (Calbiochem). Eighteen hours post-IFN-β addition, cells were harvested and lysed in extract buffer (50 mM Tris [pH 7.5], 280 mM NaCl, 0.5% Igepal, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 1 mM dithiothreitol, 1 mM sodium vanadate, and protease inhibitors [Complete; Roche]). Total cell extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were transferred to a polyvinylidene difluoride (PVDF) membrane and probed with rabbit polyclonal antibodies against human STAT1 (BD Transduction Laboratories) and β-actin (Sigma). The Western blots were developed using the Western Lightning ECL kit (Perkin-Elmer, Boston, MA) and Kodak BioMax film (Kodak, Rochester, NY).
To detect MxA in EBOV-infected Vero cells, cell lysates were frozen and decontaminated by exposure to 5 million rads of gamma radiation. Aliquots of the protein lysates were then resolved by SDS-PAGE and transferred to PVDF. The membrane was blocked in 5% milk and 0.2% Tween 20 in PBS (PBS-T) and then incubated with a mouse antibody raised against MxA diluted 1:500 (the antibody was generously provided by Georg Koch and Otto Haller, Department of Virology, University of Freiberg, Freiberg, Germany), and a mouse antibody raised against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Abcam, Cambridge, United Kingdom). After rinsing with PBS-T, the blot was incubated with an alkaline phosphatase-conjugated goat antibody raised against mouse immunoglobulin G (IgG) (Rockland, Gilbertsville, PA) and visualized with ECF reagent and a Storm 860 PhosphorImager (Amersham Biosciences, Piscataway, NJ). To detect the presence of the EBOV protein VP24, the membrane was incubated with a rabbit antibody raised against VP24 (diluted 1:2,000), rinsed with PBS-T, and incubated with a horseradish peroxidase (HRP)-conjugated goat antibody raised against rabbit IgG (Sigma, St. Louis, MO). The VP24 Western blot was developed using the Western Lightning ECL kit (Perkin-Elmer, Boston, MA) and Kodak BioMax film (Kodak, Rochester, NY).
For coimmunoprecipitation experiments, 293T cells were transfected with 1 μg of expression plasmids unless otherwise indicated. Following a 24-h incubation, cells were washed in ice-cold PBS and lysed in 500 μl of extract buffer for 10 min on ice. For Fig. 6B and C, prior to lysis, the cells were mock treated or treated with 1,000 U/ml human IFN-β (Calbiochem) for 1 h as indicated. Extracts were centrifuged at 13,000 rpm for 10 min at 4°C in a microcentrifuge, the supernatant was collected, and 20 μl of 50% slurry of M2 (anti-FLAG) monoclonal antibody cross-linked to agarose beads (Sigma) was added and incubated for at least 1 h at 4°C with rotation. The M2-agarose beads were washed three times with extract buffer and boiled in SDS-PAGE sample buffer. For VP24 coimmunoprecipitation with FLAG-karyopherin αs, the immunoprecipitated material was analyzed by SDS-PAGE and immunoblotting with rabbit polyclonal antibody against FLAG (Sigma) or against VP24. Separately, blots were probed with an anti-VP35 monoclonal antibody. For FLAG-karyopherin α1, STAT1-green fluorescent protein (GFP) and hemagglutinin (HA)-VP24 coimmunoprecipitations, the immunoprecipitated material was analyzed by SDS-PAGE and immunoblotting with rabbit polyclonal antibody against PY-STAT1 (pY701; Cell Signaling Technology) or against STAT1 (BD Biosciences) where indicated. Additionally, immunoblotting was also performed with monoclonal anti-HA and anti-FLAG antibodies (Sigma) where indicated. Whole-cell extracts (1% of total material used for immunoprecipitation) were immunoblotted with the same antibodies in parallel. The Western blots were developed using the Western lightning ECL kit (Perkin-Elmer, Boston, MA) and Kodak BioMax film (Kodak, Rochester, NY).
FIG. 6.
VP24 interacts with karyopherin α1 and inhibits karopherin α1-phospho (Tyr 701)-STAT1 interaction. (A) Coimmunoprecipitation of VP24 with karyopherin α1. 293T cells were transfected with the indicated plasmids expressing VP24, VP35, or firefly luciferase (indicated by a hyphen) in the absence or presence of plasmids expressing FLAG-tagged karyopherin α1 (FLAG-Kα1), α2 (FLAG-Kα2), α3 (FLAG-Kα3), or α4 (FLAG-Kα4). Cell extracts were prepared 1 day posttransfection, and immunoprecipitations (IP) were performed with anti-FLAG monoclonal antibody M2 bound to agarose beads. After washing, immunoprecipitates were analyzed by Western blotting (WB) with polyclonal rabbit antisera recognizing FLAG and VP24 (top panel; 50% of total material was analyzed). Whole-cell extracts (WCE) (1% of total) were also analyzed by Western blotting for karyopherin expression (anti-FLAG antibody), VP24, or VP35, as indicated. (B) 293T cells were transfected with the indicated expression plasmids. Lanes 1 and 4, FLAG-karyopherin α1 and STAT1-GFP; lanes 2 and 5, FLAG-karyopherin α1 and HA-VP24; lanes 3 and 6, FLAG-karyopherin α1, STAT1-GFP, and HA-VP24. The cells were then mock treated or treated with 1,000 U/ml human IFN-β and subsequently immunoprecipitated with anti-FLAG monoclonal antibody M2 bound to agarose beads (IP: FLAG). After washing, the immunoprecipitates were analyzed by Western blotting with a monoclonal antibody recognizing STAT1 (WB: STAT1), and monoclonal antibodies against FLAG (WB: FLAG) and HA (WB: HA) (top panel; 10% of total material analyzed). Whole-cell extracts (1% of total) were also analyzed similarly. (C) 293T cells were transfected with FLAG-karyopherin α1 (250 ng); STAT1-GFP (250 ng); or 0, 25, 250, 500, and 1,000 ng of HA-VP24 (lanes 1 to 5, respectively) expression plasmids. Lane 6 contains samples derived from cells transfected with STAT1-GFP plasmid but not with karyopherin α1 plasmid. The cells were then treated with 1,000 U/ml human IFN-β and subsequently immunoprecipitated with anti-FLAG monoclonal antibody and analyzed by Western blotting with polyclonal rabbit antisera recognizing phosphorylated STAT1 and monoclonal antibodies against FLAG and HA (top panel, 10% of total material analyzed). Whole-cell extracts (1% of total) were also analyzed. Asterisks indicate the heavy chain of the anti-FLAG monoclonal M2 antibody used for the immunoprecipitation.
Measuring the antiviral state in cells.
Vero cells were transfected by using LF2000 with 2.5 μg of the indicated expression plasmid. Twenty-four hours posttransfection, the cells were mock treated or treated with 1,000 U/ml of human IFN-β as described above. Twenty-four hours post-IFN-β treatment, the cells were infected with Newcastle disease virus expressing GFP (NDV-GFP) (6.4 turkey red blood cell hemagglutinating units/ml) (50), and after 24 h of infection GFP was visualized by fluorescence microscopy.
STAT1 and GFP-IRF-3 nuclear translocation.
