Abstract
Lipoic acid is essential for the activation of a number of protein complexes involved in key metabolic processes. Growth of Mycobacterium tuberculosis relies on a pathway in which the lipoate attachment group is synthesized from an endogenously produced octanoic acid moiety. In patients with multiple-drug-resistant M. tuberculosis, expression of one gene from this pathway, lipB, encoding for octanoyl-[acyl carrier protein]-protein acyltransferase is considerably up-regulated, thus making it a potential target in the search for novel antiinfectives against tuberculosis. Here we present the crystal structure of the M. tuberculosis LipB protein at atomic resolution, showing an unexpected thioether-linked active-site complex with decanoic acid. We provide evidence that the transferase functions as a cysteine/lysine dyad acyltransferase, in which two invariant residues (Lys-142 and Cys-176) are likely to function as acid/base catalysts. Analysis by MS reveals that the LipB catalytic reaction proceeds by means of an internal thioesteracyl intermediate. Structural comparison of LipB with lipoate protein ligase A indicates that, despite conserved structural and sequence active-site features in the two enzymes, 4′-phosphopantetheine-bound octanoic acid recognition is a specific property of LipB.
Keywords: catalytic dyad, lipoic acid, x-ray structure, thioester formation, mass spectrometry
Several multicomponent enzyme complexes that catalyze key metabolic reactions in the citric acid cycle and single-carbon metabolism are posttranslationally modified by attachment to lipoic acid (1). These systems share a domain that covalently binds lipoic acid by means of an amide bond to the ε-amino group of a conserved exposed lysine residue. In many organisms, lipoylation is catalyzed by two separate enzymes, lipoyl protein ligase A (LplA) or octanoyl-[acyl carrier protein]-protein transferase (LipB; see Fig. 5, which is published as supporting information on the PNAS web site). Although LplA uses exogenous lipoic acid, LipB transfers endogenous octanoic acid, which is attached by means of a thioester bond to the 4′-phosphopantetheine cofactor of acyl carrier protein (ACP) onto lipoyl domains (2–4). These octanoylated domains are converted into lipoylated derivatives by the S-adenosyl-l-methionine-dependent enzyme, lipoyl synthase (LipA), which catalyzes the insertion of sulfur atoms into the six- and eight-carbon positions of the corresponding fatty acid (5–7). This process bypasses the requirement for an exogenous supply of lipoic acid.
In bacteria, enzymes involved in lipoylation have gained increasing attention because of their implication in pathogenicity. For instance, a Listeria monocytogenes mutant strain lacking LplA has been found to be defective in its ability to grow in the host cytosol and is less virulent in animals because of its dependence on host-derived lipoic acid (8). Mice Lias−/− (LipA) null variants die during embryogenesis, indicating that the mammalian pathway is related to the bacterial LipB/LipA pathway and is essential for life (9). In contrast, in apicomplexan parasites, such as Plasmodium falciparum and Toxoplasma gondii, the two lipoylation pathways appear to be compartmentalized and are found either in mitochondria (LplA) or in the apicoplasts (LipA and LipB) (10, 11).
Tuberculosis remains one of the most threatening infectious diseases worldwide, with highest incidence rates in parts of Asia and Africa (12), mandating accelerated research efforts toward identification of novel drug targets. To date, LipB has emerged as one of the promising candidates, based on recent evidence suggesting that its expression is strongly up-regulated in the lungs of patients with pulmonary multiple-drug-resistant tuberculosis (13). Previous efforts to generate a knockout mutant lacking the lipB gene consistently failed (A.N.E. and J. S. Lee, unpublished data), in agreement with published findings postulating that LipB plays an essential role in the growth of Mycobacterium tuberculosis (14). Therefore, we embarked on the structural and functional characterization of LipB from M. tuberculosis (mtbLipB). Unexpectedly, the atomic-resolution structure of LipB revealed a covalent adduct that was identified as thioether-bound decanoic acid. Based on evidence by biochemical, MS, and structural data, we propose a two-step catalytic mechanism by means of a thioester acyl intermediate. Our findings are unprecedented, because they indicate that LipB functions as a cysteine/lysine acyltransferase.
Results
Atomic-Resolution Structure of the LipB–Decanoic Acid Complex.
