Abstract
RAF is a critical effector of the small GTPase RAS in normal and malignant cells. Despite intense scrutiny, the mechanism regulating RAF activation remains partially understood. Here, we show that the scaffold KSR (kinase suppressor of RAS), a RAF homolog known to assemble RAF/MEK/ERK complexes, induces RAF activation in Drosophila by a mechanism mediated by its kinase-like domain, but which is independent of its scaffolding property or putative kinase activity. Interestingly, we found that KSR is recruited to RAF prior to signal activation by the RAF-binding protein CNK (connector enhancer of KSR) in association with a novel SAM (sterile α motif) domain-containing protein, named Hyphen (HYP). Moreover, our data suggest that the interaction of KSR to CNK/HYP stimulates the RAS-dependent RAF-activating property of KSR. Together, these findings identify a novel protein complex that controls RAF activation and suggest that KSR does not only act as a scaffold for the MAPK (mitogen-activated protein kinase) module, but may also function as a RAF activator. By analogy to catalytically impaired, but conformationally active B-RAF oncogenic mutants, we discuss the possibility that KSR represents a natural allosteric inducer of RAF catalytic function.
Keywords: RAF activation, RAS/MAPK signaling, SAM domain, scaffold proteins
Signal transmission via the RAF/MEK/ERK pathway, also known as the mitogen-acitvated protein kinase (MAPK) module, is a central event triggered by the small GTPase RAS to regulate a number of basic cellular processes in metazoans, including cell proliferation, differentiation, and survival (Pearson et al. 2001). Unrestrained signaling through this pathway caused, for instance, by activating mutations in specific isoforms of either RAS or RAF, has been linked to several types of cancer in humans and, for some of these, at an impressively high frequency (Malumbres and Barbacid 2003; Wellbrock et al. 2004). Because of potential benefits to human health, extensive efforts have been devoted to describe in molecular terms the signal transfer mechanism within the RAS/MAPK pathway. Despite significant progress, a number of events have proven particularly challenging. One notable example is the mechanism leading to the activation of the RAF serine/threonine kinase.
Three RAF members have been identified in mammals (A-RAF, B-RAF, and C-RAF/Raf-1) and homologs are present in other metazoans, including Caenorhabditis elegans and Drosophila, where a single gene encoding a protein more closely related to B-RAF has been identified (Dhillon and Kolch 2002; Chong et al. 2003). RAF proteins comprise an N-terminal regulatory region, followed by a C-terminal catalytic domain. The N-terminal region includes a RAS-binding domain (RBD), a cysteine-rich domain (CRD), and an inhibitory 14–3–3-binding site encompassing Ser 259 (S259) in C-RAF. The binding of 14–3–3 to this latter site requires the phosphorylation of the S259-like residue in RAF proteins, which in turn mediates their cytoplasmic retention in unstimulated cells (Morrison and Cutler 1997). Upon receptor tyrosine kinase (RTK)-dependent activation, GTP-loaded RAS binds the RBD of RAF and facilitates the dephosphorylation of the S259-like residue, thereby releasing 14–3–3 and promoting the association of RAF to the membrane (Jaumot and Hancock 2001; Dhillon et al. 2002; Light et al. 2002). A number of phosphorylation events are then required to fully induce RAF catalytic activity (Chong et al. 2003). Although some are isozyme specific, two are probably common to all members and affect conserved serine/threonine residues (T599 and S602 in B-RAF) situated in the activation loop of the kinase domain (Zhang and Guan 2000; amino acid numbering of B-RAF is according to Wellbrock et al. 2004). Mutational analyses as well as a recent crystallographic study of the B-RAF kinase domain strongly suggest that phosphorylation of these residues plays a critical role in the final stage of activation by destabilizing an inhibitory interaction that takes place between the P loop (subdomain I) and the DFG motif (subdomain VII)/activation loop of the kinase domain (Wan et al. 2004). The mechanism and kinase(s) leading to the phosphorylation of these residues are unknown.
