Abstract
The checkpoint mediator protein Claspin is indispensable for the ATR-dependent phosphorylation of Chk1 in response to stalled DNA replication forks in Xenopus egg extracts. We show that Claspin also participates in the detection of chromosomal double-stranded DNA breaks (DSBs) in this system. Significantly, removal of Claspin from egg extracts only partially abrogates the activation of Chk1 in response to chromatin with DSBs, whereas depletion of both Claspin and BRCA1 completely abolishes this activation. The function of Claspin in this DSB-triggered pathway depends on phosphorylation of T817 and S819 by ATR. Conversely, neither phosphorylation of Claspin on these sites nor the presence of BRCA1 is necessary for activation of Chk1 in response to stalled replication forks. Thus, site-specific phosphorylation of a checkpoint mediator protein is a crucial determinant in the discrimination between various checkpoint-inducing structures. Furthermore, checkpoint mediator proteins exhibit functional overlap that varies depending on the nature of the checkpoint-triggering DNA signal.
Keywords: Claspin, ATR, BRCA1, checkpoint control, Xenopus egg extract
In eukaryotic cells, various checkpoint control mechanisms safeguard the integrity of the genomic DNA (Nyberg et al. 2002; Sancar et al. 2004). These systems monitor the fidelity of DNA replication during S phase and detect the occurrence of DNA damage throughout the cell cycle. Members of the phosphoinositide kinase-related family of proteins kinases (PIKKs) are key checkpoint regulatory enzymes (Abraham 2001). These kinases include ATM and ATR, which principally regulate cellular responses to double-stranded DNA breaks (DSBs) and aberrant DNA replication forks, respectively. Importantly, the PIKKs depend on numerous other proteins in order to phosphorylate their downstream targets (Nyberg et al. 2002; Osborn et al. 2002; Sancar et al. 2004). Among these essential regulators are proteins known as checkpoint mediators. For example, to phosphorylate the downstream checkpoint effector kinase Chk1, ATR requires the assistance of a checkpoint mediator protein called Claspin (Kumagai and Dunphy 2000; Chini and Chen 2003; Kumagai et al. 2004; Lin et al. 2004). Other checkpoint mediators include various BRCA1 C-terminal (BRCT) domain-containing proteins, such as 53BP1, Mdc1, BRCA1 itself, and possibly TopBP1 (Sancar et al. 2004).
Checkpoint mediators are large and complex proteins that execute multiple functions. For example, in addition to functioning as kinase adaptors, Claspin and its yeast homolog Mrc1 appear both to monitor the status of DNA replication forks and to promote the progression of S phase (Katou et al. 2003; Lee et al. 2003; Osborn and Elledge 2003; Lin et al. 2004; Lee et al. 2005). Furthermore, 53BP1, which has been implicated in the ATM-dependent activation of Chk2 (Wang et al. 2002), displays complex interactions with chromatin. In particular, 53BP1 specifically recognizes methylated and phosphorylated forms of histones H3 and H2AX, respectively (for review, see van Attikum and Gasser 2005). Collectively, these findings have led to the concept that checkpoint mediators also participate in some way in the recognition of specific DNA structures in the genome.
We have been using Xenopus egg extracts to examine the molecular mechanisms underlying various checkpoint responses. These extracts can be used to study checkpoint responses to both incompletely replicated DNA and various different forms of damaged DNA (Kumagai et al. 1998b; Costanzo et al. 2000; Guo and Dunphy 2000; Lupardus et al. 2002; Stokes et al. 2002). The mediator protein Claspin is essential for the activation of Xenopus Chk1 (Xchk1) by Xenopus ATR (Xatr) in egg extracts containing incompletely replicated DNA (Kumagai and Dunphy 2000). Another feature of Claspin is that it interacts specifically with chromatin during S phase and thus might help to monitor the status of DNA replication forks (Lee et al. 2003). Consistent with these observations, Claspin associates with various key replication and checkpoint proteins at replication sites, including Cdc45, DNA polymerase ε (Polε), RPA, and both the replicative and Rad17-containing complexes (Lee et al. 2005). In addition, it has been shown that Claspin is necessary for human cells to display a normal response to IR-induced DNA damage (Lin et al. 2004).
The function of Claspin is highly regulated by phosphorylation (Kumagai and Dunphy 2000, 2003; Jeong et al. 2003). For example, Claspin must be phosphorylated on both S864 and S895 in order to associate with Xchk1 and mediate its activation. Interestingly, S864 and S895 do not reside in consensus target sites for Xatr, and this kinase cannot phosphorylate these sites in vitro. Nonetheless, phosphorylation of Claspin on S864 and S895 is dependent on Xatr. These observations suggest that Xatr either phosphorylates Claspin on other critical sites and/or that Xatr regulates a pathway containing the kinase that phosphorylates S864 and S895.
For the studies in this report, we initially set out to analyze how phosphorylation of Claspin by Xatr might regulate its function. For this purpose, we prepared various mutants of Claspin in its different SQ/TQ motifs, the standard phosphorylation sites for Xatr. Intriguingly, we find that none of these motifs are required for Claspin to mediate activation of Xchk1 in response to DNA replication blocks. In contrast, phosphorylation on T817 and/or S819 in the SQ/TQ motifs of Claspin is necessary for normal activation of Xchk1 in response to damaged chromatin with DSBs. These studies indicate that the specificity of a mediator protein for a certain checkpoint pathway is at least in part determined by site-specific phosphorylation. This type of mechanism would substantially increase the functional diversity of this class of proteins.
Results
Claspin responds to DSBs in chromatin
It has previously been shown that Claspin is essential for the Xatr-dependent phosphorylation of Xchk1 in response to DNA replication blocks in both Xenopus egg extracts and human cells (Kumagai and Dunphy 2000; Chini and Chen 2003; Lee et al. 2003; Lin et al. 2004). In addition, Lin et al. (2004) recently found that phosphorylation of Chk1 in human cells following treatment with ionizing radiation (IR), which is known to create DSBs, also involves Claspin. Since our laboratory previously demonstrated that binding of Claspin to undamaged chromatin in Xenopus egg extracts requires the formation of replication structures (Lee et al. 2003), we were interested in assessing how Claspin might interact with damaged chromatin. For this purpose, we incubated demembranated Xenopus sperm chromatin in egg extracts in the presence or absence of the restriction endonuclease EcoRI, which creates double-stranded breaks in the DNA (Kobayashi et al. 2002; Yoo et al. 2004b). For comparison, we treated the extracts with aphidicolin to induce the formation of stalled DNA replication forks (Dasso and Newport 1990; Kumagai et al. 1998a). We also incubated these various extracts either with or without geminin, which inhibits the formation of replication structures by blocking assembly of the prereplication complex (pre-RC) (McGarry and Kirschner 1998). Next, we examined chromatin binding of Claspin as well as of other checkpoint and replication proteins, including Xatr, Xbrca1, Xrad17, RPA70, Xmcm2, Xcdc45, Polα, and Xorc2.
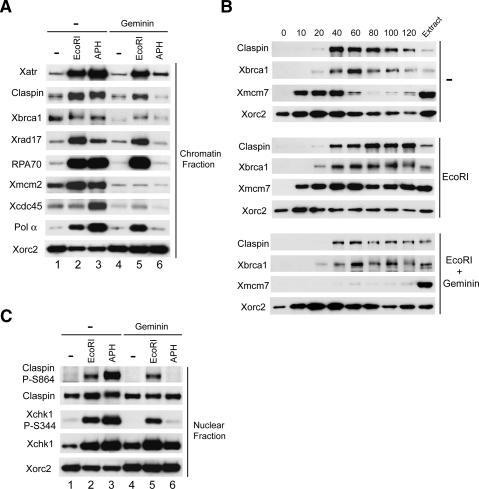
As described previously, Claspin binds in elevated amounts to aphidicolin-treated chromatin (Fig. 1A; Lee et al. 2003). We observed that Claspin likewise associates in higher amounts with EcoRI-damaged chromatin than with undamaged chromatin. As expected, geminin blocked the binding of Claspin to aphidicolin-treated chromatin (Lee et al. 2003). However, geminin caused only a partial reduction in the amount of Claspin on EcoRI-treated chromatin. We verified that the treatment with geminin was effective by showing that there was negligible association of Xmcm2 with chromatin under these conditions. The elevated binding of Xatr, Xrad17, RPA70, and Polα that typically occurs in the presence of aphidicolin was largely abolished by geminin. In contrast, geminin did not block the binding of Xatr, Xrad17, RPA70, and Polα to EcoRI-damaged chromatin. As anticipated, Xcdc45 could not be found on chromatin in significant amounts under any condition in the presence of geminin. Interestingly, Xbrca1 displayed a somewhat different behavior from these other proteins. Xbrca1 bound specifically to untreated chromatin, and its binding did not increase significantly in the presence of either aphidicolin or EcoRI. Moreover, geminin only modestly inhibited the interaction of Xbrca1 with chromatin. Therefore, Xbrca1 is capable of associating with chromatin under a wide variety of conditions.