We have determined the x-ray structure of mtbLipB at 1.08-Å resolution (Figs. 1 and 2; see Table 2 and Fig. 6, which are published as supporting information on the PNAS web site). Gel filtration data using the purified enzyme (data not shown) and the content of the LipB crystals (one molecule per asymmetric unit) indicate that mtbLipB is monomeric. The overall structure of mtbLipB consists of a central five-stranded β-sheet that is wrapped by three long helices on its two opposite faces. In addition, the structure comprises extensive loops and a small second β-sheet, referred to as the “capping sheet,” plus several short helical turns. Residues 217–230 from the very C terminus were not modeled because of insufficient electron density.
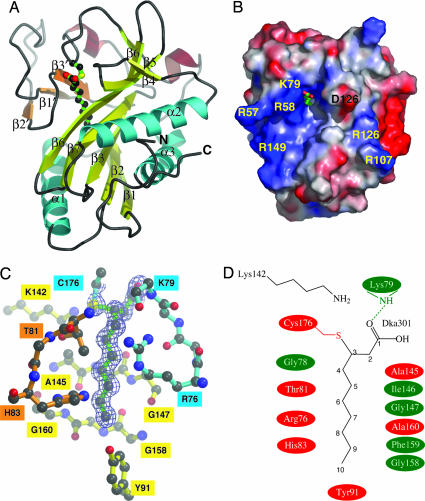
Fig. 1.
Molecular structure of mtbLipB. (A) Ribbon representation of mtbLipB. Color codes: core β-sheet, yellow; capping β-sheet, orange; α-helices, wrapping the core β-sheet, cyan; other α-helices, brown. The decanoic acid–Cys-176 adduct is shown in ball-and-stick representation, using atom-type colors and bonds in green. (B) Surface presentation of mtbLipB, showing relative electrostatic potentials, using the apbs program incorporated in pymol (see ref. 30; www.pymol.org). The visible part of the decanoid acid adduct is shown by sticks in atom-type colors. The orientation has been optimized to provide a view into the mtbLipB active site. Some charged surface residues within the vicinity of the access area into the active site are labeled. (C) mtbLipB active site in ball-and-stick representation, using the color scheme from A. The σA-weighted final 2mFo−DFc electron density covering the decanoic acid moiety is shown at a contour level of 1σ. (D) Schematic presentation of the mtbLipB active site. Specific interactions are limited to Cys-176 and Lys-79. All other interactions are shown schematically; involvement of side chains is in red, and involvement of main chain atoms only is in green.
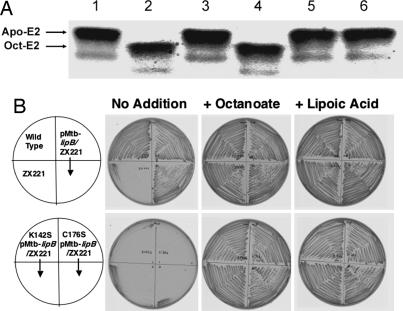
Fig. 2.
Biochemical data. (A) In vitro gel-shift assay. Lane 1, mtbLipB, expressed in E. coli, without octanoy-l-ACP; lane 2, mtbLipB, expressed in M. smegmatis; lane 3, negative control; lane 4, mtbLipB, expressed in E. coli; lane 5, mtbLipB(K142S), expressed in E. coli; lane 6, mtbLipB(C176S), expressed in E. coli. (B) Abilities of wild-type and mutant M. tuberculosis lipB genes to complement growth of an E. coli lipB null mutant strain. The upper row of plates shows the growth behavior of the wild-type E. coli strain JK1 (left top quadrant), the lipB::cat mutant strain ZX221 derived from JK1 (left bottom quadrant), and strain ZX221 carrying the plasmid encoding for the wild-type M. tuberculosis LipB protein (both right quadrants). The lower row of plates similarly shows growth of strain ZX221 carrying plasmid encoding for the mutant M. tuberculosis LipB proteins, either the K142S protein (both left quadrants) or the C176S protein (both right quadrants). Experimental procedures have been described (7).
