Abstract
Production of extended-spectrum β-lactamases and plasmid-mediated AmpC enzymes was investigated among 291 Escherichia coli and 282 Klebsiella pneumoniae isolates that showed decreased susceptibilities to extended-spectrum cephalosporins from seven Taiwanese medical centers. CTX-M-type and SHV-type enzymes were the most prevalent extended-spectrum β-lactamases. CMY-2-like and DHA-1-like β-lactamases were the most prevalent AmpC-type enzymes.
The increasing prevalence of extended-spectrum β-lactamases (ESBLs) and plasmid-mediated AmpC β-lactamases in members of the family Enterobacteriaceae is a matter of great concern worldwide (1, 2, 10). In Taiwan, TEM-, SHV-, and CTX-M-type ESBLs and CMY- and DHA-type AmpC β-lactamases have been reported in Escherichia coli and Klebsiella pneumoniae isolates from a few individual institutions (3, 17-20). The present multicenter study was conducted to determine the distribution of ESBLs and AmpC β-lactamases among clinical isolates of E. coli and K. pneumoniae that showed decreased susceptibilities to extended-spectrum cephalosporins in Taiwan.
Isolates that were suspected of ESBL production by the CLSI (formerly NCCLS) screening method (8) with disks of ceftazidime, cefotaxime, ceftriaxone, aztreonam, and/or cefpodoxime were considered to exhibit decreased susceptibilities to extended-spectrum cephalosporins and consecutively collected between March and August 2003 from seven medical centers in Taiwan. These hospitals were chosen because they represent the largest referral medical centers in each region of Taiwan. A total of 291 E. coli and 282 K. pneumoniae isolates were obtained (Table 1). Each isolate came from a unique patient.
TABLE 1.
Distribution of ESBLs and AmpC enzymes among 291 E. coli and 282 K. pneumoniae isolates that showed decreased susceptibilities to extended-spectrum cephalosporins from seven medical centers in Taiwan
| Species and ESBL or AmpC | pI of β-lactamase | No. of isolates from hospitala:
|
Total no. (%) of isolates | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N1 | N2 | C1 | C2 | C3 | S | E | |||
| E. coli | 78 | 33 | 84 | 41 | 18 | 19 | 18 | 291 | |
| CMY-2-like | 9.0 | 57 | 0 | 40 | 13 | 8 | 1 | 8 | 127 (43.6) |
| CTX-M | 21 | 28 | 47 | 28 | 8 | 17 | 9 | 158 (54.3) | |
| CTX-M-1 group | 8.4 or 8.6 | 5 | 4 | 19 | 5 | 2 | 2 | 4 | 41 (14.1) |
| CTX-M-9 group | 7.9 or 8.0 | 16 | 24 | 28 | 23 | 6 | 15 | 5 | 117 (40.2) |
| SHV-5-like | 8.2 | 10 | 3 | 5 | 4 | 2 | 3 | 0 | 27 (9.3) |
| Noneb | 1 | 3 | 4 | 1 | 3 | 0 | 2 | 14 (4.8) | |
| K. pneumoniae | 58 | 59 | 78 | 18 | 23 | 37 | 9 | 282 | |
| CMY-2-like | 9.0 | 4 | 1 | 4 | 0 | 0 | 1 | 0 | 10 (3.5) |
| DHA-1-like | 7.8 | 17 | 1 | 5 | 1 | 3 | 4 | 0 | 31 (11.0) |
| CTX-M | 21 | 21 | 67 | 15 | 16 | 10 | 5 | 155 (55.0) | |
| CTX-M-1 group | 8.4 | 12 | 7 | 44 | 8 | 14 | 8 | 5 | 98 (34.8) |
| CTX-M-9 group | 7.9 or 8.0 | 9 | 14 | 23 | 7 | 2 | 2 | 0 | 57 (20.2) |
| SHV | 34 | 39 | 18 | 1 | 7 | 32 | 4 | 135 (47.9) | |
| SHV-2-like | 8.2 | 0 | 5 | 1 | 1 | 0 | 2 | 0 | 9 (3.2) |
| SHV-5-like | 7.6 | 34 | 34 | 17 | 0 | 7 | 30 | 4 | 126 (44.7) |
| None | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 5 (1.8) | |
The seven medical centers were Triservice General Hospital (N1) and National Taiwan University Hospital (N2) in northern Taiwan, Taichung Veterans General Hospital (C1), China Medical College Hospital (C2), and Chung Shan Medical University Hospital (C3) in central Taiwan, Chi-Mei Medical Center (S) in southern Taiwan, and Buddhist Tzu Chi General Hospital (E) in eastern Taiwan. Hospitals C1 and S are district general hospitals, and the remainder are university hospitals.