Figure 1.
Activation of Xchk1 in response to DSBs in chromatin. (A) Interphase extracts containing sperm chromatin were incubated in the absence (lanes 1–3) or presence of geminin with no additional treatment (lanes 1,4), EcoRI (lanes 2,5), or aphidicolin (APH) (lanes 3,6). Chromatin fractions were isolated and immunoblotted for the indicated proteins. (B) Time courses for binding of proteins to damaged chromatin. Extracts were incubated with control buffer (top), EcoRI (middle), and EcoRI plus geminin (bottom). Chromatin fractions were isolated at the times shown and immunoblotted for the indicated proteins. (C) Activation of Xchk1 in response to different DNA templates. Nuclear fractions from the extracts in A were immunoblotted with anti-Claspin-P–S864, anti-Claspin, anti-Xchk1-P-S344, anti-Xchk1, and anti-Xorc2 antibodies.
To characterize further the binding of Claspin to EcoRI-damaged chromatin, we performed time-course experiments (Fig. 1B). As described previously, binding of Claspin to untreated, normally replicating chromatin in egg extracts begins at ∼40 min (when sealing of the nuclear membrane allows both concentration of proteins in nuclei and initiation of DNA replication) and declines by 120 min (when DNA replication typically reaches completion). In the presence of EcoRI, binding of Claspin began at a similar time, but it did not decline later. The pattern of binding for Claspin to EcoRI-damaged chromatin was similar in the presence of geminin, although the overall amount of binding was lower. In these experiments, Xbrca1 bound to chromatin with similar timing as Claspin under the three different conditions. The precise timing with which Xbrca1 associates with chromatin argues that this binding is highly specific. Taken together, these finding argue that Claspin is capable of interacting with chromatin without the formation of replication forks, but only when sites of damage have opened up access to the chromatin.
DSBs in chromatin induce Xatr-dependent phosphorylation of Xchk1
We proceeded to assess whether EcoRI-damaged chromatin could trigger the activation of Xchk1. For this question, we examined phosphorylation of Xchk1 on S344, the major phosphorylation site for Xatr (Guo et al. 2000). We also monitored phosphorylation of Claspin on S864, one of the two residues that must be phosphorylated for Claspin to interact with Xchk1 during a checkpoint response (Kumagai and Dunphy 2003). As shown in Figure 1C, there was robust phosphorylation of Xchk1 on S344 and of Claspin on S864 in extracts containing EcoRI-damaged chromatin in both the absence and presence of geminin. We also observed increased phosphorylation of Xchk1 on S344 in response to treatment with EcoRV and PstI, which leave blunt ends and 3′ overhangs, respectively, as opposed to the 5′ overhangs created by EcoRI (data not shown). These findings argue that chromatin with DSBs can induce activation of both Xchk1 and Claspin without the occurrence of semiconservative DNA replication.
In numerous systems, ATR controls the activation of Chk1 in response to replication stress (Guo et al. 2000; Hekmat-Nejad et al. 2000; Liu et al. 2000; Zhao and Piwnica-Worms 2001). However, ATM appears to be involved in the earliest responses to DSBs (Bakkenist and Kastan 2004). Therefore, we asked whether the activation of Xchk1 in response to DSBs involved Xatr and/or Xatm. Initially, we examined sensitivity to caffeine, an inhibitor of both ATR and ATM (Abraham 2001). As shown in Figure 2A, caffeine abolished the phosphorylation of Xchk1 in EcoRI-treated extracts as effectively as it did in extracts containing aphidicolin. Next, we removed either Xatm or Xatr from the egg extracts by immunodepletion with specific antibodies against either protein (Fig. 2B). We observed that removal of Xatr abolished the EcoRI-induced phosphorylation of Xchk1 on S344, whereas absence of Xatm did not appear to have any effect (Fig. 2C).
Figure 2.
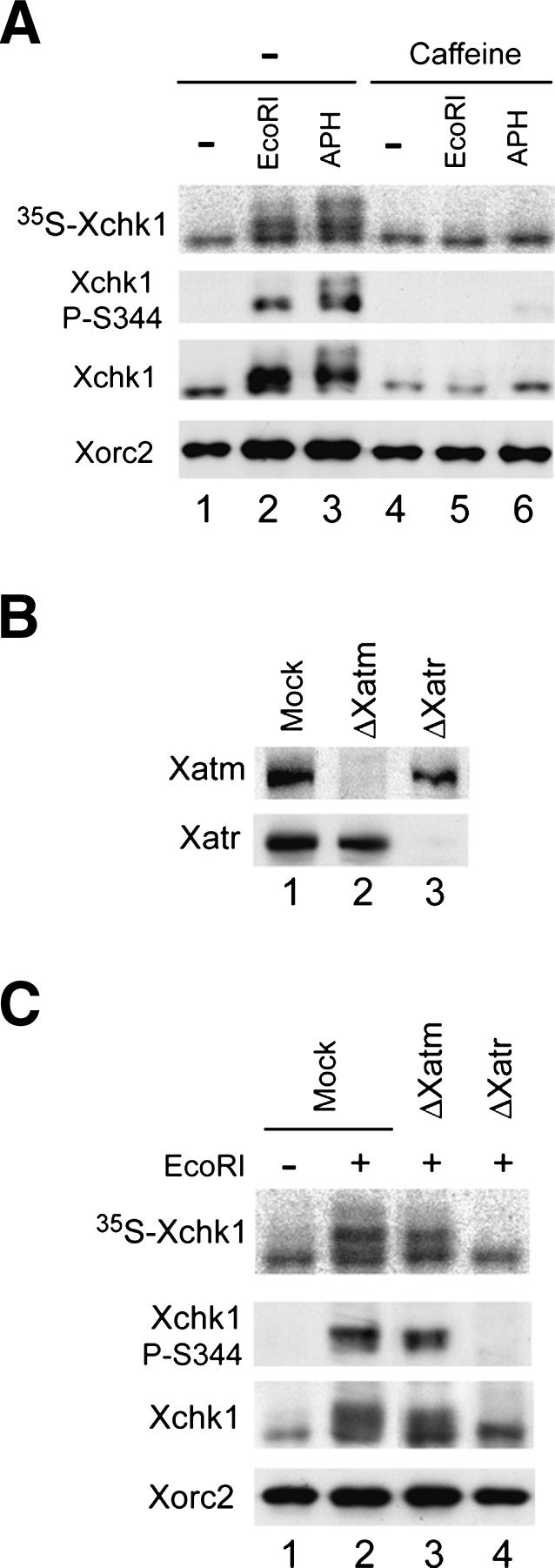
Activation of Xchk1 in response to DSBs depends on Xatr. (A) Activation of Xchk1 in response to EcoRI-damaged chromatin is sensitive to caffeine. Interphase extracts containing 35S-labeled Xchk1 were incubated with control buffer (lanes 1,4), EcoRI (lanes 2,5), and aphidicolin (lanes 3,6) in the absence (lanes 1–3) or presence (lanes 4–6) of caffeine. Nuclear fractions were immunoblotted with anti-Xchk1, anti-Xchk1-P-S344, and anti-Xorc2 antibodies. 35S-Xchk1 in nuclear fractions was detected with a PhosphorImager. (B) Immunodepletion of Xatm or Xatr. Extracts were treated with control (lane 1), anti-Xatm (lane 2), and anti-Xatr antibodies (lane 3) and immunoblotted for Xatm (top) or Xatr (bottom). (C) Mock-depleted (lanes 1,2), Xatm-depleted (lane 3), and Xatr-depleted extracts (lane 4) were incubated in the absence (lane 1) or presence (lanes 2–4) of EcoRI. Nuclear fractions were immunoblotted with anti-Xchk1, anti-Xchk1-P-S344, and anti-Xorc2 antibodies. 35S-Xchk1 was detected with a PhosphorImager.