During structural refinement, we observed additional electron density linked to Cys-176, a strictly conserved residue, which could not be accounted for by protein atoms or by ordered solvent molecules (Fig. 1C). MS analysis of purified mtbLipB revealed two peaks of similar heights (Table 1). The first peak corresponds to the theoretical mass of the mtbLipB polypeptide chain (24,308 Da). The second peak (22,478 Da) had a mass 170 Da greater and was interpreted to be an mtbLipB–decanoic acid complex. Indeed, our high-resolution x-ray data revealed the presence of decanoic acid to be covalently attached by means of a thioether bond between carbon-3 and the thiol of Cys-176 (Fig. 1 C and D). Occupancy refinement of the decanoic acid adduct, which is reliable at the resolution of the available mtbLipB diffraction data, did not indicate a substoichiometric contribution of the decanoic acid adduct. Therefore, we concluded that the crystals used for x-ray structure determination contained the mtbLipB–decanoic acid complex only. Our structural findings are supported by MS analysis demonstrating that a variant of the enzyme in which Cys-176 was mutated fails to bind decanoic acid (Table 1). Attempts to crystallize the apo form of mtbLipB failed, indicating that the presence of the covalently bound adduct was essential for crystallization (data not shown).
Table 1.
mtbLipB mass spectrometry data
| Sample | Molecular mass, Da | Interpretation |
|---|---|---|
| LipB (E. coli) | 24,307.7 | apo |
| 24,478.2 | Decanoic acid thioether adduct | |
| LipB(C176S) (E. coli) | 24,293.3 | apo |
| LIpB(K142S)(E. coli) | 24,268.1 | apo |
| LipB (M. smegmatis) | 24,304.6 | apo |
| LipB + ACP (E. coli) | 24,307.9 | apo |
| 24,434.2 | Octanoic acid thioester adduct | |
| 24,478.1 | Decanoic acid thioether adduct | |
| LipB(C176S) + ACP (E. coli) | 24,292.6 | apo |
| LlpB(K142S) + ACP (E. coli) | 24,266.8 | apo |
| 24,392.9 | Octanoic acid thioester adduct |
ACP, octanoylated ACP.
Except for its terminal carboxylate group, decanoic acid is entirely buried in a tunnel-like cavity of mtbLipB. The cavity is formed by mostly conserved residues from the central mtbLipB β-sheet, the additional small β-sheet, helix α1, and several loop segments (Fig. 1 and Fig. 7, which is published as supporting information on the PNAS web site). The carbon chain of decanoic acid is held together by van der Waals interactions with residues from different sequence segments. Several of these contacts are with aromatic side chains of highly conserved mtbLipB residues (Tyr-22, His-49, His-83, and Tyr-91). The decanoic acid cavity is also lined with several conserved glycine residues (Gly-77, Gly-78, Gly-147, and Gly-158). Cys-176, which covalently binds decanoic acid, is part of a highly conserved residue motif (PCG) within the long loop segment that connects the C-terminal strand β5 of the central β-sheet and helix α3 (Figs. 1A and 5). The ε-amino group of the invariant Lys-142 from strand β6 is in close distance (3.4 Å) to the sulfhydryl group of Cys-176 (Fig. 1 C and D), suggesting formation of a charged Lys-142–Cys-176 hydrogen bond in the absence of substrate. Such an interaction would lead to a downward shift of the pKa value of Cys-176 on the order of two pH units (15, 16).
The high level of sequence conservation of the residues that form the decanoic acid-binding site (Figs. 1 C and D, 5, and 6) suggests the site actually represents the mtbLipB active site. However, the presence of covalently bound decanoic acid would lead to an irreversible inhibition of mtbLipB, with toxic or even lethal effects for M. tuberculosis. Therefore, we hypothesized that the observed adduct could be an artifact of heterologous mtbLipB expression in Escherichia coli, an organism that has a fatty acid synthesis pathway that markedly differs from that of mycobacteria. To test this hypothesis, we expressed mtbLipB using the nonpathogenic mycobacterium Mycobacterium smegmatis as the host. Subsequent MS analysis revealed that, indeed, contrary to E. coli-expressed mtbLipB, the enzyme did not contain a covalently bound ligand adduct (Table 1), demonstrating that such irreversible inhibition is not found under the living conditions of mycobacteria. These findings may reflect differences in the fatty acid biosynthesis pathways in E. coli and mycobacteria. For instance, the decanoic acid adduct found in E. coli-expressed LipB seems likely to be the result of a Michael addition of the Cys-176 thiol to the unsaturated bond of trans-2 decanoyl-ACP or cis-3-decanoyl-ACP, a key intermediate in E. coli fatty acid biosynthesis (17).