A number of scaffold proteins have also been suggested to regulate RAF activity (Kolch 2000). However, their mode of action and functional interdependency is poorly understood. One example corresponds to the kinase suppressor of RAS (KSR) members, which are known to assemble RAF, MEK, and MAPK into functional complexes (Morrison and Davis 2003). Interestingly, these proteins are structurally related to RAF, although they have some key differences. For instance, they do not contain an RBD, but comprise a conserved region called CA1 that was found in Drosophila to mediate an interaction between KSR and RAF (Roy et al. 2002). Further, they possess a kinase-like domain that constitutively binds MEK, but which appears to be devoid of kinase activity (Morrison and Davis 2003). While the function of KSR as a scaffold of the MAPK module has been convincingly documented, genetic and biochemical characterization of the single Drosophila KSR isoform suggested that its activity is also required upstream of RAF (Therrien et al. 1995; Anselmo et al. 2002). This other role, however, has not been determined.
Connector enhancer of KSR (CNK) is another scaffold protein acting as a putative regulator of RAF activity. As for KSR, its activity is essential for multiple RTK signaling events, where it appears to regulate the MAPK module at the level of RAF (Therrien et al. 1998). CNK homologs have been identified in other metazoans and evidence gathered in mammalian cell lines supports their participation in the regulation of B-RAF and C-RAF (Lanigan et al. 2003; Bumeister et al. 2004; Ziogas et al. 2005). A similar conclusion was also recently reached in C. elegans (Rocheleau et al. 2005). In flies, CNK associates directly with the catalytic domain of RAF through a short amino acid sequence called the RAF-interacting motif (RIM) and modulates RAF activity according to the RTK signaling status (Douziech et al. 2003; Laberge et al. 2005). In the absence of RTK signals, CNK-bound RAF is inhibited by a second motif adjacent to the RIM, called the inhibitory sequence (IS). In contrast, upon RTK activation, CNK integrates RAS and Src activity, which in turn leads to RAF activation. The ability of RAS to promote RAF activation was found to strictly depend on two domains: a sterile α motif (SAM) domain and the so-called conserved region in CNK (CRIC) located in the N-terminal region of CNK (Douziech et al. 2003). The molecular role of these domains is currently unknown. On the other hand, the binding of a Src family kinase, Src42, to an RTK-dependent phospho-tyrosine residue (pY1163) located C-terminal to the IS motif appears to release the inhibitory effect that the IS motif imposes on RAF catalytic function (Laberge et al. 2005).
In this report we investigated the role of the SAM and CRIC domains of CNK during RAS-dependent RAF activation in Drosophila S2 cells. Strikingly, we found that their activity is mediated by KSR and that KSR stimulates RAF catalytic function independently of its capacity to bridge RAF and MEK. This effect occurs at a step upstream of the activation loop phosphorylation, but downstream of the dephosphorylation of the S259-like residue, thus indicating that it regulates the final stage of RAF activation. While the catalytically devoid KSR kinase domain appears to be the primary effector of this event, CNK participates in at least two ways: (1) It assembles a KSR/RAF complex in vivo by interacting separately with the kinase domains of KSR and RAF through its SAM domain and RIM element, respectively, and (2) its CRIC region promotes CNK-bound KSR activity toward RAF in a RAS-dependent manner. Finally, we found that the KSR/CNK interaction depends on a novel and evolutionarily conserved SAM domain-containing protein, Hyphen, whose presence is essential for RAS-induced signaling through the MAPK module at a step upstream of RAF. Together, this work unveils a network of interacting scaffolds that regulates the RAS-dependent catalytic function of RAF.