Activation of Xchk1 in response to DSBs in chromatin depends on both Claspin and Xbrca1
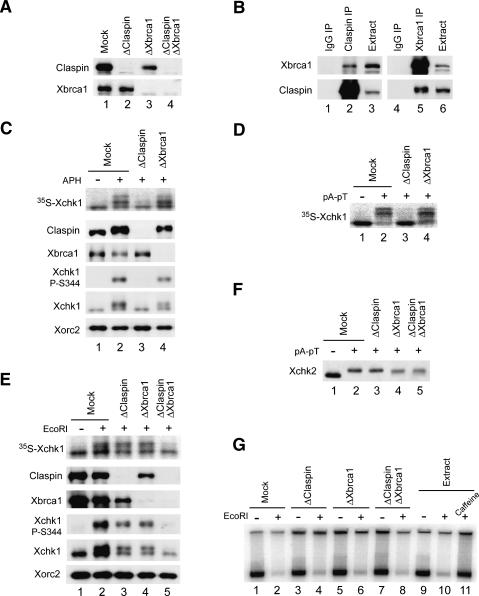
Claspin is essential for the Xatr-dependent activation of Xchk1 in response to DNA replication blocks (Kumagai and Dunphy 2000). On the other hand, BRCA1 appears to play a role in the activation of Chk1 in response to IR in human cells (Yarden et al. 2002; Lin et al. 2004). Therefore, we explored whether Claspin and/or Xbrca1 would be involved in the response of egg extracts to EcoRI-generated DSBs. To examine this issue, we removed Claspin and/or Xbrca1 from the extracts by immunodepletion (Fig. 3A). Elimination of Claspin did not affect the level of Xbrca1 in the Claspin-depleted extracts. However, there was a noticeable reduction in the amount of Claspin in Xbrca1-depleted extracts. In reciprocal immunoprecipitation experiments, we were able to show that Claspin and Xbrca1 associate specifically with one another (Fig. 3B), which is consistent with observations in human cells (Lin et al. 2004).
Figure 3.
Activation of Xchk1 in response to EcoRI-damaged chromatin is dependent on both Claspin and Xbrca1. (A) Immunodepletion of Claspin, Xbrca1, or both. Interphase extracts were treated with control (lane 1), anti-Claspin (lane 2), anti-Xbrca1 (lane 3), or both anti-Claspin and anti-Xbrca1 antibodies (lane 4) and immunoblotted for Claspin (top) or Xbrca1 (bottom). (B) Claspin interacts specifically with Xbrca1. Interphase extracts were immunoprecipitated with control (lanes 1,4), anti-Claspin (against residues 1–464) (lane 2), or anti-Xbrca1 (lane 5) antibodies. The immunoprecipitates were immunoblotted with anti-Xbrca1 (top) and anti-Claspin antibodies (bottom). Lanes 3 and 6 show untreated extracts. (C) Activation of Xchk1 in response to DNA replication blocks depends entirely on Claspin. Mock-depleted (lanes 1,2), Claspin-depleted (lane 3), and Xbrca1-depleted (lane 4) extracts were incubated with 35S-Xchk1 in the absence (lane 1) or presence (lanes 2–4) of aphidicolin. Nuclear fractions were immunoblotted with anti-Claspin, anti-Xbrca1, anti-Xchk1-P-S344, anti-Xchk1, and anti-Xorc2 antibodies. 35S-Xchk1 was detected with a PhosphorImager. (D) Activation of Xchk1 in response to pA-pT is completely dependent on Claspin. Mock-depleted (lanes 1,2), Claspin-depleted (lane 3), and Xbrca1-depleted (lane 4) extracts were incubated with 35S-Xchk1 in the absence (lane 1) or presence (lanes 2–4) of pA-pT. Extracts were subjected to SDS-PAGE, and 35S-Xchk1 was detected with a PhosphorImager. (E) Activation of Xchk1 in response to treatment with EcoRI depends on both Claspin and Xbrca1. Mock-depleted extracts (lanes 1,2), Claspin-depleted extracts (lane 3), Xbrca1-depleted extracts (lane 4), and extracts depleted of both Claspin and Xbrca1 (lane 5) were incubated with 35S-Xchk1 in the absence (lane 1) or presence (lanes 2–5) of EcoRI. Nuclear fractions were immunoblotted with anti-Claspin, anti-Xbrca1, anti-Xchk1-P-S344, anti-Xchk1, and anti-Xorc2 antibodies. 35S-Xchk1 was detected with a PhosphorImager. (F) Neither Claspin nor Xbrca1 is required for activation of Xchk2 in response to pA-pT. Mock-depleted extracts (lanes 1,2), Claspin-depleted extracts (lane 3), Xbrca1-depleted extracts (lane 4), and extracts depleted of both Claspin and Xbrca1 (lane 5) were incubated in the absence (lane 1) or presence (lane 2–5) of pA-pT and immunoblotted for Xchk2. (G) Depletion of both Claspin and Xbrca1 does not rescue the arrest of DNA replication in EcoRI-damaged chromatin. The indicated extracts from A were incubated with control buffer (lanes 1,3,5,7,9), EcoRI (lanes 2,4,6,8,10), or EcoRI plus caffeine (lane 11). DNA replication was determined by incorporation of 32P into chromatin as described in Materials and Methods.
We proceeded to compare checkpoint responses to different DNA templates in the various depleted extracts. As expected, phosphorylation of Xchk1 was abolished in Claspin-depleted extracts containing sperm chromatin with aphidicolin-stalled DNA replication forks (Fig. 3C). There was also no phosphorylation of Xchk1 in Claspin-depleted egg extracts containing annealed polynucleotides comprised of (dA)70-(dT)70 (hereafter referred to as pA-pT), a model checkpoint-inducing DNA template (Fig. 3D; Kumagai and Dunphy 2000). Conversely, there was no reduction in the phosphorylation of Xchk1 in Xbrca1-depleted extracts that were treated with either aphidicolin or pA-pT (Fig. 3C,D). These findings argue that Xbrca1 does not play an essential role in the Claspin-dependent pathway for the checkpoint response to either type of DNA template.
Next, we examined responses to EcoRI-damaged chromatin in the various depleted extracts. Removal of Claspin reduced the phosphorylation of Xchk1 to approximately half of its normal level (Fig. 3E). Likewise, immunodepletion of Xbrca1 caused a similar reduction in the phosphorylation of Xchk1. Significantly, concurrent removal of both Claspin and Xbrca1 abolished the phosphorylation of Xchk1 almost entirely. We similarly observed roles for both Claspin and Xbrca1 in the EcoRI-induced phosphorylation of Xchk1 in geminin-treated extracts (data not shown). Taken together, these results indicate that both Claspin and Xbrca1 function in the response to EcoRI-induced DSBs in chromatin in both the presence and absence of DNA replication.
Overall, these observations indicate that the dependency of Xchk1 on mediator proteins for its activation varies with the nature of the checkpoint-inducing DNA structure. In particular, activation of Xchk1 in response to stalled replication forks and a simple double-stranded template (pA-pT) is completely dependent on Claspin. The template pA-pT, which contains two annealed strands only 70 nucleotides (nt) in length, would be too small to mediate the assembly of nucleosomes, the basic structural unit of chromatin. On the other hand, the response to a more complex version of template with exposed double-stranded DNA ends (e.g., sperm chromatin that has been cleaved with EcoRI) involves both Claspin and Xbrca1.
DSBs are known to elicit several other effects besides activation of Xchk1 in egg extracts. These responses include activation of Xenopus Chk2 (Xchk2) and inhibition of chromosomal DNA replication (Costanzo et al. 2000; Guo and Dunphy 2000; Kobayashi et al. 2002; Yoo et al. 2004b). The pA-pT template is also an excellent activator of Xchk2 (Guo and Dunphy 2000; Li et al. 2004). However, as shown in Figure 3F, removal of Claspin and/or Xbrca1 had no discernible effect on the activation of Xchk2 following addition of pA-pT. Treatment of egg extracts with EcoRI causes a very pronounced inhibition of DNA replication of the chromatin in these extracts. This inhibition is dependent on both Xatr and Xatm and thus can be alleviated by treatment with caffeine (Yoo et al. 2004b). Nonetheless, immunodepletion of Claspin or Xbrca1 or both did not cause any restoration of DNA replication in EcoRI-treated extracts (Fig. 3G). Taken together, these results imply that activation of Xchk2 and inhibition of chromosomal DNA replication in response to DSBs involve mediator proteins distinct from Claspin and Xbrca1 or, alternatively, do not depend on such mediator proteins.