The modified mtbLipB adduct, nonetheless, turned out to serve as an excellent substrate analog model for further investigations into the mtbLipB catalyzed reaction using structure-based biochemistry experiments (Fig. 2). Because the LipB substrate, ACP-4′phosphopantetheine-linked decanoic acid or octanoic acid, differs by two methylene groups, it is plausible to assume that the eight-carbon substrate should superimpose with carbon positions C3–C10 of decanoic acid (Fig. 1 C and D). This model suggests that Cys-176, which is involved in the adduct thioether linkage in the crystal structure, functions as a catalytic nucleophile on the carboxylic acid carbon during the acyltransferase reaction. Moreover, the presence of decanoic acid allowed us to model the linkage of the ACP-4′-phosphopantetheinyl-bound substrate with mtbLipB. In this scenario, the thioester linkage of 4′-phosphopantetheine to the carboxyl group of octanoic acid is expected to superimpose onto the C1 and C2 positions of the decanoic acid adduct (Fig. 1 C and D). The inherent flexibility of the 4′-phosphopantetheine cofactor, however, does not allow the building of a precise model of the rest of the cofactor. Nevertheless, the identification of the mtbLipB substrate access site provides an idea of the putative involvement of additional surface residues (Fig. 1B). We note a predominance of positively charged residues (Arg-58, Arg-79, Arg-130, and Arg-149) surrounding the access site; however, these are less conserved than most of the active-site residues forming the decanoic binding site. Similar observations have been made for the ACP docking site of β-ketoacyl-ACP synthase III, in which a cluster of positively charged residues is also found (18).
LipB Functions as a Cysteine/Lysine Acyltransferase.
To further assess the role of the strictly conserved residue pair Cys-176 and Lys-142 during mtbLipB catalysis, we mutated both residues to serines (K142S and C176S). We measured the in vitro enzymatic activity of the different mtbLipB versions by a gel-shift assay, using purified octanoyl-ACP and PDC apo-E2 domain as donor and acceptor, respectively (Fig. 2A). Although wild-type mtbLipB, expressed in either E. coli or M. smegmatis, was active, neither of the mtbLipB mutants allowed considerable octanoylation of the PDC E2 domain, thereby supporting our assumption that Lys-142 and Cys-176 are catalytic residues. Moreover, when we incubated mtbLipB with octanoyl-ACP in the absence of the PDC apo-E2 acceptor domain, MS analysis of the reaction mixture revealed an additional peak with a molecular mass 126 Da greater than that of the mtbLipB sequence, which corresponds to the mass of an octanoyl moiety (Table 1). In contrast, the adduct has not been observed for the mtbLipB C176S mutant. We interpret this finding as a thioester-bound acyl enzyme intermediate by means of Cys-176 that becomes trapped in the absence of the acceptor protein (Fig. 2A and Table 1).
To further validate our in vitro mtbLipB data, we assessed the in vivo activities of the mutant enzymes by measuring their ability to support the growth of a lipB-null mutant E. coli strain (Fig. 2B). In this assay, only the plasmid encoding for the wild-type mtbLipB supported the growth of the E. coli lipB strain on minimal medium. The growth rate of the complemented strain was comparable to that of a wild-type E. coli LipB strain, whereas the strains expressing the two mutant proteins failed to grow in this medium. Thus, mtbLipB can functionally replace E. coli lipB, with residues Lys-142 and Cys-176 being essential for the biological activity of mtbLipB.
Discussion
LipB and LplA Share Similar Active Sites Despite Unrelated Catalytic Mechanisms.
Recent sequence comparisons revealed distant relations among biotin protein ligase (BirA), LplA, and LipB, establishing a family of biotin/lipoyl/octanoyl attachment enzymes with a common ancestry (1, 19). The LplA and LipB products, lipoylated or octanoylated lipoyl acceptor domains, are closely related and can be interconverted by LipA (5, 6). Yet the presentation of their substrates is unrelated. The LplA and BirA reactions proceed by a noncovalently bound acyl adenylate intermediate, whereas LipB forms a covalently bound acyl thioester intermediate. On the other hand, although substrate adenylation is common to the BirA- and LplA-catalyzed reactions, suggesting similar catalytic mechanisms, the enzymes act on distinct acceptor domains.