ESBLs and AmpC-like enzymes not detected.
All isolates were tested for ESBL production by the CLSI-recommended disk diffusion confirmatory test (8), and 171 E. coli and 260 K. pneumoniae isolates were considered ESBL producers. All putative non-ESBL producers were tested further by the double-disk synergy test with ceftazidime, cefotaxime, aztreonam, and cefepime disks placed 20 mm from an amoxicillin-clavulanate disk (13). The method has been shown to be capable of detecting false negatives which are caused by coproduction of different classes of β-lactamases in the CLSI method (20). Five E. coli and five K. pneumoniae isolates gave a positive result in the double-disk synergy test. Thus, ESBL production was detected in 176 (60.5%) of the 291 E. coli isolates and 265 (94.0%) of the 282 K. pneumoniae isolates.
The expression of β-lactamases was detected by isoelectric focusing as described previously (6, 18). PCR was performed with the previously reported oligonucleotide primers to detect blaTEM (5), blaSHV (9), and bla genes related to blaCTX-M-1 (12), blaCTX-M-9 (12), blaCMY-1 (17), blaCMY-2 (16), blaDHA-1 (4), and blaOXA-10 (14). The PCR-NheI method was used to discriminate between blaSHV-ESBL and blaSHV-non-ESBL genes (9).
Overall, the production of ESBLs and AmpC-like enzymes was confirmed in 60.5% (176 isolates) and 43.6% (127 isolates), respectively, of the 291 E. coli isolates, and in 94.0% (265 isolates) and 14.5% (41 isolates), respectively, of the 282 K. pneumoniae isolates (Table 1). Thirty-five (12.0%) E. coli isolates and 46 (16.3%) K. pneumoniae isolates harbored two or three β-lactamases involved in extended-spectrum cephalosporin resistance. CTX-M-type ESBLs and CMY-2-related AmpC enzymes were predominant in E. coli, while SHV- and CTX-M-type ESBLs, and DHA-1-like AmpC enzymes were most prevalent in K. pneumoniae (Table 1). Among the CTX-M-type ESBLs, the CTX-M-1 and CTX-M-9 groups were predominant in K. pneumoniae and in E. coli, respectively. All the above enzymes showed a nationwide distribution.
Among the 10 ESBL-producing isolates negative for the CLSI confirmatory method, all five E. coli isolates coproduced a CMY-2-related enzyme and an SHV-5-related ESBL, and all five K. pneumoniae isolates coproduced a DHA-1-related enzyme and an SHV-5-related ESBL. The presence of additional β-lactamases (non-ESBL, non-AmpC-like) was also suggested by PCR and isoelectric focusing. They were pI 5.4 TEM-1-like (238 E. coli isolates and 194 K. pneumoniae isolates), pI 6.1 OXA-10-like (5 K. pneumoniae isolates), and pI 7.6 SHV-1-like (all 282 K. pneumoniae isolates) β-lactamases. No β-lactamase was detected in three E. coli isolates, and no CMY-1-related enzymes were detected in any isolate.
PCR products from randomly selected isolates were sequenced on both strands. The sequences of selected amplicons of blaDHA-1-, blaCMY-2-, and blaOXA-10-related genes were all identical to those of blaDHA-1 (23 isolates), blaCMY-2 (16 isolates), and blaOXA-10 (5 isolates), respectively. The results of sequence analysis of 251 blaCTX-M-positive isolates are shown in Table 2. Seven blaCTX-M subtypes were identified, which were blaCTX-M-3, blaCTX-M-9, blaCTX-M-14, blaCTX-M-15, blaCTX-M-17, blaCTX-M-19, and an unpublished blaCTX-M-9-related gene, blaCTX-M-38 (GenBank accession no. AY822595). blaCTX-M-3 and blaCTX-M-14 were the most common blaCTX-M subtypes in K. pneumoniae and E. coli, respectively.