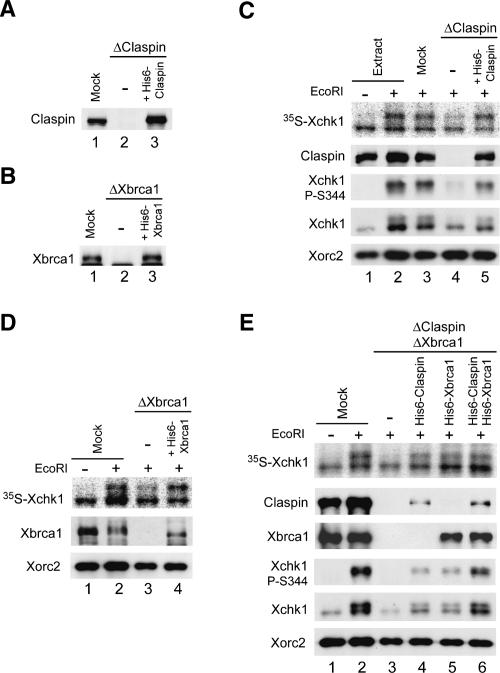
To characterize further the roles of Claspin and Xbrca1 in the chromosomal DSB-induced phosphorylation of Xchk1, we asked whether recombinant Claspin and Xbrca1 could rescue the checkpoint defect that was observed in the absence of either or both of these proteins. For this purpose, we used a previously described version of full-length Claspin (His6-Claspin) and a His6-tagged version of full-length Xbrca1 (His6-Xbrca1) that we prepared for these experiments. As shown in Figure 4, A and B, we were able to restore Claspin-depleted and Xbrca1-depleted extracts with His6-Claspin and His6-Xbrca1, respectively, at levels similar to those of the endogenous proteins. In both cases, addition of recombinant Claspin or Xbrca1 rescued the partial defect in phosphorylation of Xchk1 in response to EcoRI-damaged chromatin (Fig. 4C,D). Finally, we added His6-Claspin, His6-Xbrca1, or both to extracts that had been depleted of both endogenous Claspin and Xbrca1. Addition of either His6-Claspin or His6-Xbrca1 alone resulted in significant restoration of the phosphorylation of Xchk1 on S344, while both proteins together induced even stronger phosphorylation of Xchk1 (Fig. 4E). Overall, these experiments indicate that both Claspin and Xbrca1 operate in pathways that lead to the phosphorylation of Xchk1 in response to DSBs in chromatin.
Figure 4.
Recombinant Claspin and Xbrca1 can restore the checkpoint defects in depleted extracts. (A,B) Immunodepletion of Claspin or Xbrca1. Egg extracts were mock-depleted with control antibodies (A,B, lane 1) or immunodepleted with either anti-Claspin (A, lanes 2,3) or anti-Xbrca1 (B, lanes 2,3) antibodies. Subsequently, no recombinant protein (A,B, lane 2), wild-type His6-Claspin (A, lane 3), and His6-Xbrca1 (B, lane 3) were added back to the depleted extracts. Extracts were immunoblotted for Claspin (A) or Xbrca1 (B). (C) The indicated extracts from A were incubated with 35S-Xchk1 in the absence (lane 1) or presence (lanes 2–5) of EcoRI. Nuclear fractions were immunoblotted with anti-Claspin, anti-Xchk1-P-S344, anti-Xchk1, and anti-Xorc2 antibodies. 35S-Xchk1 was detected with a PhosphorImager. (D) The indicated extracts from B were incubated with 35S-Xchk1 in the absence (lane 1) or presence (lanes 2–4) of EcoRI. Nuclear fractions were immunoblotted for Xbrca1 and Xorc2. 35S-Xchk1 was detected with a PhosphorImager. (E) Add-back of recombinant Claspin and Xbrca1 to depleted extracts. Mock-depleted extracts (lanes 1,2) and extracts depleted of both Claspin and Xbrca1 (lanes 3–6) were supplemented with His6-Claspin (lane 4), His6-Xbrca1 (lane 5), or both (lane 6). Extracts were incubated with 35S-Xchk1 in the absence (lane 1) or presence (lanes 2–6) of EcoRI. Nuclear fractions were immunoblotted with antibodies against Claspin, Xbrca1, Xchk1, anti-Xchk1-PS344, and Xorc2. 35S-Xchk1 was detected with a PhosphorImager.
Phosphorylation of Claspin on T817 and S819 is necessary for its response to DSBs but not to DNA replication blocks
In budding yeast, elimination of all of the canonical phosphorylation sites for Mec1/ATR (SQ/TQ motifs) on Mrc1, a homolog of Claspin, severely compromises its checkpoint-dependent phosphorylation and blocks activation of the downstream effector kinase Rad53 (Osborn and Elledge 2003). Claspin requires phosphorylation on both S864 and S895 in order to associate with and mediate the activation of Xchk1 (Kumagai and Dunphy 2003). However, these two residues do not sit in consensus target sites for Xatr. Moreover, these residues appear not to be direct targets of Xatr, although their phosphorylation does depend on Xatr.
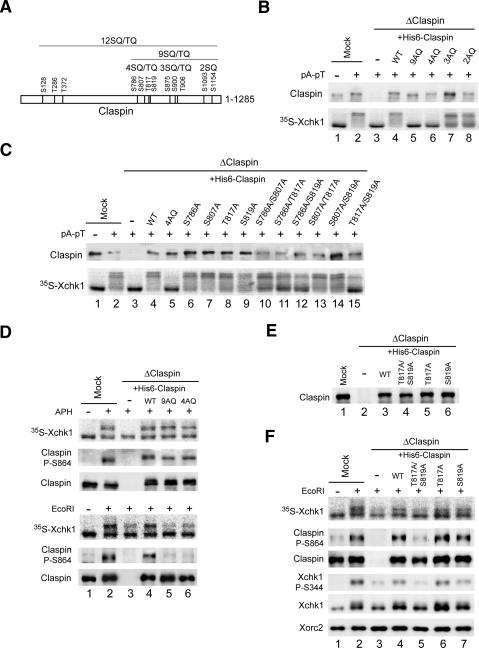
To probe if Xatr might regulate Claspin directly, we engaged in a systematic analysis of the potential functional role of the various SQ/TQ motifs in Claspin. Initially, we focused on the C-terminal domain of Claspin (residues 774–1285), which contains nine SQ/TQ motifs (Fig. 5A). This region of Claspin is sufficient for the Xatr-dependent phosphorylation of Xchk1 (Lee et al. 2005). These nine motifs reside in three clusters that contain four (S786, S807, T817, and S819), three (S875, S900, and T906), and two (S1093 and S1154) SQ and/or TQ sites, respectively. We prepared various mutants in which all of the residues in each cluster or all nine residues were changed to alanine. Initially, we tested the abilities of these mutants to mediate activation of Xchk1 in response to the model DNA template pA-pT. For this question, we immunodepleted the endogenous Claspin from egg extracts and replaced it with similar amounts of the various mutant proteins. As shown in Figure 5B, we found that elimination of the sites in the first cluster (in the 4AQ mutant) abolished phosphorylation of Xchk1 in response to pA-pT. Consistent with this finding, the 9AQ mutant also was defective in this response. In contrast, mutants of the second and third clusters of SQ/TQ sites did not have any obvious defect. We proceeded further by eliminating single residues or combinations of two residues in the 4SQ/TQ cluster (Fig. 5C). The results clearly indicated that removal of both T817 and S819 abolished the ability of Claspin to mediate activation of Xchk1 in response to pA-pT. The single T817A and S819A mutants displayed good activity for mediating activation of Xchk1, which suggests that phosphorylation at one site is sufficient for significant function.
Figure 5.