Vero cells were plated onto 12-mm-diameter glass coverslips and transfected with empty vector (pCAGGS), pCAGGS-FLAG-VP24, or pCAGGS-FLAG-VP35 using Lipofectamine 2000 following the manufacturer's recommended protocol. Twenty-four hours posttransfection, the cells were serum starved for 4 h and then either mock treated or treated with 1,000 U/ml of human IFN-β or human IFN-γ for 30 min at 37°C. After rinsing three times with PBS containing 1 mM each of calcium chloride and magnesium chloride (PBS-CM), the cells were placed on ice and fixed with −20°C methanol for 10 min. After rehydrating in PBS-CM, the cells were blocked in PBS-CM containing 4% normal goat serum, 1% BSA, and 0.075% saponin for 45 min at ambient temperature. The coverslips were incubated with a mouse antibody raised against STAT1 (5 μg/ml; BD Bioscience, Franklin Lakes, NJ) and a rabbit antibody raised against the FLAG epitope (4 μg/ml; Sigma, St. Louis, MO) in 1% BSA-Tris-buffered saline (TBS) for 1 h, followed by three washes of PBS. The coverslips were incubated with an Alexa 488-conjugated goat antibody raised against mouse IgG and an Alexa 594-conjugated antibody raised against rabbit IgG (0.5 μg/ml each; Molecular Probes, Eugene, OR) and Hoechst 33342 (0.1 μg/ml; Molecular Probes, Eugene, OR) for 30 min. After rinsing, the coverslips were mounted on slides and imaged on a Zeiss Axiophot 2 equipped with a Ratiga charge-coupled device (CCD) camera. Individual color channels were acquired as 8-bit monochrome images and pseudocolored using ImageJ (NIH, Bethesda, MD). To quantify the number of cells expressing nuclear STAT1, at least three independent fields (at least 36 cells) for each experimental condition were acquired. Cells that demonstrated colocalization of Hoechst and STAT1 staining were considered to have nuclear STAT1.
GFP-IRF-3 translocation assays were performed in Vero cells following a previously described procedure (5).
Detection of tyrosine-phosphorylated STAT1 in IFN-β-treated Vero cells. (i) Immunofluorescence microscopy.
Vero cells were plated on 12-mm-diameter glass coverslips and cultured in 10% FBS in DMEM. Before treatment with IFN-β, cells were rinsed twice with Optimem medium (Invitrogen) and incubated in the absence of serum for 4 h. Human IFN-β was added (1,000 U/ml in DMEM), and the cells were cultured for 30 min. The cells were then fixed and blocked as described above for STAT1 staining. The cells were rinsed three times with 1% BSA in TBS and incubated with a rabbit antibody raised against a synthetic phosphopeptide (keyhole limpet hemocyanin coupled) corresponding to residues surrounding Tyr 701 of human Stat1 (0.2 μg/ml; Cell Signaling, Beverly, MA) and a mouse antibody raised against the FLAG epitope (4 μg/ml; Sigma, St. Louis MO) in 1% BSA-TBS for 1 h, followed by three washes of PBS. For cells infected with EBOV under BSL-4 conditions, coverslips were decontaminated at this stage by incubation with 4% paraformaldehyde in PBS for 48 h before transferring them to a BSL-2 environment.
The coverslips were incubated with Alexa 594-conjugated goat antibodies raised against mouse IgG (0.5 μg/ml; Molecular Probes, Eugene, OR), Alexa 488-conjugated antibodies raised against rabbit IgG (0.5 μg/ml each; Molecular Probes, Eugene, OR) and Hoechst 33342 (0.1 μg/ml; Molecular Probes, Eugene, OR) for 30 min. After rinsing, the coverslips were mounted on slides and imaged on a Zeiss Axiophot 2 equipped with a Ratiga CCD camera. Individual color channels were acquired as 8-bit monochrome images and pseudocolored using ImageJ (NIH, Bethesda, MD). To quantify the number of cells expressing nuclear PY-STAT1, four independent fields of Ebola virus-infected cells were acquired. Cells that demonstrated colocalization of Hoechst and STAT1 staining were considered to have nuclear STAT1. Cells demonstrating staining for the Ebola virus VP35 protein were considered to be infected.
(ii) To detect the presence of tyrosine-phosphorylated STAT1 in EBOV-infected Vero cells, extracts were prepared using the Pierce NE-PER kit (Pierce, Rockford, IL) according to the manufacturer's recommendation. The lysates were frozen and decontaminated with 5 million rads of gamma radiation. Prior to analysis, the nuclear and cytoplasmic fractions were combined to produce a total lysate. The protein samples were resolved on a 10% polyacrylamide gel by SDS-PAGE and transferred to PVDF. After blocking in 5% nonfat milk and 0.2% Tween 20 in TBS (TBS-T), the membrane was probed by incubation with a rabbit antibody raised against tyrosine-phosphorylated STAT1 (0.02 μg/ml; Cell Signaling, Beverly, MA) and a rabbit antibody raised against GAPDH (Abcam, Cambridge, United Kingdom). After rinsing, the blot was incubated with an alkaline phosphatase-conjugated goat antibody raised against rabbit IgG (Rockland, Gilbertsville, PA) and visualized using using ECF reagent and a Storm 860 PhosphorImager (Amersham Biosciences, Piscataway, NJ).
RESULTS
EBOV VP24 inhibits induction of gene expression by IFN-β and IFN-γ.
In order to identify an EBOV protein(s) capable of inhibiting cellular responses to IFN treatment, cDNAs encoding individual EBOV proteins were screened for their ability to inhibit the IFN-β-induced expression of an IFN-responsive, ISG54 promoter-driven CAT reporter gene. Only the VP24 protein was able to efficiently and reproducibly inhibit gene activation in this assay (data not shown). The ability of VP24 to inhibit IFN-β signaling is demonstrated in Fig. 1A. In this experiment, Vero cells were cotransfected with the ISG54-CAT reporter and an empty expression plasmid or expression plasmids for the Nipah virus W protein, a previously demonstrated inhibitor of IFN signaling (50, 56), FLAG-tagged VP24, an untagged VP24 protein, or VP35, the EBOV protein previously implicated as an inhibitor of IFN-α/β production (5, 7) (Fig. 1A). An ∼30-fold induction in CAT activity was seen in IFN-β-treated samples transfected with empty vector (as compared with an empty vector-transfected, mock-treated sample). This activation was inhibited in cells expressing either tagged or untagged VP24 protein and in cells expressing Nipah W, whereas a different EBOV protein, VP35, did not inhibit IFN-β-induced gene expression (Fig. 1A).
FIG. 1.