Results
CNK and KSR are required for activation of the catalytic domain of RAF
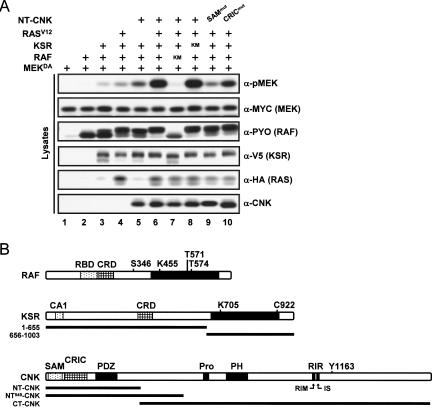
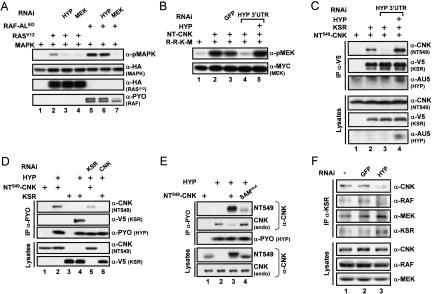
RAS-dependent activation of the MAPK module in Drosophila S2 cells was previously found to depend on two domains (SAM and CRIC) located in the N-terminal portion of CNK (Fig. 1), and this requirement occurred at a step upstream of RAF (Douziech et al. 2003). To investigate this event at the molecular level, we took advantage of a KSR-dependent MEK activation assay based on Drosophila proteins that was previously used to demonstrate KSR’s ability to facilitate MEK phosphorylation by RAF (Roy et al. 2002). In that assay, coexpression of wild-type variants of epitope-tagged KSR, RAF, and kinase-inactivated MEKDA is sufficient to induce MEK phosphorylation on activating residues S237 and S241 (Fig. 1A, cf. lanes 1–3), which could be further enhanced by coexpression of RASV12 (Fig. 1A, lane 4). Interestingly, coexpression of NT-CNK (amino acid position 2–384) (Fig. 1B) along with KSR, RAF, and MEK also enhanced MEK activation (Fig. 1A, lane 5), thus indicating that the effect of NT-CNK is integrated by the KSR/RAF/MEK complex. Consistent with the cooperation observed between RASV12 and NT-CNK (Douziech et al. 2003), their coexpression with KSR, RAF, and MEK resulted in a synergistic activation of MEK (Fig. 1A, cf. lanes 4 and 6, or 5 and 6). The specificity of this assay still critically depended on RAF catalytic function, as a kinase-mutant RAF variant (RAFKM) did not support MEK activation (Fig. 1A, lane 7). In contrast, although the presence of KSR was absolutely essential for MEK phosphorylation (see below), its function did not appear to depend on a putative kinase activity, as a presumed catalytically impaired mutant was as active as wild-type KSR (Fig. 1A, cf. lanes 6 and 8; see below). Finally, and most importantly, NT-CNK activity depended on the integrity of its SAM and CRIC domains (Fig. 1A, lanes 9,10).
Figure 1.
The N-terminal portion of CNK augments KSR-induced MEK phosphorylation by RAF. (A) S2 cells were transfected using the indicated plasmid combinations. RAF activity was assessed by determining phospho-MEK levels in cell lysates by immunoblotting (α-pMEK). Protein levels were verified using the antibodies indicated to the right. Results presented here and thereafter are representative of at least three similar experiments. RAF and KSR kinase-mutant (KM) variants have a K455S and a K705M change, respectively. As previously reported (Douziech et al. 2003), coexpression of full-length CNK along with RASV12 is inhibitory. This is caused by the RIR and is naturally alleviated by RTK-induced Src42 binding to the Y1163 region located C-terminal to the RIR (Laberge et al. 2005). To facilitate the characterization of the positive effect of the SAM and CRIC domains with respect to RAS-dependent RAF activation, we used NT-CNK constructs instead of full-length CNK, thereby bypassing the requirement in additional RTK signals. The SAMmut NT-CNK variant has a L71K mutation, while the CRICmut variant has a three-amino-acid deletion (Del A162-H163-R164) that is similar to a mutation found in a cnk loss-of-function allele (Therrien et al. 1998). The difference between the effects produced by the mutations within the SAM domain and the CRIC region may be due to the fact that the CRIC mutation is hypomorphic, while the L71K mutation obliterates normal SAM domain function. Alternatively, the involvement of the CRIC region