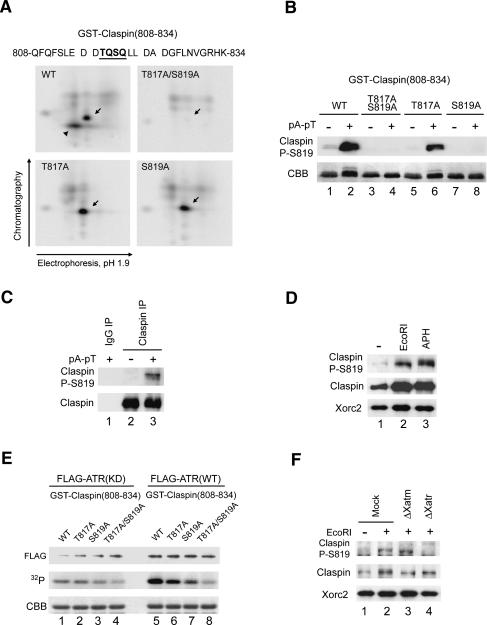
T817A/S819A mutants of Claspin are defective in mediating the activation of Xchk1 in response to DSBs. (A) Diagram of the 12SQ/TQ motifs in Claspin. Different clusters of SQ/TQ motifs are indicated. (B) Mock-depleted (lanes 1,2) and Claspin-depleted (lanes 3–8) extracts containing the indicated AQ mutants of His6-Claspin were incubated with 35S-Xchk1 in the absence (lane 1) or presence (lanes 2–8) of pA-pT. (Bottom) Extracts were subjected to SDS-PAGE, and 35S-Xchk1 was detected with a PhosphorImager. (Top) Both endogenous and recombinant Claspin were detected by immunoblotting with anti-Claspin antibodies. (C) The S817A/T819A mutant of Claspin is defective in mediating the activation of Xchk1 in response to pA-pT. The experiment was performed in the same manner as in B except that indicated single (lanes 6–9) or double (lanes 10–15) mutants of Claspin were added back to Claspin-depleted extracts. (D) Function of Claspin-AQ mutants in extracts treated with aphidicolin or EcoRI. Extracts were subjected to mock immunodepletion (lanes 1,2) or immunodepletion of Claspin (lanes 3–6). Control buffer (lane 3), His6-Claspin (lane 4), His6-Claspin-9AQ (lane 5), and His6-Claspin-4AQ (lane 6) were added back to the Claspin-depleted extracts. The extracts were supplemented with 35S-Xchk1 and incubated with sperm nuclei in the absence (lane 1) or presence of either aphidicolin (top three panels, lanes 2–6) or EcoRI (bottom three panels, lanes 2–6). Nuclear fractions were immunoblotted with anti-Claspin-P–S864 and anti-Claspin antibodies. 35S-Xchk1 was detected with a PhosphorImager. (E) Egg extracts were depleted with control (lane 1) or anti-Claspin (lanes 2–6) antibodies. To the Claspin-depleted extracts, no recombinant protein (lane 2), wild-type His6-Claspin (lane 3), His6-Claspin–T817A/S819A (lane 4), His6-Claspin–T817A (lane 5), and His6-Claspin–S819A (lane 6) were added back. Aliquots of the extracts were immunoblotted for Claspin. (F) T817A/S819A mutants of Claspin are defective for Xchk1 activation in response to EcoRI. The indicated extracts from E-containing sperm nuclei were incubated with 35S-Xchk1 in the absence (lane 1) or presence (lanes 2–7) of EcoRI. Nuclear fractions were immunoblotted with anti-Claspin-P–S864, anti-Claspin, anti-Xchk1-P-S344, anti-Xchk1, and anti-Xorc2 antibodies. 35S-Xchk1 was detected with a PhosphorImager.
Next, we examined the responses of the phosphorylation site mutants of Claspin to other checkpoint-inducing templates (e.g., sperm chromatin in egg extracts that had been treated with either aphidicolin or EcoRI). Interestingly, we did not observe any reduction in the phosphorylation of Xchk1 in aphidicolin-treated extracts containing the 4AQ or 9AQ mutants of Claspin (Fig. 5D). Similarly, there was no reduction in the phosphorylation of the Claspin-9AQ mutant on S864 in response to aphidicolin. We observed similar results with the 12AQ mutant of Claspin, in which S128, T286, and T372 were also changed to alanine (Supplementary Fig. S1). Therefore, the SQ/TQ residues of Claspin appear not to be critical for its response to aphidicolin.
In contrast, we did observe that elimination of SQ/TQ residues from Claspin had an effect on the response to EcoRI-damaged chromatin. As shown in Figure 5D, the partial reduction in the phosphorylation of Xchk1 in Claspin-depleted extracts that were treated with EcoRI could not be restored by adding back either the 4AQ or 9AQ mutants of Claspin. Consistent with these results, the 12AQ mutant was also inactive in rescuing the EcoRI-dependent phosphorylation of Xchk1 (Supplementary Fig. S1). Furthermore, the EcoRI-dependent phosphorylation of Claspin on S864 did not occur in extracts containing the 4AQ, 9AQ, or 12AQ mutants (Fig. 5D; Supplementary Fig. S1). To pursue these observations further, we removed endogenous Claspin and replaced it with equivalent amounts of wild-type, T817A/S819A, T817A, and S819A forms of Claspin (Fig. 5E). We observed that adding back the wild-type but not the T817A/S819A mutant version of full-length Claspin could alleviate the reduced phosphorylation of Xchk1 on S344 (and of Claspin on S864) in response to EcoRI-damaged chromatin (Fig. 5F). Furthermore, both the single T817A and S819A mutants were able to rescue to a significant extent the phosphorylation of Xchk1 and Claspin on S344 and S864, respectively. Taken together, these results indicate that Claspin must be phosphorylated on T817 and/or S819 in order to mediate activation of Xchk1 following the occurrence of DSBs in chromatin.
Xatr is responsible for phosphorylation of T817 and S819 of Claspin in egg extracts
To examine whether T817 and S819 actually undergo phosphorylation in egg extracts, we first performed phosphopeptide mapping of a GST fusion peptide containing residues 808–834 of Claspin (Fig. 6A). For this analysis, the peptide was incubated in egg extracts containing pA-pT in the presence of 32P-orthophosphate. The peptide was reisolated and digested with endoproteinase Asp-N, and the resulting digest was subjected to two-dimensional thin-layer chromatography and electrophoresis. The results of these experiments indicated that both T817 and S819, indeed, undergo phosphorylation in egg extracts (Fig. 6A).
Figure 6.
ATR phosphorylates T817 and S819 of Claspin. (A) Phosphopeptide maps of wild-type and mutant forms (T817A, S819A, and T817A/S819A) of a GST fusion protein containing residues 808–834 of Claspin. The sequence of this region is indicated in the top of the panel, and spaces denote positions of cleavage by Asp-N. The fusion proteins were radiolabeled in 32P-containing egg extracts as described in Materials and Methods. Arrows indicate the position of a phosphopeptide containing phosphorylated T817 or S819. The arrowhead in the panel for the wild-type peptide indicates the doubly phosphorylated species. (B) Specificity of anti-Claspin-P–S819 antibodies. Interphase extracts containing no DNA (lanes 1,3,5,7) or pA-pT (lanes 2,4,6,8) were incubated with GST-Claspin(808–834) (lanes 1,2), GST-Claspin(808–834)–T817A/S819A (lanes 3,4), GST-Claspin(808–834)–T817A (lanes 5,6), and GST-Claspin(808–834)–S819A (lanes 7,8). The GST-tagged Claspin fragments were isolated with glutathione-agarose beads and subjected to SDS-PAGE. GST-Claspin fragments were probed by immunoblotting with anti-Claspin-P–S819 antibodies (top) and stained with Coomassie brilliant blue (CBB) (bottom). (C) Phosphorylation of S819 in extracts containing pA-pT. Interphase extracts lacking (lane 2) or containing (lanes 1,3) pA-pT were immunoprecipitated with control (lane 1) or anti-Claspin (lanes 2,3) antibodies. Immunoprecipitates were immunoblotted with anti-Claspin-P–S819 and anti-Claspin antibodies. (D) Phosphorylation of Claspin on S819 is induced by both EcoRI and aphidicolin. Extracts were incubated with sperm nuclei alone (lane 1), sperm nuclei plus EcoRI (lane 2), or sperm nuclei plus aphidicolin (lane 3). Nuclear fractions were immunoblotted with anti-Claspin-P–S819, anti-Claspin, and anti-Xorc2 antibodies. (E) Phosphorylation of Claspin on T817 and S819 by ATR. Wild-type (WT) and kinase-deficient (KD) Flag-ATR were isolated from 293T cells as described in Materials and Methods. GST-Claspin(808–834) (lanes 1,5), GST-Claspin(808–834)–T817A (lanes 2,6), GST-Claspin(808–834)–S819A (lanes 3,7), and GST-Claspin(808–834)–T817A/S819A (lanes 4,8) were incubated in the presence of 32P-ATP with Flag-ATR(KD) (lanes 1–4) and Flag-ATR(WT) (lanes 5–8). The samples were subjected to SDS-PAGE. GST-tagged Claspin fragments were visualized by Coomassie blue staining (CBB) (bottom), and incorporation of 32P was detected with a PhosphorImager (middle). (Top) Recombinant ATR was immunoblotted with anti-Flag antibodies. (F) Phosphorylation of Claspin on S819 in egg extracts depends on Xatr. Mock-depleted (lanes 1,2), Xatm-depleted (lane 3), and Xatr-depleted (lane 4) extracts were incubated in the absence (lane 1) or presence (lanes 2–4) of EcoRI. Nuclear fractions were immunoblotted with anti-Claspin-P–S819, anti-Claspin, and anti-Xorc2 antibodies.