Ebola virus VP24 inhibits IFN-β and -γ induced gene expression. (A) IFN-β-induced reporter gene activation in Vero cells was measured. Cells were cotransfected with the ISG54-CAT reporter plasmid, a constitutively expressed Renilla luciferase reporter plasmid and empty vector (Vec) or with plasmids expressing Nipah virus W protein (W), Flag-tagged EBOV VP24 (F-24), untagged VP24 (24), or EBOV VP35 (35). Twenty-four hours posttransfection, the cells either were mock treated or were treated with 1,000 U of human IFN-β/ml for 24 h, harvested, and then assayed for CAT and luciferase activities. The data are presented as the activation (fold) relative to empty vector, mock-treated controls. Error bars indicate means ± the standard deviation of three experiments. (B) Levels of STAT1 gene expression in 293T cells transfected with empty vector (Vec) or plasmids expressing the indicated protein are shown. Twenty-four hours posttransfection, the cells were treated with 1,000 U of IFN-β/ml, and 18 h posttreatment, the cells were lysed and subjected to Western blot analysis with anti-STAT1 (αSTAT1) and anti-β-actin (α β-actin) antibodies. (C) Levels of IFN-γ-induced reporter gene activation in Vero cells transfected with empty vector (Vec) or with plasmids expressing EBOV VP35 (35) or FLAG-tagged EBOV VP24 (24). Vero cells were transfected with the IFN-γ-responsive IRF-1-promoter luciferase reporter and a constitutively expressed Renilla luciferase reporter. Twenty-four hours posttransfection, the cells were either mock treated or treated with 1,000 U/ml of IFN-γ, and 24 h posttreatment, reporter gene activity was measured. IFN-γ-induced reporter values were normalized to the Renilla luciferase reporter. Results are presented as induction (fold) of the IRF-1-luciferase reporter relative to an empty vector-transfected, mock-treated control. Error bars indicate the mean ± the standard deviation of three experiments.
The ability of VP24 to inhibit IFN-induced expression of an endogenous gene, STAT1, was also assessed. (STAT1 is a key transcription factor in the IFN signaling pathways, and its expression is upregulated upon IFN-β treatment [15, 37].) After 18 h of treatment with IFN-β, a clear increase in STAT1 protein expression was observed in cells transfected with empty vector or with VP35 expression plasmid (Fig. 1B, compare empty vector-IFN-β- to empty vector-transfected, mock-treated cells). In contrast, either an untagged or a FLAG-tagged VP24 plasmid was able to inhibit IFN-β-induced STAT1 expression (Fig. 1B).
Given that EBOV infection is reported to block not only IFN-α/β but also IFN-γ signaling, we asked whether the antagonistic effect of VP24 extended to the IFN-γ pathway. For this purpose, a luciferase reporter under control of the IFN-γ-responsive IRF-1 promoter, which contains GAS elements was used. IFN-γ treatment resulted in reporter activation in empty vector-transfected cells. VP35, which is unable to inhibit IFN-α/β signaling, was also unable to inhibit IFN-γ-mediated signaling. In contrast, VP24 was able to inhibit reporter gene activation (Fig. 1C).
Each of the experiments described in Fig. 1 included a constitutively expressed reporter plasmid to which the levels of IFN-induced gene expression were normalized. When the expression of a constitutively-expressed Renilla luciferase gene was analyzed over the course of four independent transfections, luciferase values in IFN-β-treated cells were not statistically different in VP24-expressing versus VP35- or VP40-expressing cells (data not shown). This suggests that VP24 is not exerting a nonspecific, global effect upon gene expression. In addition, when VP24 was transfected at a ratio of 5:1 with a GFP expression plasmid and GFP-expressing cells sorted by fluorescence-activated cell sorting were analyzed for their uptake of 7-AAD, a vital dye, we did not detect any evidence that VP24 was cytotoxic (data not shown).
VP24 counteracts the antiviral effects of IFN-β.
The results obtained above demonstrate the ability of VP24 to inhibit the IFN-induced activation of specific IFN-responsive promoters and the IFN-induced expression of the STAT1 gene. We next assessed the ability of VP24 to counteract the antiviral effects of IFN-β. Vero cells were transfected with either empty vector, VP24 expression plasmid, or, as a positive control, Nipah virus W expression plasmid. Nipah W was included because it was previously demonstrated to overcome the antiviral effects of IFN (50, 56). Twenty-four hours posttransfection, the cells were either mock treated or treated with IFN-β and after an additional 24 h the cells were infected with NDV-GFP (50). As expected, IFN-β treatment exerted an antiviral effect and inhibited virus replication and thus GFP expression in cells transfected with empty vector; whereas mock-treated, empty vector-transfected cells were able to support NDV-GFP replication (Fig. 2, compare vector ± IFN-β treatment). As previously demonstrated, Nipah W expression restored growth of NDV-GFP in IFN-β-treated cells. Similarly, VP24 was able to restore growth of NDV-GFP in cells treated with IFN-β (Fig. 2). Thus, expression of VP24 is sufficient to prevent the establishment, in Vero cells, of an IFN-β-induced antiviral state.
FIG. 2.
Ebola virus VP24 rescues growth of NDV-GFP in cells pretreated with IFN-β. Vero cells were transfected with the empty vector (Vector) or plasmids expressing the Nipah virus W (Nipah W) or EBOV VP24 (VP24) proteins. Twenty-four hours posttransfection, the cells were mock treated (−IFNβ) or treated with 1,000 U/ml of IFN-β (+IFNβ) as indicated. Twenty-four hours post-IFN-β treatment, the cells were infected with NDV-GFP. The green fluorescence was visualized 24 h postinfection under a fluorescence microscope.
VP24 prevents IFN-induced nuclear accumulation of STAT1.
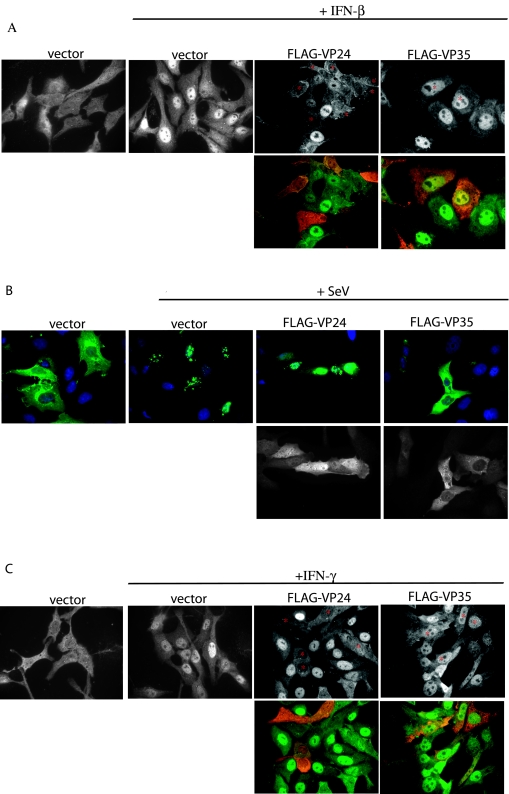
The ability of VP24 to inhibit both IFN-α/β and IFN-γ signaling suggests that it targets a factor common to both pathways. We therefore assessed the impact of VP24 upon STAT1 (the activation of which is central to both the IFN-α/β and IFN-γ signaling pathways). We first asked whether VP24 expression would affect, upon IFN treatment, the nuclear accumulation of STAT1. In mock-treated cells, immunofluorescence analysis of STAT1 revealed that the protein was distributed in a diffuse pattern within cells. In the empty vector-transfected cells treated with IFN-β, we observed prominent nuclear STAT1 in 63% of the cells (n = 95). Transfection of VP35 did not alter the ability of STAT1 to translocate into the nucleus. Greater than 90% of the cells expressing VP35 (n = 41) exhibited prominent nuclear STAT1. However, when VP24 was expressed, STAT1 failed to concentrate in the nucleus following IFN-β treatment (Fig. 3A). In cells expressing VP24, only 11% were observed to have nuclear STAT1 (n = 48). To determine whether VP24 might globally block nuclear import, we determined whether it affected the localization of a GFP-IRF-3 fusion, which accumulates in the nucleus following Sendai virus infection. In contrast to the data obtained with STAT1, VP24 did not inhibit the Sendai virus-induced nuclear localization of GFP-IRF-3, although as was previously reported, VP35 did block GFP-IRF-3 nuclear localization (5) (Fig. 3B). Finally, the ability of VP24 expression to inhibit IFN-γ-induced nuclear accumulation of STAT1 was also demonstrated (Fig. 3C). After IFN-γ treatment, 95% of empty vector-transfected cells (n = 78) and 92% of VP35-expressing cells (n = 36) exhibited nuclear STAT1. In contrast, only 8% of VP24-expressing cells (n = 38) exhibited nuclear STAT1. These observations suggest that VP24 blocks IFN signaling by inhibiting STAT1 nuclear accumulation.