TABLE 2.
blaCTX subtypes detected in randomly selected E. coli (n = 128) and K. pneumoniae (n = 123) isolates from seven medical centers in Taiwan
| Species and blaCTX-M | No. of isolates from hospitala:
|
Total no. of isolates | ||||||
|---|---|---|---|---|---|---|---|---|
| N1 | N2 | C1 | C2 | C3 | S | E | ||
| E. coli | ||||||||
| blaCTX-M-1 group | ||||||||
| blaCTX-M-3 | 4 | 3 | 15 | 4 | 2 | 2 | 3 | 33 |
| blaCTX-M-15 | 1 | 1 | 2 | |||||
| blaCTX-M-9 group | ||||||||
| blaCTX-M-9 | 1 | 1 | 2 | 4 | ||||
| blaCTX-M-14 | 12 | 17 | 21 | 14 | 3 | 14 | 7 | 88 |
| blaCTX-M-19 | 1 | 1 | ||||||
| K. pneumoniae | ||||||||
| blaCTX-M-1 group | ||||||||
| blaCTX-M-3 | 8 | 6 | 41 | 4 | 14 | 5 | 4 | 82 |
| blaCTX-M-9 group | ||||||||
| blaCTX-M-14 | 5 | 7 | 12 | 4 | 2 | 3 | 33 | |
| blaCTX-M-17 | 1 | 2 | 1 | 1 | 5 | |||
| blaCTX-M-19 | 2 | 2 | ||||||
| blaCTX-M-38 | 1 | 1 | ||||||
See Table 1, footnote a.
blaCTX-M-17, blaCTX-M-19, and blaCTX-M-38 were first identified in Taiwan. The sequence of blaCTX-M-38 first deposited in the GenBank database was obtained from a K. pneumoniae strain from China. blaCTX-M-38 differs from blaCTX-M-14 by a C-to-G change at position 669 of the structural gene, leading to a Ser220Arg substitution. In the susceptibility test by the standard agar dilution method (7), our blaCTX-M-38-positive isolate showed high-level resistance to cefotaxime (MIC, >128 μg/ml) and decreased susceptibilities to ceftazidime (MIC, 8 μg/ml), aztreonam (MIC, 32 μg/ml), and cefepime (MIC, 16 μg/ml) and was susceptible to cefoxitin (MIC, 16 μg/ml) and imipenem (MIC, 1 μg/ml). The resistance phenotype is similar to those of reported CTX-M-14-producing strains (1).
Isolates carrying blaCTX-M-3, blaCTX-M-14, or blaCMY-2 were randomly selected from the seven hospitals for enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) analysis with primer ERIC2 (15). Each unique pattern was defined as an ERIC-PCR type. Thirteen, 11, and 10 types were observed among 20 CTX-M-14-producing isolates, 16 CTX-M-3-producing isolates, and 15 CMY-2-producing isolates, respectively, in E. coli, and 11 and 14 types were observed among 16 CTX-M-14-producing isolates and 21 CTX-M-3-producing isolates, respectively, in K. pneumoniae. Clonal relationships between isolates with the same ERIC-PCR type were further confirmed by pulsed-field gel electrophoresis with chromosomal DNA restricted by XbaI as described previously (18). No interhospital clonal dissemination of strains carrying the three most common resistance genes was observed (data not shown).
Selected isolates belonging to different lineages (as per the results of ERIC-PCR typing) were subjected to the liquid mating-out assay as described previously (11, 19). Transconjugants were selected on tryptic soy agar plates containing streptomycin (512 μg/ml) and ceftazidime (2 μg/ml) or cefotaxime (2 μg/ml). Thirteen, 18, and 17 transconjugants were obtained from 15 CMY-2-producing E. coli isolates, 20 CTX-M-14-producing E. coli isolates, and 21 CTX-M-3-producing K. pneumoniae isolates, respectively.
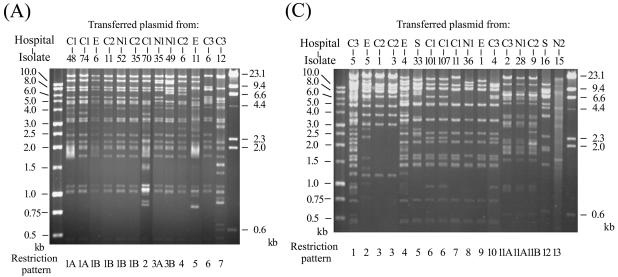
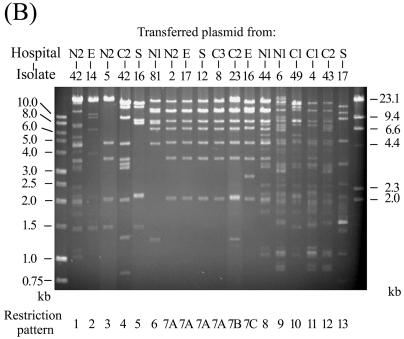
The presence of transferred relevant β-lactamase genes in transconjugants was confirmed by PCR and isoelectric focusing. Plasmids from the transconjugants were extracted and analyzed by agarose gel electrophoresis after digestion with the endonuclease EcoRI. Identical or similar restriction patterns were observed among the transferred plasmids from the isolates collected from different hospitals (Fig. 1), suggesting that horizontal transfer of similar resistance plasmids could play an important role in the wide spread of the resistance genes in Taiwan.