We also prepared antibodies against a short peptide containing phosphorylated S819. To characterize the specificity of these antibodies, we incubated various forms of the GST-Claspin(808–834) in egg extracts in the presence and absence of pA-pT. As shown in Figure 6B, these antibodies recognized the wild-type and T817A versions of this peptide from extracts that were treated with pA-pT, but not the S819A and T817A/S819A versions of this peptide. Next, we examined phosphorylation of the full-length version of Claspin with the anti-phospho-S819 antibodies. We found that these antibodies could react well with Claspin in extracts that had been treated with pA-pT, aphidicolin, or EcoRI (Fig. 6C,D). Therefore, although Claspin exhibits different dependencies on phosphorylation of S819 (and T817) for mediating activation of Xchk1 in response to these templates, it appears that S819 (and most likely T817) becomes well phosphorylated under all three conditions.
Next, we asked if ATR is directly involved in the phosphorylation of T817 and S819. For this question, we first prepared wild-type (WT) and kinase-deficient (KD) forms of Flag-tagged recombinant human ATR and incubated these proteins with a GST fusion protein containing residues 808–834 of Claspin. As shown in Figure 6E, Flag-ATR(WT) phosphorylated the GST-Claspin(808–834) substrate to an extent that was significantly higher than the background level that was observed with Flag-ATR(KD). Moreover, the T817A and S819A mutants underwent significant phosphorylation by active ATR. Phosphorylation of the T817A mutant was typically higher, which suggests that S819 is a better substrate for ATR. There was no phosphorylation of the T817A/S819A double mutant over background. In combination with these experiments, we also carried out immunodepletion of Xatr (and Xatm) from egg extracts. Immunoblotting with the anti-phosphopeptide antibodies indicated that removal of Xatr (but not Xatm) abolished the phosphorylation of Claspin on S819 following treatment with EcoRI (Fig. 6F). Taken together, these results suggest that S819 and most likely T817 are physiological substrates of Xatr.
Phosphorylation on T817 and S819 is necessary for the function of Claspin in the Xbrca1-independent, DSB-triggered checkpoint pathway
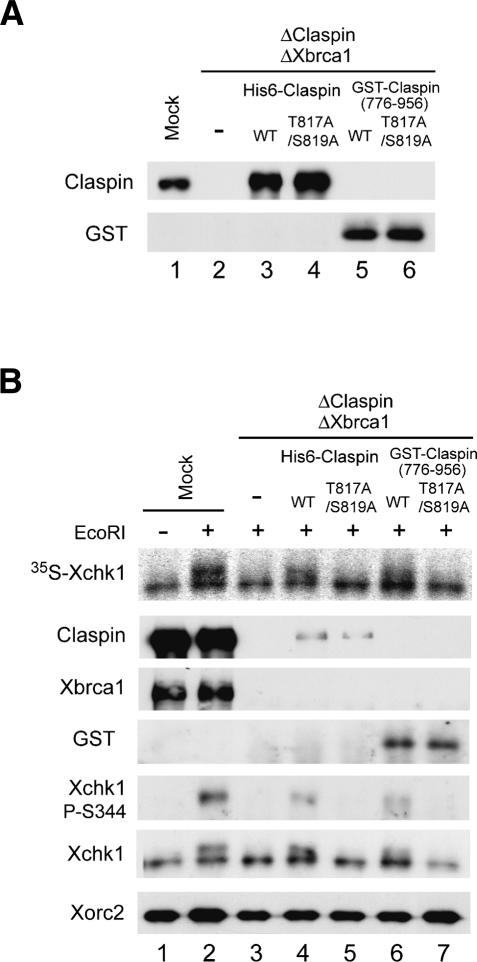
As described above, we have shown that the activation of Xchk1 in response to EcoRI-induced DSBs involves phosphorylation of Claspin on T817/S819. Furthermore, both Claspin and Xbrca1 function in the EcoRI-induced phosphorylation of Xchk1. Therefore, we wished to examine whether Claspin must be phosphorylated on T817/S819 in order to mediate the EcoRI-induced phosphorylation of Xchk1 in the absence of Xbrca1. For this question, we removed both Claspin and Xbrca1 from egg extracts and added back wild-type or T817A/S819A versions of Claspin (Fig. 7A). As expected from the results described above, there was no phosphorylation of Xchk1 on S344 in EcoRI-treated extracts lacking both Claspin and Xbrca1 (Fig. 7B). Addition of wild-type recombinant Claspin restored significant phosphorylation of Xchk1, but the T817A/S819A mutant was ineffective at rescuing this defect.
Figure 7.
Activation of Xchk1 in response to chromosomal DSBs is completely dependent on T817/S819 of Claspin in the absence of Xbrca1. (A) Egg extracts were mock-depleted with control antibodies (lane 1) or immunodepleted with both anti-Claspin and anti-Xbrca1 antibodies (lanes 2–6). Subsequently, control buffer (lane 2), wild-type His6-Claspin (lane 3), His6-Claspin–T817A/S819A (lane 4), GST-NLS-Claspin(776–956) (lane 5), and GST-NLS-Claspin(776–956)–T817A/S819A (lane 6) were added back to the extracts that were depleted of both Claspin and Xbrca1. Aliquots of the extracts were immunoblotted with anti-Claspin and anti-GST antibodies. (B) Mock-depleted extracts (lanes 1,2) and extracts depleted of both Claspin and Xbrca1 (lanes 3–7) were supplemented with control buffer (lane 3), wild-type His6-Claspin (lane 4), His6-Claspin–T817A/S819A (lane 5), GST-NLS-Claspin(776–956) (lane 6), and GST-NLS-Claspin(776–956)–T817A/S819A (lane 7). The extracts were incubated with 35S-Xchk1 in the absence (lane 1) or presence (lanes 2–7) of EcoRI. Nuclear fractions were immunoblotted with anti-Claspin, anti-Xbrca1, anti-GST, anti-Xchk1-P-S344, anti-Xchk1, and anti-Xorc2 antibodies. 35S-Xchk1 was detected with a PhosphorImager.
Interestingly, although we added back both wild-type and T817A/S819A mutant Claspin to the level of endogenous Claspin, the nuclear accumulation of these recombinant proteins was significantly lower than that of endogenous Claspin in mock-depleted extracts. We have not observed this effect following depletion of Claspin alone, which suggests that Xbrca1 contributes in some manner to the nuclear uptake and/or retention of Claspin. To control for this effect on nuclear accumulation, we made use of a GST fusion protein containing an ectopic NLS and residues 776–956 of Claspin, which contain its Chk1-activating domain (Lee et al. 2005). The wild-type and T817A/S819A mutant versions of GST-NLS-Claspin(776–956) accumulated to normal levels in nuclei from extracts that had been depleted of both Claspin and Xbrca1 (data not shown). As depicted in Figure 7B, the T817A/S819A mutant of this peptide still could not rescue phosphorylation of Xchk1, whereas the wild-type peptide restored significant phosphorylation. Taken together, these results indicate that the activation of Xchk1 in response to DSBs depends entirely on the phosphorylation of Claspin on T817/S819 when Xbrca1 is missing from the system.
Discussion
Although the role of Claspin in the checkpoint response to incompletely replicated DNA is understood in some detail, comparatively little is known about the role of this protein in DNA damage responses. In this report, we have examined how phosphorylation contributes to the regulation of Claspin in various checkpoint pathways. We have found that Claspin is a versatile protein that can participate in checkpoint responses to both incompletely replicated and damaged DNA by relying differently on phosphorylation of its side chains.
Many checkpoint-regulatory proteins are controlled by reversible phosphorylation. In the case of Claspin, it has been shown that phosphorylation on both its S864 and S895 residues is necessary for the binding to Xchk1 (Kumagai and Dunphy 2003). Both of these phosphorylations are also necessary for the ability of Claspin to mediate the activation of Xchk1 in response to stalled DNA replication forks. Interestingly, although Xatr is a pivotal upstream regulator of this pathway, it appears not to be the kinase that is directly responsible for phosphorylation of these residues. The sequence around these residues does not resemble the consensus for phosphorylation by Xatr, and we have not been able to observe in vitro phosphorylation of these residues by Xatr. One possible explanation is that initial phosphorylation of Claspin by Xatr on another site(s) is necessary for a distinct kinase to subsequently phosphorylate S864/S895. Another, not mutually exclusive, possibility is that the kinase that directly phosphorylates S864/S895 is itself regulated by Xatr.