FIG. 3.
EBOV protein VP24 prevents IFN-mediated nuclear translocation of STAT1. (A) IFN-β treatment (30 min) of Vero cells causes STAT1 to relocate from the cytoplasm (vector) to the nucleus (vector + IFN-β). In cells expressing FLAG-VP24 (FLAG-VP24 + IFN-β; relevant cells marked with an asterisk), STAT1 fails to relocate to the nucleus after IFN-β treatment. IFN-β treatment of FLAG-VP35-expressing cells (FLAG-VP35 + IFN-β; relevant cells marked with an asterisk) causes STAT1 to relocate to the nucleus. Upper panels show only STAT1 images. Lower panels show the STAT1 images (green) merged with images of FLAG-tagged Ebola virus proteins (red). (B) Vero cells express GFP-IRF-3 in the cytoplasm in the absence of viral infection (vector), but translocate GFP-IRF-3 to the nucleus when infected with Sendai virus (vector + SeV). Coexpression of FLAG-VP35 prevents translocation of GFP-IRF-3 (FLAG-VP35 + SeV), but FLAG-VP24-expressing cells are still able to traffic GFP-IRF-3 to the nucleus (FLAG-VP24 + SeV). Upper panels show the GFP-IRF-3 (green) merged with Hoechst nuclear staining (blue). Lower panels show the FLAG-tagged Ebola virus proteins. (C) In Vero cells cotransfected with empty vector, STAT1 is predominately cytoplasmic in the absence of IFN-γ treatment (vector), but STAT1 concentrates in the nucleus after a 30-min treatment with IFN-γ (vector + IFN-γ). In the presence of FLAG-VP24, IFN-γ treatment fails to relocate STAT1 to the nucleus (FLAG-VP24 + IFN-γ).
VP24 prevents nuclear translocation of tyrosine-phosphorylated STAT1.
Experiments were then performed to determine whether VP24 expression affects the IFN-induced tyrosine phosphorylation of STAT1 or the nuclear localization of PY-STAT1. We first examined, in the presence or absence of VP24, the location of endogenous PY-STAT1 by staining cells with a phospho-specific antibody. In control cells (Vero cells transfected with FLAG-tagged VP35), PY-STAT1 (green) appears in the nucleus after 30 min of IFN-β treatment (Fig. 4A). However, FLAG-VP24-expressing cells display PY-STAT1 in the cytoplasm (Fig. 4A). To control for the specificity of this staining, we confirmed that no PY-STAT1 signal could be detected in the Vero cells in the absence of IFN treatment (data not shown). In a separately performed transfection, 77 of 86 (89%) mock-transfected, IFN-treated cells displayed nuclear tyrosine-phosphorylated STAT1 30 min post-IFN addition. Similarly, 30 of 42 (71%) FLAG-VP35-transfected, IFN-treated cells displayed nuclear tyrosine-phosphorylated STAT1. For these VP35 cells, there was a minority of cells which displayed high levels of PY-STAT1 staining that had either cytoplasmic PY-STAT1 or at least PY-STAT1 that was not in a clearly defined nucleus. In contrast, for the IFN-treated, FLAG-VP24 transfectants, in a total of 107 expressing cells, only 8 (7.5%) had nuclear PY-STAT1 (data not shown).
FIG. 4.
VP24 prevents the nuclear translocation of phosphorylated STAT1 (Tyr 701). (A) After treatment with IFN-β for 30 min, cells expressing FLAG-VP35 (top row, red channel) are able to translocate PY-STAT1 (P-STAT1; green channel) to the nucleus. In cells expressing FLAG-VP24 (bottom row, red channel) PY-STAT1 is located in the cytoplasm. (B) VP24 expression does not detectably inhibit the tyrosine phosphorylation of STAT1. 293T cells were transfected with expression plasmids encoding: pCAGGS empty vector (vector), luciferase, Nipah virus V (NipV), VP35, VP24, or FLAG-VP24. After treatment with IFN-β (or mock treatment), the cells were lysed and analyzed for the presence of PY-STAT1 by Western blotting.
Since an effect of VP24 expression upon levels of PY-STAT1 might be difficult to detect using fluorescence microscopy, we also examined STAT1 tyrosine phosphorylation by Western blotting extracts from transfected, IFN-β-treated 293T cells, 30 min following IFN-β-treatment. Both FLAG-VP35- and FLAG-VP24-transfected cells contained amounts of PY-STAT1 similar to that seen in cells expressing no recombinant protein or an irrelevant protein, luciferase (Fig. 4B). In a control sample, expression of the Nipah virus V protein, which has been shown to prevent STAT1 phosphorylation (52, 56), reduced the amount of PY-STAT1 (Fig. 4B). We conclude that VP24 inhibits transcriptional responses to IFN, at least in part, by preventing the normal trafficking of PY-STAT1 to the nucleus.
Expression of supraphysiological intracellular concentrations of VP24 could conceivably lead to effects of VP24 not seen in EBOV-infected cells. To address this concern, Western blots were performed on cell lysates prepared from EBOV-infected and VP24-transfected cells. The lysates for this experiment were derived from Vero cells infected such that 50% of cells stained positive for viral antigen or Vero cells transfected with FLAG-VP24 such that approximately 40% of cells were stained by anti-FLAG antibody. A significantly stronger VP24 signal was detected from the infected cell lysate versus the transfected cell lysate (data not shown). Thus, the transfection experiments described above appear to involve biologically relevant levels of VP24.
EBOV infection inhibits both IFN-β-induced gene expression and nuclear accumulation of PY-STAT1.