In support of distant similarities among the different family members, an invariant lysine (Lys-142 in mtbLipB) was identified. Mutational analysis of the corresponding residue in BirA and LplA indeed reveals that the lysine side chain is essential for the function of all members of the biotin/lipoyl/octanoyl attachment enzyme family (Y. Jiang and J.E.C., unpublished data) and thus can be regarded as family marker. However, given that probably only LipB catalysis proceeds by means of a thioester intermediate, whereas LplA and BirA, because of the lack of a conserved active site cysteine, are likely to use a different catalytic mechanism, we hypothesize that the function of the conserved lysine differs among members of this family.
Whereas the first BirA structure became available more than a decade ago (20, 21), structures of LplA from E. coli and Thermoplasma acidophilum were published only recently (22, 23). The LplA structures were determined in the presence of lipoic acid (22) as well as in the presence of lipoyl-AMP and ATP (23). To date, no structural information on any LipB representative has been made available. Comparison of our mtbLipB structure with the available LplA structures indeed reveals distant structural similarity, and the protein structures can be superimposed by using all matching C-α positions with an rms deviation of ≈2.5 Å (Fig. 3A). The topology of most of the secondary structural elements match in LplA and LipB. At the very N terminus, the mtbLipB structure contains 10 additional residues that interact with helices α2 and α3, whereas the N terminus of LplA corresponds to the N terminus of the first strand of the mtbLipB central β-sheet. In LplA, the central β-sheet is wrapped by two long α-helices on each side, whereas the LplA C-terminal helical region (223–257) is missing in mtbLipB, reducing the overall number of wrapping helices to three. In addition, there are significant variations in some loop regions, generating three insertions in mtbLipB and another three insertions in LplA.
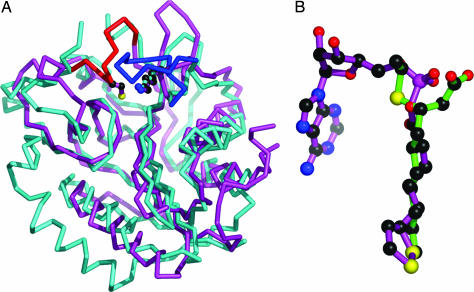
Fig. 3.
Comparison of the structures of LipB and LplA. (A) Superimposed Cα traces of structures of LipB from M. tuberculosis (cyan) and LplA from T. acidophilum (magenta). Enzyme-specific loops are shown in red and blue, respectively. Lys-142 and Cys-176 from LipB and Lys-145 from LplA are shown in ball-and-stick representation. (B) Superposition of decanoid acid (LipB) and lipoyl–AMP (LplA), based on the superimposed protein coordinates from A.
Despite these differences in the active-site structures of mtbLipB and LplA, we noticed that the corresponding active-site ligands in mtbLipB (decanoic acid) and in one of the two LplA structures (lipoyl–AMP; see ref. 23) are shared (Fig. 3B). Within the superimposed structures, the positions of the hydrocarbon tails of decanoic acid (LipB) and lipoyl-AMP (LplA) are in register. The terminal methyl group of decanoate and the dithiolane ring of lipoic acid superimpose on each other, demonstrating that the maximum length of the hydrocarbon portion is determined by the shapes of the respective binding sites. In addition, the invariant Lys-142 (Lys-145 in LplA) is found in an identical position with respect to the overall enzyme folds (Fig. 3A) and their bound ligands, suggesting a similar function in catalysis. In contrast, the AMP moiety of lipoyl–AMP in LplA is sterically blocked by a mtbLipB-specific loop comprising Cys-176, which is invariant among the available LipB sequences but is absent from any LplA sequences. In the LplA sequence, this loop comprises a 10-residue insertion, which is, however, disordered in the available structures (22, 23). Moreover, a long less-conserved loop that functions as a lid of the active site in LplA (127–137) is absent in mtbLipB, rendering its active site more accessible to substrates than that of LplA. Despite these structural differences, which account for the different substrate preferences of LipB and LplA, and in support of a related active-site architecture, it was possible to rescue growth of an E. coli lipB null mutant on lipoate by overexpressing LplA. The in vitro activity for lipoyl-ACP is ≈16,000 lower than that for lipoic acid plus ATP (4).
The structural similarity between mtbLipB and the available BirA structures is ≈2.5 Å for matching Cα positions and thus in the same order of magnitude as that seen for LplA (Fig. 3A). Previous findings on similarities between lipoate protein ligase A (LplA) and BirA (22, 23) are confirmed by our data.