We have investigated the potential regulation of Claspin by Xatr by systematically mutating its various SQ/TQ motifs (12 total). Intriguingly, the 12AQ mutant of Claspin does not display any obvious defect in its ability to mediate the activation of Xchk1 in response to aphidicolin-induced replication blocks. This finding suggests that either Xatr does not regulate Claspin directly in the aphidicolin-triggered pathway or perhaps that Xatr phosphorylates critical sites on Claspin that do not fit the typical SQ/TQ consensus.
On the other hand, we have been able to detect a direct role for Xatr in the regulation of Claspin in response to other DNA structures. For example, a simple double-stranded DNA template such as pA-pT requires Xatr-catalyzed phosphorylation on either T817 or S819. This template, in which each strand is only 70 nt long, is too small to support the assembly of nucleosomes and thus would not be able to form chromatin. Furthermore, a genuine chromatin template with exposed double-stranded ends (e.g., sperm chromatin that has been digested with the restriction endonuclease EcoRI) also triggers a Claspin-dependent checkpoint response that involves T817 and S819. However, in this case, the regulation is more elaborate in that removal of Claspin from egg extracts containing EcoRI-damaged chromatin only partially abrogates the phosphorylation of Xchk1. This observation suggested that some other factor(s) might share a function with Claspin.
We pursued this possibility by examining the potential role of Xbrca1. In human cells, BRCA1 has been implicated in the regulation of Chk1 following IR (Yarden et al. 2002; Lin et al. 2004). Indeed, we found that immunodepletion of Xbrca1 from egg extracts also partially reduced phosphorylation of Xchk1 in response to chromatin with DSBs, while removal of both Claspin and Xbrca1 appeared to eliminate this phosphorylation almost entirely. Our observations regarding the response of Xenopus egg extracts to EcoRI-damaged chromatin differ somewhat from those of Lin et al. (2004). These investigators reported that depletion of either Claspin or BRCA1 from human cells by treatment with small interfering RNA (siRNA) reduced the phosphorylation of Chk1 in response to IR. However, depletion of both Claspin and BRCA1 did not result in a further reduction in the phosphorylation of Chk1. The reason for this distinction is not clear, but it may be related to differences between species, embryonic versus somatic systems, or the methods for induction of DNA damage.
Overall, our results suggest that phosphorylation of a checkpoint mediator protein can have a crucial impact on its specificity for a given checkpoint pathway. In the case of Claspin, phosphorylation of T817/S819 is not necessary for a response to stalled replication forks, but is important for a proper reaction to DSBs. Furthermore, by modulating the specificity of a checkpoint mediator protein, site-specific phosphorylation can have critical implications for functional relationships among checkpoint mediator proteins. For instance, when Xbrca1 is absent from the egg extract system, phosphorylation of Xchk1 in response to chromosomal DSBs comes to depend entirely on phosphorylation of T817/S819 of Claspin.
Another aspect of this work involves the relationship between DNA replication and initiation of checkpoint signaling. It was originally shown that activation of Xchk1 occurs in response to treatment of Xenopus egg extracts (containing sperm chromatin) with either aphidicolin or UV (Kumagai et al. 1998a). It had been well established that aphidicolin induces the formation of stalled replication forks with primed DNA structures (Mahbubani et al. 1997). However, it was not clear whether UV damage led directly to the activation of Xchk1 or instead also caused the formation of stalled replication forks, which in turn elicited this activation. Subsequent work supported the latter model (Lupardus et al. 2002; Stokes et al. 2002). Nonetheless, it appears that DNA damage can also elicit a checkpoint response without the need for replication under certain circumstances. For example, Costanzo et al. (2003) showed that treatment with agents that elicit the formation of single-stranded DNA gaps in DNA (e.g., etoposide and exonuclease III) caused replication-independent recruitment of RPA to chromatin. In this study, we have shown that introduction of DSBs into chromatin causes the activation of Xchk1 even when chromosomal DNA replication is prevented by treatment with geminin. We have also observed that Claspin can still interact with EcoRI-damaged chromatin in the presence of geminin. Our laboratory had initially found that geminin blocked almost totally the binding of Claspin to chromatin during a normal S phase (Lee et al. 2003). As shown in this study, however, it appears that Claspin can associate with chromatin by a replication-independent mechanism when damage creates access to the DNA.
In conclusion, we have shown that site-specific phosphorylation is a crucial determinant of how a checkpoint mediator protein participates in checkpoint responses. Thus, post-translational modification can increase the functional diversity of this important class of regulatory proteins.
Materials and methods
Xenopus egg extracts
Xenopus egg extracts were prepared as described (Yoo et al. 2004a). Extracts were treated with 50 μg/mL pA-pT to induce checkpoint responses as before (Kumagai and Dunphy 2000). To prepare extracts containing chromatin with DSBs or DNA replication blocks, demembranated sperm nuclei (1000–3000/μL) were incubated in extracts containing 0.05 U/μL EcoRI or 50 μg/mL aphidicolin, respectively. Caffeine was added at a final concentration of 5 mM to override checkpoint responses (Yoo et al. 2004b).
Antibodies
Affinity-purified antibodies against Claspin (either residues 1–464 or 232–606), P-S864 of Claspin, Xorc2, Xatr, Xatm, p70 of Polα, Xcdc45, Xchk1, and Xchk2 were described previously (Kumagai and Dunphy 2000, 2003; Lee et al. 2003, 2005; Yoo et al. 2004b). Antibodies against residues 1001–1192 of Xenopus BRCA1 (Xbrca1) were raised as described (Joukov et al. 2001). Anti-Xmcm7 antiserum was generously supplied by J. Blow (University of Dundee, Dundee, UK). For anti-P-S819 antibodies to phosphorylated Claspin, the peptide CLEDDTQSQLLDA (containing residues 813–824 of Claspin) was synthesized with a phosphate on S819 and a cysteine at the N-terminal end for conjugation to keyhole limpet hemocyanin. Anti-phosphopeptide antibodies were purified as described (Yoo et al. 2004b). Antibodies that recognize phospho-S344 of Xchk1 were purchased from Cell Signaling Technology. Anti-human Mcm2 (BM28), anti-Flag, and control rabbit antibodies (IgG fraction) were obtained from BD Transduction Laboratories, Sigma, and Zymed, respectively.
Immunoprecipitation and immunodepletion
For immunoprecipitations, egg extracts (100 μL) were incubated with Affiprep-protein A beads (Bio-Rad) containing anti-Claspin (3 μg) or anti-Xbrca1 antibodies (3 μg) for 45 min at 4°C. The beads were washed three times with buffer A (10 mM HEPES-KOH at pH 7.5, 150 mM NaCl, 0.5% NP-40, 2.5 mM EGTA, and 20 mM β-glycerolphosphate) and twice with HEPES-buffered saline (HBS; 10 mM HEPES-KOH at pH 7.5 and 150 mM NaCl), and subjected to SDS-PAGE and immunoblotting. For immunodepletion of Xbrca1, interphase extracts (100 μL) were incubated with 25 μg of anti-Xbrca1 antibodies bound to 25 μL of Affiprep protein A beads at 4°C for 45 min. The same amount of control rabbit IgG was used for mock depletion. After incubation, the beads were removed by centrifugation, and the supernatants were treated again for a second round of depletion. Claspin, Xatm, and Xatr were immunodepleted as described previously (Kumagai and Dunphy 2000; Yoo et al. 2004b).
Production of recombinant proteins in insect cells and bacteria
Single or multiple mutants in the SQ/TQ motifs of Claspin were produced using the QuikChange kit with pFastBacHT-Claspin-Flag as template (Lee et al. 2005). The PCR-generated Xenopus BRCA1 gene was cloned into the pFastBacHT vector to express His6-Xbrca1 protein. Recombinant baculoviruses were generated with the Bac-to-Bac system (Invitrogen). Recombinant baculovirus-expressed proteins were produced in Sf9 insect cells and purified using nickel-agarose beads (Yoo et al. 2004a). To prepare GST-tagged peptide fragments of Claspin, PCR-generated DNA fragments encoding amino acids 808–834 and 776–956 of Claspin from either pFastBac-His6-Claspin-Flag wild-type or other mutants were cloned into pGEX-2T and pGEX-NLS vectors (Lee et al. 2005). GST-tagged proteins were produced and purified as described (Yoo et al. 2004a). 35SLabeled proteins were synthesized in vitro with the TnT system (Promega). Geminin was prepared as described (Lee et al. 2003).