If VP24 contributes to the inhibition of IFN signaling by EBOV, EBOV infection should inhibit the nuclear transport of PY-STAT1. To address this hypothesis, we infected Vero cells with EBOV (MOI = 1) and, 24 h postinfection, treated them overnight with IFN-β. By Western blotting, MxA, a protein whose expression is induced by interferon (1), was found to be up-regulated following IFN-β treatment in mock-infected cells, but this up-regulation was absent in EBOV-infected cells (Fig. 5A). Infection of the cells was confirmed by examining expression of the VP24 protein (Fig. 5A). These data are consistent with previous reports that EBOV infection blocks IFN signaling (27). In a separate experiment, Vero cells were grown on glass coverslips, infected with EBOV (MOI = 0.5), and, 24 h postinfection, treated with IFN-β for 30 min. After fixation, the cells were processed for immunofluorescence analysis of PY-STAT1 and of viral antigen (the VP35 protein). Greater than 90% of cells lacking significant viral antigen staining (n = 74) demonstrated nuclear STAT1 (Fig. 5B, upper panel). Consistent with the data obtained when VP24 was expressed by transfection, we observed that only 21% of EBOV-infected cells (n = 80) accumulated PY-STAT1 in their nucleus (Fig. 5B, lower panel). It should be noted that, in these experiments, cells were infected at an MOI of 0.5 and examined 48 h postinfection. Under these conditions, it is likely that more than one round of infection was initiated in these cultures, and thus some of the infected cells were probably expressing only low levels of VP24.
FIG. 5.
EBOV infection inhibits IFN-β-induced expression of MxA and prevents nuclear translocation of PY-STAT1. (A) Mock-infected Vero cells express MxA in response to overnight treatment with IFN-β (lane 2). However, EBOV-infected cells (MOI = 1) do not produce MxA in response to IFN-β (lane 4). Lanes 1 and 2 were mock infected. Lanes 3 and 4 were infected with EBOV. Lanes 2 and 4 were treated with IFN-β, while lanes 1 and 3 were mock treated. (Top panel) Western blot to detect MxA and GAPDH. (Bottom panel) Western blot to detect VP24. (B) Mock-infected Vero cells translocate PY-STAT1 to the nucleus after 30 min of IFN-β treatment (top panel), but EBOV-infected cells retain PY-STAT1 in the cytoplasm (bottom panel). Red represents viral antigen (VP35), and green represents PY-STAT1.
VP24 interacts with the STAT1 nuclear localization signal receptor, karyopherin α1, and inhibits VP24-PY-STAT1 interaction.
PY-STAT1 is reported to accumulate in the nucleus via interaction with a specific nuclear localization signal receptor, karyopherin α1 (45, 48, 55). Given the ability of VP24 to prevent the nuclear accumulation of STAT1, we asked whether VP24 might interact with this or other nuclear localization signal receptors. FLAG-tagged karyopherin α1, α2, α3, or α4 expression plasmid was cotransfected with empty plasmid, untagged VP24 expression plasmid, or VP35 expression plasmid. One day posttransfection, immunoprecipitations were performed with an anti-FLAG monoclonal antibody, and the precipitated material was analyzed by Western blotting with antibodies recognizing FLAG, VP24, or VP35. VP24 coprecipitated exclusively with karyopherin α1 (Fig. 6A). In contrast, VP35 did not coprecipitate with any of the karyopherin αs (data not shown).
To determine if the interaction of VP24 with karyopherin α1 might account for the loss of STAT1 nuclear accumulation, we asked if VP24 is able to disrupt the STAT1-karyopherin α1 interaction. FLAG-tagged karyopherin α1 was coexpressed with or without STAT1-GFP in the presence and absence of VP24. In the absence of IFN-β (Fig. 6B, lanes 1 to 3), no STAT1-GFP was coprecipitated with the karyopherin α1, despite the presence of STAT1-GFP in the whole-cell lysates (Fig. 6B, lanes 1 and 3). However, following addition of IFN-β to the cells (Fig. 6B, lanes 4 to 6), STAT1-GFP coprecipitated with karyopherin α1 (Fig. 6B, lane 4). VP24 was once again coprecipitated with karyopherin α1, regardless of whether or not IFN-β was added (Fig. 6B, lanes 2, 3, 5, and 6). Most significantly, STAT1-GFP did not associate with karyopherin α1 when VP24 was present (Fig. 6B, lane 6). To determine whether there is a dose-dependent effect of V24 on karyopherin α1-STAT1 interaction, we coexpressed STAT1-GFP with karyopherin α1 (Fig. 6C, lanes 1 to 5) or without karyopherin α1 (Fig. 6C, lane 6). VP24 either was not expressed (Fig. 6C, lane 1) or was expressed in increasing amounts (lanes 2 to 5). The cells were treated with IFN-β, and immunoprecipitation of the FLAG-karyopherin α1 was performed. In this experiment, detection of STAT1-GFP was performed with anti-PY-STAT1 antibody, which, in our hands, is more sensitive than the Western blot for total STAT1. Western blotting was also performed to detect FLAG-karyopherin α1 and VP24. As was seen previously, the activated STAT1-GFP coprecipitated with karyopherin α1 when VP24 was absent (Fig. 6C, lane 1). In contrast, no STAT1-GFP was precipitated with anti-FLAG antibody when the Flag-karyopherin α1 was omitted (Fig. 6C, lane 6). When a low level of VP24 was present in the precipitation, less PY-STAT1-GFP was coprecipitated (Fig. 6C, lane 2). When higher levels of VP24 were present, the level of STAT-GFP coprecipitated was further decreased (Fig. 6C, lanes 2 to 5). These data suggest that the interaction of VP24 with karyopherin α1 inhibits karyopherin α1-PY-STAT1 association, thus providing an explanation as to how VP24 may inhibit STAT1 nuclear accumulation.
DISCUSSION
The data described above provide a molecular explanation for the impaired cellular responses to IFN-α/β and to IFN-γ in EBOV-infected cells and shed further light on the ability of this highly lethal pathogen to modulate the host IFN system. Previously, it was demonstrated that EBOV-infected human umbilical vein endothelial cells did not respond to either IFN-α treatment or IFN-γ treatment (27). Specifically, the up-regulation of several IFN-α/β- or IFN-γ-induced genes was absent or reduced in the infected cells (27). However, IL-1β-induced expression of IL-6 and ICAM-1 was intact, demonstrating that infection did not globally block transcriptional responses to all cytokines (27). Electrophoretic mobility shift assays on nuclear extracts further demonstrated that in EBOV-infected cells, IFN-α- and IFN-γ-induced nuclear transcription factor complexes were absent or were present in greatly reduced amounts (27). The present study more clearly defines the EBOV-imposed block to the IFN-α/β and IFN-γ signaling pathways and identifies a single viral protein, VP24, capable of exerting this function. EBOV infection and VP24 expression appear to have equivalent effects upon these signaling pathways. In both cases, the tyrosine phosphorylation of STAT1 in response to IFN remains intact. Rather, the critical step blocked by either EBOV infection or by VP24 expression, is the nuclear accumulation of STAT1 (Fig. 4 and 5). Of note, VP24 does not globally inhibit nuclear translocation of proteins, as it did not prevent the Sendai virus-induced nuclear accumulation of GFP-IRF-3 (Fig. 3). The latter observation is consistent with the view that EBOVs utilize a second protein, VP35, to block activation of IRF-3 (5-8, 28, 51).