FIG. 1.
EcoRI restriction patterns of conjugative plasmids transferred from 13 CMY-2-producing E. coli isolates (A), 18 CTX-M-14-producing E. coli isolates (B), and 17 CTX-M-3-producing K. pneumoniae isolates (C). The numbers designating the donor isolates are shown above the gels, and numbers designating restriction patterns are shown below the gels.
In conclusion, this study demonstrated the widespread occurrence of ESBLs and plasmid-mediated AmpC enzymes in E. coli and K. pneumoniae in Taiwan and particularly points out the high prevalence and increasing diversity of CTX-M-type ESBLs. Our results underline the need for continuous surveillance of the prevalence and evolution of these enzymes in Taiwan.
Acknowledgments
We thank George A. Jacoby, Lahey Clinic, for confirming the formal assignment of blaCTX-M-38.
This work was supported by grant NSC94-2320-B-006-048 from the National Science Council, Taiwan.
REFERENCES
- 1.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, F.-Y., L. K. Siu, C.-P. Fung, M.-H. Huang, and M. Ho. 2001. Diversity of SHV and TEM β-lactamases in Klebsiella pneumoniae: gene evolution in northern Taiwan and two novel β-lactamases, SHV-25 and SHV-26. Antimicrob. Agents Chemother. 45:2407-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fortineau, N., L. Poirel, and P. Nordmann. 2001. Plasmid-mediated and inducible cephalosporinase DHA-2 from Klebsiella pneumoniae. J. Antimicrob. Chemother. 47:207-210. [DOI] [PubMed] [Google Scholar]
- 5.Mabilat, C., and S. Goussard. 1993. PCR detection and identification of genes for extended-spectrum β-lactamases, p. 553-559. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 6.Matthew, M., M. Harris, M. J. Marshall, and G. W. Rose. 1975. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 7.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 8.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial disk susceptibility tests, 8th ed. Approved standard M2-A8. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 9.Nüesch-Inderbinen, M. T., H. Hächler, and F. H. Kayser. 1996. Detection of genes coding for extended-spectrum SHV beta-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur. J. Clin. Microbiol. Infect. Dis. 15:398-402. [DOI] [PubMed] [Google Scholar]
- 10.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Provence, D. L., and R. Curtiss III. 1994. Gene transfer in gram-negative bacteria, p. 319-347. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 12.Saladin, M., V. T. B. Cao, T. Lambert, J.-L. Donay, J.-L. Herrmann, Z. Ould-Hocine, C. Verdit, F. Delisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M β-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 13.Tzelepi, E., P. Giakkoupi, D. Sofianou, V. Loukova, A. Kemeroglou, and A. Tsakris. 2000. Detection of extended-spectrum β-lactamases in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J. Clin. Microbiol. 38:542-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vahaboglu, H., R. Ozturk, H. Akbal, S. Saribas, O. Tansel, and F. Coşkunkan. 1998. Practical approach for detection and identification of OXA-10-derived ceftazidime-hydrolyzing extended-spectrum β-lactamases. J. Clin. Microbiol. 36:827-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winokur, P. L., A. Brueggemann, D. L. Desalvo, L. Hoffmann, M. D. Apley, E. K. Uhlenhopp, M. A. Pfaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC β-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan, J. J., S. M. Wu, S. H. Tsai, J. J. Wu, and I. J. Su. 2000. Prevalence of SHV-12 among clinical isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamase and identification of a novel AmpC enzyme (CMY-8) in southern Taiwan. Antimicrob. Agents Chemother. 44:1438-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan, J. J., W. C. Ko, S. H. Tsai, H. M. Wu, Y. T. Jin, and J. J. Wu. 2000. Dissemination of CTX-M-3 and CMY-2 β-lactamases among clinical isolates of Escherichia coli in southern Taiwan. J. Clin. Microbiol. 38:4320-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan, J. J., W. C. Ko, Y. C. Jung, C. L. Chuang, and J. J. Wu. 2002. Emergence of Klebsiella pneumoniae isolates producing inducible DHA-1 β-lactamase in a university hospital in Taiwan. J. Clin. Microbiol. 40:3121-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan, J. J., W. C. Ko, H. M. Wu, S. H. Tsai, C. L. Chuang, and J. J. Wu. 2004. Complexity of Klebsiella pneumoniae isolates resistant to both cephamycins and extended-spectrum cephalosporins at a teaching hospital in Taiwan. J. Clin. Microbiol. 42:5337-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]