Isolation of nuclear and chromatin fractions
Egg extracts (100–200 μL) containing 3000 sperm nuclei per microliter were incubated under the indicated conditions, overlaid on a 1-mL sucrose cushion containing chromatin isolation buffer (20 mM HEPES-KOH at pH 7.6, 1 M sucrose, 80 mM KCl, 25 mM K-gluconate, and 10 mM Mg-gluconate), and centrifuged at 6100g for 5 min. The pellets were washed twice with chromatin isolation buffer. After centrifugation, the pellets (nuclear fractions) were boiled in gel sample buffer and subjected to SDS-PAGE. For preparation of chromatin fractions, the pellets were washed once more with chromatin isolation buffer containing 0.5% NP-40. The supernatant was removed, and the chromatin pellets were processed for SDS-PAGE.
Replication assays
To monitor DNA replication, interphase extracts were incubated with sperm nuclei (1000–3000/μL) and 100 μg/mL cycloheximide in the presence of [α-32P]dATP as described (Yoo et al. 2004b).
Phosphopeptide mapping
GST-Claspin(808–834) and its T817A, S819A, and T817A/S819A mutants (1 μg each) were incubated for 100 min in interphase extracts (100 μL) containing 50 μg/mL pA-pT, 3 μM okadaic acid, 100 μg/mL cycloheximide, and 0.5 mCi 32P-orthophosphate. GST-tagged Claspin fragments were reisolated with glutathione beads and digested overnight at 37°C with endoproteinase Asp-N (Roche). Phosphopeptide mapping was performed as described previously (Kumagai and Dunphy 2003).
Kinase assays
Wild-type and kinase-deficient versions of recombinant human Flag-tagged ATR were produced in human 293T cells and assayed for kinase activity as described (Canman et al. 1998).
Acknowledgments
We are grateful to Drs. D. Livingston (Harvard University) and J. Blow (University of Dundee) for providing the plasmid encoding GST-Xbrca1(1001–1192) and anti-Xmcm7 antiserum, respectively. We also thank our colleagues in the laboratory for sharing reagents and for helpful comments on the manuscript. This work was supported by NIH grants GM043974 and GM070891 to W.G.D.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1398806
References
- Abraham R.T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Bakkenist C.J., Kastan M.B. Initiating cellular stress responses. Cell. 2004;118:9–17. doi: 10.1016/j.cell.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Canman C.E., Lim D.S., Cimprich K.A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M.B., Siliciano J.D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Chini C.C., Chen J. Human Claspin is required for replication checkpoint control. J. Biol. Chem. 2003;278:30057–30062. doi: 10.1074/jbc.M301136200. [DOI] [PubMed] [Google Scholar]
- Costanzo V., Robertson K., Ying C.Y., Kim E., Avvedimento E., Gottesman M., Grieco D., Gautier J. Reconstitution of an ATM-dependent checkpoint that inhibits chromosomal DNA replication following DNA damage. Mol. Cell. 2000;6:649–659. doi: 10.1016/s1097-2765(00)00063-0. [DOI] [PubMed] [Google Scholar]
- Costanzo V., Shechter D., Lupardus P.J., Cimprich K.A., Gottesman M., Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell. 2003;11:203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- Dasso M., Newport J.W. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: Studies in Xenopus. Cell. 1990;61:811–823. doi: 10.1016/0092-8674(90)90191-g. [DOI] [PubMed] [Google Scholar]
- Guo Z., Dunphy W.G. Response of Xenopus Cds1 in cell-free extracts to DNA templates with double-stranded ends. Mol. Biol. Cell. 2000;11:1535–1546. doi: 10.1091/mbc.11.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Kumagai A., Wang S.X., Dunphy W.G. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes & Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Nejad M., You Z., Yee M., Newport J.W., Cimprich K.A. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr. Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- Jeong S.-Y., Kumagai A., Lee J., Dunphy W.G. Phosphorylated Claspin interacts with a phosphate-binding site in the kinase domain of Chk1 during ATR-mediated activation. J. Biol. Chem. 2003;278:46782–46788. doi: 10.1074/jbc.M304551200. [DOI] [PubMed] [Google Scholar]
- Joukov V., Chen J., Fox E.A., Green J.B., Livingston D.M. Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc. Natl. Acad. Sci. 2001;98:12078–12083. doi: 10.1073/pnas.211427098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., Ashikari T., Sugimoto K., Shirahige K. S-Phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Tada S., Tsuyama T., Murofushi H., Seki M., Enomoto T. Focus-formation of replication protein A, activation of checkpoint system and DNA repair synthesis induced by DNA double-strand breaks in Xenopus egg extract. J. Cell Sci. 2002;115:3159–3169. doi: 10.1242/jcs.115.15.3159. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W.G. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W.G. Repeated phosphopeptide motifs in Claspin mediate the regulated binding of Chk1. Nat. Cell Biol. 2003;5:161–165. doi: 10.1038/ncb921. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Guo Z., Emami K.H., Wang S.X., Dunphy W.G. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J. Cell Biol. 1998a;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Yakowec P.S., Dunphy W.G. 14–3–3 proteins act as negative regulators of the mitotic inducer Cdc25 in Xenopus egg extracts. Mol. Biol. Cell. 1998b;9:345–354. doi: 10.1091/mbc.9.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A., Kim S.-M., Dunphy W.G. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. J. Biol. Chem. 2004;279:49599–49608. doi: 10.1074/jbc.M408353200. [DOI] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W.G. Claspin, a Chk1-regulatory protein, monitors DNA replication on chromatin independently of RPA, ATR, and Rad17. Mol. Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Lee J., Gold D.A., Shevchenko A., Shevchenko A., Dunphy W.G. Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol. Biol. Cell. 2005;16:5269–5282. doi: 10.1091/mbc.E05-07-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Kim S.M., Lee J., Dunphy W.G. Absence of BLM leads to accumulation of chromosomal DNA breaks during both unperturbed and disrupted S phases. J. Cell Biol. 2004;165:801–812. doi: 10.1083/jcb.200402095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y., Li K., Stewart G.S., Elledge S.J. Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc. Natl. Acad. Sci. 2004;101:6484–6489. doi: 10.1073/pnas.0401847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Guntuku S., Cui X.S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes & Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Lupardus P.J., Byun T., Yee M.C., Hekmat-Nejad M., Cimprich K.A. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes & Dev. 2002;16:2327–2332. doi: 10.1101/gad.1013502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahbubani H.M., Chong J.P., Chevalier S., Thommes P., Blow J.J. Cell cycle regulation of the replication licensing system: Involvement of a Cdk-dependent inhibitor. J. Cell Biol. 1997;136:125–135. doi: 10.1083/jcb.136.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry T.J., Kirschner M.W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Nyberg K.A., Michelson R.J., Putnam C.W., Weinert T.A. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- Osborn A.J., Elledge S.J. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes & Dev. 2003;17:1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn A.J., Elledge S.J., Zou L. Checking on the fork: The DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L.A., Ünsal-Kaçmaz K., Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Stokes M.P., Van Hatten R., Lindsay H.D., Michael W.M. DNA replication is required for the checkpoint response to damaged DNA in Xenopus egg extracts. J. Cell Biol. 2002;158:863–872. doi: 10.1083/jcb.200204127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Gasser S.M. The histone code at DNA breaks: A guide to repair? Nat. Rev. Mol. Cell Biol. 2005;6:757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- Wang B., Matsuoka S., Carpenter P.B., Elledge S.J. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- Yarden R.I., Pardo-Reoyo S., Sgagias M., Cowan K.H., Brody L.C. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat. Genet. 2002;30:285–289. doi: 10.1038/ng837. [DOI] [PubMed] [Google Scholar]
- Yoo H.Y., Kumagai A., Shevchenko A., Shevchenko A., Dunphy W.G. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell. 2004a;117:575–588. doi: 10.1016/s0092-8674(04)00417-9. [DOI] [PubMed] [Google Scholar]
- Yoo H.Y., Shevchenko A., Shevchenko A., Dunphy W.G. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J. Biol. Chem. 2004b;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- Zhao H., Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]