The nuclear localization of STAT1 (reviewed in references 47 and 49) is triggered by its tyrosine phosphorylation-induced, SH2 domain-dependent dimerization (23, 54, 57, 58). In its inactive state, STAT1 shuttles between the cytoplasm and nucleus via an energy-independent mechanism involving direct interactions with nucleoporins (44). Tyrosine phosphorylation results in STAT1 heterodimerization with STAT2 (following IFN-α/β treatment) or STAT1 homodimerization (following IFN-γ treatment) (reviewed in reference 38). Dimerization of STAT1 results in conformational changes revealing an atypical NLS. This NLS directly interacts with the carboxy-terminal armadillo repeats of karyopherin α1 (importin α5). Given that activated STAT1 did not detectably interact with other karyopherin α proteins, including karyopherin α2 (importin α1, Rch1) and karyopherin α4 (importin α3, Qip1), or importin α7, it may be only karyopherin α1 which mediates nuclear accumulation of PY-STAT1 (45, 48, 55). Following its nuclear accumulation, DNA containing STAT1 binding sequences can compete with karyopherin α1 for binding to STAT1 homodimers, allowing STAT1 to interact with its target promoters (17, 45). Ultimately, nuclear STAT1 is dephosphorylated, dissociates from DNA, and is exported to the cytoplasm in a CRM1-dependent manner (29, 30, 44, 46, 47). Our data do not provide any evidence that VP24 prevents the tyrosine phosphorylation of STAT1, but they do demonstrate an inhibition of STAT1 nuclear import.
It was interesting to note, given its ability to inhibit nuclear accumulation of STAT1, that VP24 is reported to exhibit a largely perinuclear localization in EBOV-infected cells (25). Thus, VP24 would seem appropriately positioned to influence nuclear/cytoplasmic transport. We therefore tested VP24 for the ability to interact with karyopherin α proteins. VP24 selectively interacted, in coimmunoprecipitation studies, with karyopherin α1, the previously identified NLS receptor for STAT1 (45, 48, 55). That this interaction may mediate the observed defect in STAT1 nuclear import is supported by the ability of VP24 to prevent the association of an activated STAT1-GFP protein with karyopherin α1 (Fig. 6). This selective interaction of VP24 with karyopherin α1 likely also explains why VP24 does not inhibit the nuclear import of IRF-3, as the nuclear import of this protein has been attributed to karyopherin α3 and α4 (36) (Fig. 3). However, further studies will be required to address the possibility that VP24 may also influence the nuclear import of other molecules that use karyopherin α1.
EBOV is an enveloped, nonsegmented negative-strand RNA virus with a genome of approximately 19 kb, and VP24 is one of eight major EBOV-encoded proteins (53). Although several functions have previously been attributed to the EBOV VP24 protein, its precise role in viral replication remains ambiguous and somewhat controversial. Early studies suggested that VP24 is found in EBOV virions and, as a consequence, VP24 was postulated to be a “minor matrix protein” or to function in viral uncoating (16, 53). Although the primary EBOV matrix protein is VP40, and VP40 appears to be the principal force driving the budding of EBOV particles, recent studies do support a possible role for VP24 in viral budding. In particular, VP24 oligomerizes, is hydrophobic, and associates with cellular membranes, all properties characteristic of viral matrix proteins (25). In addition, VP24 itself appears to bud at some level from cells (25). However, coexpression of VP24 with VP40 did not appear to enhance budding by VP40 (40). In addition, VP24 may play an important role in assembly of viral nucleocapsids as EBOV nucleocapsids could be reconstituted by coexpression of the EBOV nucleoprotein (NP), VP35, and VP24 (31). VP24 could be coimmunoprecipitated with NP and VP35 (31); however, VP24 did not cosediment with NP and VP35 in density gradients, suggesting that, while it promotes nucleocapsid formation, it may not be an essential component of the nucleocapsid structure (31). In a different study, VP24 was not required for either the formation or the infectivity of Ebola virus-like particles carrying an EBOV minigenome (63). More recently, the use of small interfering RNAs targeting Marburg virus VP24 supports a role for this protein in filovirus assembly and release from infected Vero cells (3). Given this array of potential functions, it will be of interest to determine which of these is influenced by the ability of VP24 to block IFN signaling pathways or to interact with karyopherin α1.
Understanding the determinants of EBOV virulence in different species may provide insights into EBOV pathogenesis in humans and may suggest novel therapeutic approaches. The IFN system has been clearly implicated in the susceptibility of mice to EBOV disease (9, 12, 42), and the sequence of VP24, along with that of several other genes, changed following the adaptation, by serial passage, of Zaire EBOV to mice (10). (The full sequence of the mouse-adapted EBOV is available under GenBank accession no. AF499101.) It is therefore intriguing that adaptation of EBOV to another host, guinea pigs, is also associated with changes in VP24. Specifically, adaptation of a Zaire EBOV from a nonlethal form to a form lethal to guinea pigs was associated with 5 amino acid changes, one each in the nucleoprotein and L (polymerase), and 3 amino acid changes in VP24 (61). Sequence analysis of the VP24 gene from an independently adapted EBOV also identified an amino acid change in VP24, and it was suggested, based on these observations, that changes in VP24 are likely to play an important role in guinea pig adaptation (61). Although STAT1 and karyopherin α1 are highly conserved between human and mouse, with each protein displaying greater than 90% amino acid identity between the two species, it will be of interest to determine whether these changes in VP24 result in an enhanced ability to counteract interferon responses in these new hosts and whether the adaptation of EBOV to the new species correlates with an increased affinity of VP24 for karyopherin α molecules of the new host.
At present, no vaccines against EBOV are licensed for use in humans, although experimental vaccines that are effective in nonhuman primates have been described (34, 59, 60). In addition, no antivirals are available to treat these severe infections, although an inhibitor of tissue factor was found to offer some protection to experimentally infected nonhuman primates (19). The ability of VP24 to block IFN signaling pathways has obvious implications for the efficacy of IFN as an anti-EBOV therapy. Although 200 IU/ml of IFN-α2b was found to inhibit EBOV replication 100-fold in Vero cells (33), daily intramuscular treatment of EBOV-infected monkeys with a relatively high dose of IFN-α2b beginning 18 h postinoculation merely delayed viremia and death by about 1 day (33). These data suggest that IFNs have limited efficacy against EBOV and fit with the view that VP24 expression renders EBOV relatively insensitive to IFN. Despite these observations, the mouse model of infection suggests that IFNs might be employed to effectively control EBOV infection. For example, mouse-adapted Zaire EBOV is lethal to mice following intraperitoneal infection but not following subcutaneous infection (42). Subcutaneous administration of the mouse-adapted virus conferred protection from challenge with an otherwise lethal intraperitoneal dose, even at 48 h postinoculation (42). This early time point, prior to the development of EBOV-specific adaptive immune responses, correlated with peak induction of IFN-α levels, suggesting that IFN-α/β can have a therapeutic effect on EBOV infection (42). Additionally, the adenosine analog 3-deazaneplanocin A was found to protect mice from illness and death following infection with mouse-adapted EBOV (32). Protection by this drug appears to involve the induction of large quantities of IFN-α (12). Our data suggest that the anti-EBOV efficacy of IFNs might be augmented by inhibitors of VP24. Additional studies to further define the mechanism by which VP24 functions should facilitate the development of such therapies.
Acknowledgments
This work was supported by grants to C.F.B. from the National Institutes of Health and grants to V.E.V. from INSERM, Deutsche Forschungsgemeinschaft (SFB 593), and from French Ministère de la Recherche (04G537). C.F.B. is an Ellison Medical Foundation New Scholar in Global Infectious Diseases.