Proposed Two-Step LipB Reaction Mechanism by Means of a Conserved Cysteine/Lysine Dyad.
Direct evidence from the structure of the mtbLipB–decanoic acid adduct, from our biochemical data and from a comparison of our findings with previous structural data on BirA and LplA, leaves little doubt that the carbon C3 position of decanoic acid mimics the thioester carbon of octanoic acid where the LipB transferase reactions take place. Our MS analysis (Table 1) demonstrates that the mtbLipB-catalyzed reaction involves a thioester intermediate in which the thiol group is donated by the invariant Cys-176. In light of the observed thioether bond formation of the decanoic acid adduct in our structure, it is plausible to model a thioester intermediate of octanoic acid (Fig. 4). In summary, our experimental and modeling data support a catalytic mechanism, assigning LipB an acyltransferase function, in which the ACP-linked 4′-phosphopantetheine thioester is replaced by a mtbLipB Cys-176 octanoyl-thioester.
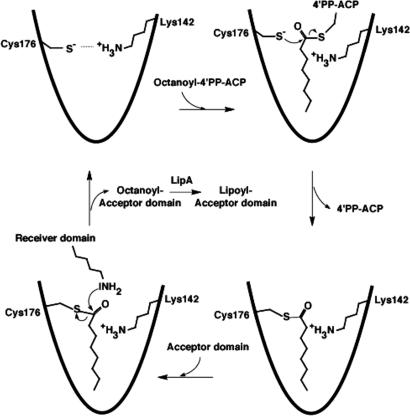
Fig. 4.
Structure-based proposal for a LipB reaction mechanism by means of a covalent Cys-176–acyl intermediate. The ε-amino group of the lysine from the lipoyl domain to which octanoate is covalently attached needs to be deprotonated to be catalytically active as a nucleophile. Given that there are neither possible candidate residues, except Lys-142, nor ordered solvent molecules near the active-site center in the LipB structure, we hypothesize there may be proton shuttling between Lys-142 from LipB and the lipoyl domain active lysine (not shown).
In addition, our structural and mutagenesis data strongly suggest involvement of the invariant Lys-142 in catalysis. Modeling of the apo mtbLipB structure by a conservative approach (energy minimization) indicates that, in the absence of a substrate, this lysine probably forms a charged residue pair with Cys-176, promoting its thiolate form as nucleophilic group for acyltransferase catalysis. The model is based on previous data from cysteine/histidine dyad proteases in which a similar residue pair was observed in the active site (24). Moreover, a lysine/serine catalytic dyad has been found in a number of proteases, such as type I signal peptidase, Tsp protease, and the autoproteolytic activity of LexA repressor (25). As seen with LipB, the reactions catalyzed by these proteases proceed by means of an acyl-enzyme intermediate, and the catalytic activity can be retained when the catalytic serine is mutated to a cysteine (26).
By analogy to the catalytic dyads found in the lysine/serine dyad proteases, in LipB, Lys-142/Cys-176 could be assigned a plausible role as base/acid catalysts. This analogy, in turn, would render LipB a cysteine/lysine acyltransferase, which, to our knowledge, is unprecedented in the literature. In line with previous data on cysteine/histidine proteases, our data suggest that Lys-142 may be involved in activating the octanoyl moiety for Cys-176 thioester formation. This model is supported by recent measurements of the pH-rate catalysis profile of LipB from E. coli, revealing the involvement of residues with pKa values of ≈6.4 and 8.4 (27). The only candidate residues near the active center are Lys-142 and Cys-176, suggesting that the pKa values of both residues may be substantially lowered during catalysis. In contrast to previous structural findings in cysteine/histidine proteases, for instance, there is no structural evidence for ordered solvent-assisted catalysis in mtbLipB.
The conserved Cys-176 is present neither in the BirA nor in any LplA sequences, suggesting that the invariant lysine (Lys-142) may serve other functions during catalysis by these enzymes. Given that this lysine is conserved among all members of the biotin/lipoyl protein ligase family, it is plausible to assume it may be involved in the product amide bond formation with the ε-amino group of an exposed lysine residue from either lipoyl or biotinyl acceptor domains, in addition to its specific role in LipB activation of the conserved Cys-176 by promoting its thiolate form. The most plausible common model suggests an involvement of Lys-142 to promote nucleophilic attack of the ε-amino group from the external lipoyl domain, which ultimately leads to a stable amide-like octanoyl–protein product. This hypothesis is supported by recent structural data from a complex of BirA from Pyrococcus horikoshii complexes with biotinyl-5′-AMP (28). In this complex, the equivalent invariant lysine is within hydrogen-bonding distance of one of the carboxylate oxygen atoms in the biotin moiety, thereby increasing the polarization of the reactive group to allow ε-amino amide bond formation with the biotinyl acceptor domain.
Conclusion
Our data demonstrate that mtbLipB exhibits promising target properties, in terms of its altered expression pattern during disease and its peculiar structural properties, revealing a large active-site cavity for its substrate, ACP–octanoic acid. LipB appears to be the only member from the family of biotin/lipoyl/octanoyl attachment enzymes that uses a catalytic reaction mechanism by means of a cysteine/lysine dyad and an intermediate thioester.
Materials and Methods
Sample Preparation and Structural Analysis.
The ORF of the M. tuberculosis strain H37Rv lipB gene (Rv2217) was amplified by PCR from genomic DNA and cloned into the expression vector pETM11. Different versions of LipB (wt, K142S, and C176S) were overexpressed in the E. coli Rosetta (DE3) plysS strain and purified by means of Ni-NTA affinity chromatography, N-terminal polyhistidine-tag cleavage, and gel filtration. For functional studies, mtbLipB was also overexpressed in the M. smegmatis mc2155 strain. Crystals were grown by the hanging-drop vapor diffusion method by mixing 1 μl of concentrated mtbLipB solution in 10 mM sodium Hepes (pH 7.5)/50 mM NaCl/1 mM DTT/1 μl of well solution, containing 0.1 mM Hepes (pH 6.8), 0.2 mM MgCl2, and 20% (wt/vol) polyethylene glycol-3350. The x-ray structure was determined by the single isomorphous replacement combined with anomalous scattering (SIRAS) method at 1.08-Å resolution, using x-ray diffraction data from a bromide-soaked crystal (Table 2).
Functional in Vitro and in Vivo Analysis.
In vitro mtbLipB enzymatic activity was measured by a gel-shift assay (29). In vivo, the enzyme activity was monitored by the ability to complement a lipB-deficient E. coli strain (7). The molecular mass of different samples was determined by electrospray ionization MS. Additional details are described in Supporting Text, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank Anja Bathke and Dr. Yanfang Jiang for assistance during the experimental work. The initial work was inspired by the associate membership of M.W. in the U.S. Mycobacterium tuberculosis Structural Genomics Consortium. This work was funded by German Ministry for Science and Education Consortium Grant BIO/0312992 (to M.W. and S.H.E.K.) and National Institutes of Health Grant AI15650 (to J.E.C.).
Abbreviations
- ACP
acyl carrier protein
- BirA
biotin protein ligase
- LipA
lipoyl synthase
- LipB
octanoyl-[acyl carrier protein]-protein transferase
- LplA
lipoyl protein ligase A
- mtbLipB
LipB from Mycobacterium tuberculosis.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1w66).