We thank Luis Martínez-Sobrido and Adolfo García-Sastre (Mount Sinai School of Medicine) for providing the GFP-IRF-3 and IRF-9 expression plasmids; Georg Kochs and Otto Haller (University of Freiberg) for providing anti-MxA antibody and the IRF-1-luciferase reporter plasmid; and David E. Levy (New York University) for providing the ISG54-CAT reporter plasmid.
REFERENCES
- 1.Aebi, M., J. Fäh, N. Hurt, C. E. Samuel, D. Thomis, L. Bazzigher, J. Pavlovic, O. Haller, and P. Staeheli. 1989. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol. Cell. Biol. 9:5062-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baize, S., E. M. Leroy, E. Mavoungou, and S. P. Fisher-Hoch. 2000. Apoptosis in fatal Ebola infection. Does the virus toll the bell for immune system? Apoptosis 5:5-7. [DOI] [PubMed] [Google Scholar]
- 3.Bamberg, S., L. Kolesnikova, P. Möller, H.-D. Klenk, and S. Becker. 2005. VP24 of Marburg virus influences formation of infectious particles. J. Virol. 79:13421-13433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reference deleted.
- 5.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Mühlberger, M. Bray, H.-D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basler, C. F., and P. Palese. 2004. Modulation of innate immunity by filoviruses, p. 305-349. In H.-D. Klenk and H. Feldmann (ed.), Molecular and cellular biology of Marburg and Ebola viruses. Horizon Scientific Press, Norfolk, United Kingdom.
- 7.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosio, C. M., M. J. Aman, C. Grogan, R. Hogan, G. Ruthel, D. Negley, M. Mohamadzadeh, S. Bavari, and A. Schmaljohn. 2003. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J. Infect. Dis. 188:1630-1638. [DOI] [PubMed] [Google Scholar]
- 9.Bray, M. 2001. The role of the type I interferon response in the resistance of mice to filovirus infection. J. Gen. Virol. 82:1365-1373. [DOI] [PubMed] [Google Scholar]
- 10.Bray, M., K. Davis, T. Geisbert, C. Schmaljohn, and J. Huggins. 1998. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J. Infect. Dis. 178:651-661. [DOI] [PubMed] [Google Scholar]
- 11.Bray, M., and T. W. Geisbert. 2005. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int. J. Biochem. Cell. Biol. 37:1560-1566. [DOI] [PubMed] [Google Scholar]
- 12.Bray, M., J. L. Raymond, T. Geisbert, and R. O. Baker. 2002. 3-Deazaneplanocin A induces massively increased interferon-alpha production in Ebola virus-infected mice. Antivir. Res. 55:151-159. [DOI] [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 2005. Outbreak of Marburg virus hemorrhagic fever—Angola, October 1, 2004-March 29, 2005. Morb. Mortal. Wkly. Rep. 54:308-309. [PubMed] [Google Scholar]
- 14.Chan, S. Y., M. C. Ma, and M. A. Goldsmith. 2000. Differential induction of cellular detachment by envelope glycoproteins of Marburg and Ebola (Zaire) viruses. J. Gen. Virol. 81:2155-2159. [DOI] [PubMed] [Google Scholar]
- 15.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 16.Elliott, L. H., M. P. Kiley, and J. B. McCormick. 1985. Descriptive analysis of Ebola virus proteins. Virology 147:169-176. [DOI] [PubMed] [Google Scholar]
- 17.Fagerlund, R., K. Melen, L. Kinnunen, and I. Julkunen. 2002. Arginine/lysine-rich nuclear localization signals mediate interactions between dimeric STATs and importin alpha 5. J. Biol. Chem. 277:30072-30078. [DOI] [PubMed] [Google Scholar]
- 18.Geisbert, T. W., L. E. Hensley, T. R. Gibb, K. E. Steele, N. K. Jaax, and P. B. Jahrling. 2000. Apoptosis induced in vitro and in vivo during infection by Ebola and Marburg viruses. Lab. Investig. 80:171-186. [DOI] [PubMed] [Google Scholar]
- 19.Geisbert, T. W., L. E. Hensley, P. B. Jahrling, T. Larsen, J. B. Geisbert, J. Paragas, H. A. Young, T. M. Fredeking, W. E. Rote, and G. P. Vlasuk. 2003. Treatment of Ebola virus infection with a recombinant inhibitor of factor VIIa/tissue factor: a study in rhesus monkeys. Lancet 362:1953-1958. [DOI] [PubMed] [Google Scholar]
- 20.Geisbert, T. W., L. E. Hensley, T. Larsen, H. A. Young, D. S. Reed, J. B. Geisbert, D. P. Scott, E. Kagan, P. B. Jahrling, and K. J. Davis. 2003. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 163:2347-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisbert, T. W., H. A. Young, P. B. Jahrling, K. J. Davis, E. Kagan, and L. E. Hensley. 2003. Mechanisms underlying coagulation abnormalities in Ebola hemorrhagic fever: overexpression of tissue factor in primate monocytes/macrophages is a key event. J. Infect. Dis. 188:1618-1629. [DOI] [PubMed] [Google Scholar]
- 22.Gibb, T. R., M. Bray, T. W. Geisbert, K. E. Steele, W. M. Kell, K. J. Davis, and N. K. Jaax. 2001. Pathogenesis of experimental Ebola Zaire virus infection in BALB/c mice. J. Comp. Pathol. 125:233-242. [DOI] [PubMed] [Google Scholar]
- 23.Greenlund, A. C., M. A. Farrar, B. L. Viviano, and R. D. Schreiber. 1994. Ligand-induced IFN gamma receptor tyrosine phosphorylation couples the receptor to its signal transduction system (p91). EMBO J. 13:1591-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta, M., S. Mahanty, R. Ahmed, and P. E. Rollin. 2001. Monocyte-derived human macrophages and peripheral blood mononuclear cells infected with Ebola virus secrete MIP-1alpha and TNF-alpha and inhibit poly-IC-induced IFN-alpha in vitro. Virology 284:20-25. [DOI] [PubMed] [Google Scholar]
- 25.Han, Z., H. Boshra, J. O. Sunyer, S. H. Zwiers, J. Paragas, and R. N. Harty. 2003. Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding. J. Virol. 77:1793-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harcourt, B. H., A. Sanchez, and M. K. Offermann. 1998. Ebola virus inhibits induction of genes by double-stranded RNA in endothelial cells. Virology 252:179-188. [DOI] [PubMed] [Google Scholar]
- 27.Harcourt, B. H., A. Sanchez, and M. K. Offermann. 1999. Ebola virus selectively inhibits responses to interferons, but not to interleukin-1β, in endothelial cells. J. Virol. 73:3491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartman, A. L., J. S. Towner, and S. T. Nichol. 2004. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology 328:177-184. [DOI] [PubMed] [Google Scholar]
- 29.Haspel, R. L., and J. E. Darnell, Jr. 1999. A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc. Natl. Acad. Sci. USA 96:10188-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haspel, R. L., M. Salditt-Georgieff, and J. E. Darnell, Jr. 1996. The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends upon a protein tyrosine phosphatase. EMBO J 15:6262-6268. [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, Y., L. Xu, Y. Sun, and G. Nabel. 2002. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell 10:307. [DOI] [PubMed] [Google Scholar]