References
- 1.Perham R. N. Annu. Rev. Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- 2.Morris T. W., Reed K. E., Cronan J. E., Jr. J. Bacteriol. 1995;177:1–10. doi: 10.1128/jb.177.1.1-10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordan S. W., Cronan J. E., Jr. Methods Enzymol. 1997;279:176–183. doi: 10.1016/s0076-6879(97)79021-9. [DOI] [PubMed] [Google Scholar]
- 4.Jordan S. W., Cronan J. E., Jr. J. Bacteriol. 2003;185:1582–1589. doi: 10.1128/JB.185.5.1582-1589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller J. R., Busby R. W., Jordan S. W., Cheek J., Henshaw T. F., Ashley G. W., Broderick J. B., Cronan J. E., Jr., Marletta M. A. Biochemistry. 2000;39:15166–15178. doi: 10.1021/bi002060n. [DOI] [PubMed] [Google Scholar]
- 6.Cicchillo R. M., Iwig D. F., Jones A. D., Nesbitt N. M., Baleanu-Gogonea C., Souder M. G., Tu L., Booker S. J. Biochemistry. 2004;43:6378–6386. doi: 10.1021/bi049528x. [DOI] [PubMed] [Google Scholar]
- 7.Zhao X., Miller J. R., Jiang Y., Marletta M. A., Cronan J. E. Chem. Biol. 2003;10:1293–1302. doi: 10.1016/j.chembiol.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 8.O’Riordan M., Moors M. A., Portnoy D. A. Science. 2003;302:462–464. doi: 10.1126/science.1088170. [DOI] [PubMed] [Google Scholar]
- 9.Yi X., Maeda N. Mol. Cell. Biol. 2005;25:8387–8392. doi: 10.1128/MCB.25.18.8387-8392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomsen-Zieger N., Schachtner J., Seeber F. FEBS Lett. 2003;547:80–86. doi: 10.1016/s0014-5793(03)00673-2. [DOI] [PubMed] [Google Scholar]
- 11.Wrenger C., Muller S. Mol. Microbiol. 2004;53:103–113. doi: 10.1111/j.1365-2958.2004.04112.x. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann S. H., McMichael A. J. Nat. Med. 2005;11:S33–S44. doi: 10.1038/nm1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rachmann H., Strong M., Ulrichs T., Grode L., Schuchhardt J., Mollenkopf H., Kosmiadi G. A., Eisenberg D., Kaufmann S. H. Infect. Immun. 2006;74:1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sassetti C. M., Boyd D. H., Rubin E. J. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 15.Alexov E. G., Gunner M. R. Biophys. J. 1997;72:2075–2093. doi: 10.1016/S0006-3495(97)78851-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgescu R. E., Alexov E. G., Gunner M. R. Biophys. J. 2002;83:1731–1748. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell J. W., Cronan J. E., Jr. J. Bacteriol. 2001;183:5982–5990. doi: 10.1128/JB.183.20.5982-5990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y. M., Rao M. S., Heath R. J., Price A. C., Olson A. J., Rock C. O., White S. W. J. Biol. Chem. 2001;276:8231–8238. doi: 10.1074/jbc.M008042200. [DOI] [PubMed] [Google Scholar]
- 19.Reche P. A. Protein Sci. 2000;9:1922–1929. doi: 10.1110/ps.9.10.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver L. H., Kwon K., Beckett D., Matthews B. W. Proc. Natl. Acad. Sci. USA. 2001;98:6045–6050. doi: 10.1073/pnas.111128198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson K. P., Shewchuk L. M., Brennan R. G., Otsuka A. J., Matthews B. W. Proc. Natl. Acad. Sci. USA. 1992;89:9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujiwara K., Toma S., Okamura-Ikeda K., Motokawa Y., Nakagawa A., Taniguchi H. J. Biol. Chem. 2005;280:33645–33651. doi: 10.1074/jbc.M505010200. [DOI] [PubMed] [Google Scholar]
- 23.Kim D. J., Kim K. H., Lee H. H., Lee S. J., Ha J. Y., Yoon H. J., Suh S. W. J. Biol. Chem. 2005;280:38081–38089. doi: 10.1074/jbc.M507284200. [DOI] [PubMed] [Google Scholar]
- 24.Storer A. C., Menard R. Methods Enzymol. 1994;244:486–500. doi: 10.1016/0076-6879(94)44035-2. [DOI] [PubMed] [Google Scholar]
- 25.Paetzel M., Dalbey R. E. Trends Biochem. Sci. 1997;22:28–31. doi: 10.1016/s0968-0004(96)10065-7. [DOI] [PubMed] [Google Scholar]
- 26.Tschantz W. R., Sung M., Delgado-Partin V. M., Dalbey R. E. J. Biol. Chem. 1993;268:27349–27354. [PubMed] [Google Scholar]
- 27.Nesbitt N. M., Baleanu-Gogonea C., Cicchillo R. M., Goodson K., Iwig D. F., Broadwater J. A., Haas J. A., Fox B. G., Booker S. J. Protein Expr. Purif. 2005;39:269–282. doi: 10.1016/j.pep.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 28.Bagautdinov B., Kuroishi C., Sugahara M., Kunishima N. J. Mol. Biol. 2005;353:322–333. doi: 10.1016/j.jmb.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 29.Ali S. T., Moir A. J., Ashton P. R., Engel P. C., Guest J. R. Mol. Microbiol. 1990;4:943–950. doi: 10.1111/j.1365-2958.1990.tb00667.x. [DOI] [PubMed] [Google Scholar]
- 30.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.