- 32.Huggins, J. W., Z. X. Zhang, and T. I. Monath. 1995. Inhibition of Ebola virus replication in vitro and in vivo in a SCID mouse model by S-adenosylhomocysteine hydrolase inhibitors. Antivir. Res. Suppl. 1:122. [Google Scholar]
- 33.Jahrling, P. B., T. W. Geisbert, J. B. Geisbert, J. R. Swearengen, M. Bray, N. K. Jaax, J. W. Huggins, J. W. LeDuc, and C. J. Peters. 1999. Evaluation of immune globulin and recombinant interferon-alpha2b for treatment of experimental Ebola virus infections. J. Infect. Dis. 179(Suppl. 1):S224-S234. [DOI] [PubMed] [Google Scholar]
- 34.Jones, S. M., H. Feldmann, U. Stroher, J. B. Geisbert, L. Fernando, A. Grolla, H. D. Klenk, N. J. Sullivan, V. E. Volchkov, E. A. Fritz, K. M. Daddario, L. E. Hensley, P. B. Jahrling, and T. W. Geisbert. 2005. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 11:786-790. [DOI] [PubMed] [Google Scholar]
- 35.King, P., and S. Goodbourn. 1994. The beta-interferon promoter responds to priming through multiple independent regulatory elements. J. Biol. Chem. 269:30609-30615. [PubMed] [Google Scholar]
- 36.Kumar, K. P., K. M. McBride, B. K. Weaver, C. Dingwall, and N. C. Reich. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 20:4159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehtonen, A., S. Matikainen, and I. Julkunen. 1997. Interferons up-regulate STAT1, STAT2, and IRF family transcription factor gene expression in human peripheral blood mononuclear cells and macrophages. J. Immunol. 159:794-803. [PubMed] [Google Scholar]
- 38.Levy, D. E., and J. E. Darnell, Jr. 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell. Biol. 3:651-662. [DOI] [PubMed] [Google Scholar]
- 39.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 40.Licata, J. M., R. F. Johnson, Z. Han, and R. N. Harty. 2004. Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J. Virol. 78:7344-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahanty, S., and M. Bray. 2004. Pathogenesis of filoviral haemorrhagic fevers. Lancet Infect. Dis. 4:487-498. [DOI] [PubMed] [Google Scholar]
- 42.Mahanty, S., M. Gupta, J. Paragas, M. Bray, R. Ahmed, and P. E. Rollin. 2003. Protection from lethal infection is determined by innate immune responses in a mouse model of Ebola virus infection. Virology 312:415-424. [DOI] [PubMed] [Google Scholar]
- 43.Mahanty, S., K. Hutchinson, S. Agarwal, M. McRae, P. E. Rollin, and B. Pulendran. 2003. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J. Immunol. 170:2797-2801. [DOI] [PubMed] [Google Scholar]
- 44.Marg, A., Y. Shan, T. Meyer, T. Meissner, M. Brandenburg, and U. Vinkemeier. 2004. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J. Cell Biol. 165:823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McBride, K. M., G. Banninger, C. McDonald, and N. C. Reich. 2002. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO J. 21:1754-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McBride, K. M., C. McDonald, and N. C. Reich. 2000. Nuclear export signal located within the DNA-binding domain of the STAT1 transcription factor. EMBO J. 19:6196-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McBride, K. M., and N. C. Reich. 2003. The ins and outs of STAT1 nuclear transport. Sci. STKE 2003:RE13. [Online.] doi: 10.1126/stke.2003.195.re13. [DOI] [PubMed] [Google Scholar]
- 48.Melen, K., R. Fagerlund, J. Franke, M. Kohler, L. Kinnunen, and I. Julkunen. 2003. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem. 278:28193-28200. [DOI] [PubMed] [Google Scholar]
- 49.Meyer, T., and U. Vinkemeier. 2004. Nucleocytoplasmic shuttling of STAT transcription factors. Eur. J. Biochem. 271:4606-4612. [DOI] [PubMed] [Google Scholar]
- 50.Park, M.-S., M. L. Shaw, J. Muñoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reid, S. P., W. B. Cardenas, and C. F. Basler. 2005. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology 341:179-189. (First published 10 August 2005; doi: 10.1016/j.virol.2005.06.044.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez, J. J., J.-P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanchez, A., A. S. Khan, S. R. Zaki, G. J. Nabel, T. G. Ksiazek, and C. J. Peters. 2001. Filoviridae: Marburg and Ebola viruses, p. 1279-1304. In B. N. Fields and D. M. Knipe (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams and Wilkins, Philadelphia, Pa. [Google Scholar]
- 54.Schindler, C., K. Shuai, V. R. Prezioso, and J. E. Darnell, Jr. 1992. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science 257:809-813. [DOI] [PubMed] [Google Scholar]
- 55.Sekimoto, T., N. Imamoto, K. Nakajima, T. Hirano, and Y. Yoneda. 1997. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 16:7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw, M. L., A. Garcia-Sastre, P. Palese, and C. F. Basler. 2004. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 78:5633-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shuai, K., C. M. Horvath, L. H. Huang, S. A. Qureshi, D. Cowburn, and J. E. Darnell, Jr. 1994. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 76:821-828. [DOI] [PubMed] [Google Scholar]
- 58.Shuai, K., C. Schindler, V. R. Prezioso, and J. E. Darnell, Jr. 1992. Activation of transcription by IFN-gamma: tyrosine phosphorylation of a 91-kD DNA binding protein. Science 258:1808-1812. [DOI] [PubMed] [Google Scholar]
- 59.Sullivan, N. J., T. W. Geisbert, J. B. Geisbert, L. Xu, Z. Y. Yang, M. Roederer, R. A. Koup, P. B. Jahrling, and G. J. Nabel. 2003. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature 424:681-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sullivan, N. J., A. Sanchez, P. E. Rollin, Z. Y. Yang, and G. J. Nabel. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 408:605-609. [DOI] [PubMed] [Google Scholar]
- 61.Volchkov, V. E., A. A. Chepurnov, V. A. Volchkova, V. A. Ternovoj, and H. D. Klenk. 2000. Molecular characterization of guinea pig-adapted variants of Ebola virus. Virology 277:147-155. [DOI] [PubMed] [Google Scholar]
- 62.Volchkov, V. E., V. A. Volchkova, E. Muhlberger, L. V. Kolesnikova, M. Weik, O. Dolnik, and H. D. Klenk. 2001. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science 291:1965-1969. [DOI] [PubMed] [Google Scholar]
- 63.Watanabe, S., T. Watanabe, T. Noda, A. Takada, H. Feldmann, L. D. Jasenosky, and Y. Kawaoka. 2004. Production of novel Ebola virus-like particles from cDNAs: an alternative to Ebola virus generation by reverse genetics. J. Virol. 78:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang, Z. Y., H. J. Duckers, N. J. Sullivan, A. Sanchez, E. G. Nabel, and G. J. Nabel. 2000. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 6:886-889. [DOI] [PubMed] [Google Scholar]