may not be as decisive as the SAM domain. (B) Schematic of the main Drosophila RAF, KSR, and CNK constructs used in this study. Full-length RAF contains an RBD, a CRD, and a kinase domain (black box). The relative positions of the S346 (S259-like), K455 (critical lysine in subdomain II), and T571–T574 (phospho-accepting sites in the activation loop) residues are also shown. Full-length KSR comprises the so-called conserved area 1 (CA1) (Therrien et al. 1995), a CRD, and a putative kinase domain (black box). The conserved lysine residue of subdomain II (K705) and the C922 residue that is critical for MEK binding (Roy et al. 2002) are also represented. Schematic of the N-terminal (1–655) and C-terminal (656–1003) KSR constructs are indicated as tick lines. Full-length CNK includes a SAM domain, a CRIC, a PDZ domain, a proline-rich strech (Pro), a PH domain, and a RIR that includes two jointly required elements: a RIM and an IS. NT-CNK (position 2–384), NT549-CNK (position 2–549), and CT-CNK (position 382–1557) constructs are depicted as thick lines.
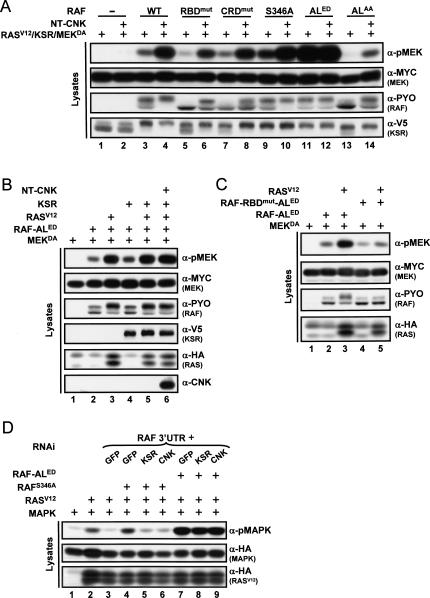
The assay presented above provided us with a molecularly tractable system to narrow down RAF regulatory event(s) that is/are positively modulated by CNK. To address this point, we independently mutagenized four regions within RAF known to carry out distinct function and evaluated how this affected the response to NT-CNK. These regions included the RBD, the CRD, the S259-like residue (S346) within the N-terminal 14–3–3-binding site and the two phospho-accepting threonine residues (T571 and T574) within the activation loop of the kinase domain (Fig. 1B). To ensure that endogenous RAF did not influence the output, we took advantage of an RNA interference (RNAi)-based strategy that we previously used to deplete the endogenous contribution of a given transcript while concomitantly restoring it exogenously using a nontargetable construct (Douziech et al. 2003). Briefly, we depleted endogenous RAF (Supplementary Fig. S1A) by transfecting a double-stranded RNA (dsRNA) encompassing exclusively RAF 3′ untranslated region (UTR) sequences and exogenously restored RAF activity by cotransfecting a RAF cDNA that had its 3′UTR sequences replaced by those of the Drosophila alcohol dehydrogenase (ADH) gene, thereby producing RAF transcripts insensitive to the dsRAF3′UTR RNA (Supplementary Fig. S1B). As shown in Figure 2A, cotransfection of RASV12 with KSR and MEKDA (±NT-CNK) in the absence of RAF did not induce MEK phosphorylation (Fig. 2A, lanes 1,2), while similar cotransfections that included wild-type RAF (WT) restored MEK phosphorylation (Fig. 2A, lane 3) as well as the response to NT-CNK (Fig. 2A, lane 4). Inactivation of the RBD, CRD, and 14–3–3-binding site worked as expected; both the RBD and CRD mutations decreased RAF activity, whereas the S346A mutant was more active than wild type (Fig. 2A, cf. lanes 3,5,7,9). Interestingly, none of those variants impeded the response to NT-CNK (Fig. 2A, lanes 5–10), thus indicating that NT-CNK is probably not regulating the ability of the RAF N-terminal domain to associate with either RAS, the plasma membrane, or 14–3–3. Nonetheless, it is remarkable that even though the RAFRBDmut variant no longer associates with RAS, the effect of NT-CNK on this mutant still depends on RASV12 (Supplementary Fig. S2A), thus further supporting the RAS dependency of NT-CNK activity. Next, we evaluated the effect of oppositely acting mutations within the activation loop (AL) of the kinase domain. Mutations changing T571 and T574 to phospho-mimetic residues (ALED) resulted in a strongly activated RAF variant (Fig. 2A, lane 11). Surprisingly, this mutant no longer responded to NT-CNK (Fig. 2A, lane 12). To ensure that the levels of MEKDA were not limiting under those conditions, we conducted similar experiments using decreasing amounts of RAFED and obtained similar results (Supplementary Fig. S2B). Conversely, changing the same threonine residues to alanine (ALAA) produced a potent loss of function that yet retained some capacity to respond to NT-CNK (Fig. 2A, lanes 13,14). Taken together, these results suggest that NT-CNK activity controls a step leading to T571–T574 phosphorylation and that this activity either is distinct or has broader consequences than the phosphorylation event per se, as the ALAA mutant still responded to NT-CNK.
Figure 2.
CNK and KSR function upstream of the RAF activation loop phosphorylation event. S2 cells were transfected using the indicated plasmid combinations. (A–C) In each condition, dsRNA (500 ng) targeting the 3′UTR of endogenous RAF was also transfected. The RBDmut, CRDmut, ALED, and ALAA amino acid changes correspond to R174L, C249S–C252S, T571E–T574D, and T571A–T574A, respectively. (D) S2 cells were plated with or without RAF 3′UTR dsRNA (10 μg/mL) in combination with the other indicated dsRNAs (RNAi; 10 μg/mL each) and cultured for 24 h prior to transfection (500 ng of the same dsRNAs was also added to the transfection mixture). RAF activity was evaluated by determining phospho-MAPK levels. GFP dsRNA is used as a negative control for RNAi. Addition of NT-CNK further reduced the mobility of RAF (cf. lanes 3 and 4, α-PYO in A). This event is most likely due to phosphorylation, as the mobility shifts could be eliminated by in vitro phosphatase treatment (data not shown). Although the phosphorylation sites and their functional relevance are unknown, there is a tight correlation between the positive effects of NT-CNK and RAF mobility shift, which we used as a second readout of NT-CNK effect on RAF.
The inability of RAFED to respond to NT-CNK prompted us to determine whether this was specific to CNK or whether this mutant also no longer integrated RAS and KSR activities. Interestingly, although RAFED activity could be further increased by RASV12 (Fig. 2B, cf. lanes 2 and 3), it was not influenced by KSR expressed either alone (Fig. 2B, lane 4) or in combination with RAS and/or NT-CNK (Fig. 2B, lanes 5,6; data not shown). Given the presence of an intact RBD in RAFED, we verified whether it accounted for its responsiveness to RAS even if the RAS-dependent NT-CNK effect did not modulate RAFED. As shown in Figure 2C, this appeared to be the case as a RAFRBDmut-ED double mutant lost the capacity to respond to RASV12 (lane 5). This finding therefore provides further evidence that the RBD-dependent and the NT-CNK-dependent steps are two independent RAS-mediated events. Curiously and in sharp contrast to RAFWT or any of the other RAF mutants that have been tested so far, the fact that RAFED could not be modulated by KSR expression suggests that the RAF–MEK bridging effect of KSR is not critical for the ability of RAFED to phosphorylate MEK even if, as RAFWT, RAFED does not stably associate with MEK (data not shown). Together, these results suggest that the scaffolding function of KSR is secondary to a more critical role played by KSR that influences the activation loop of RAF.
To determine whether endogenous CNK and KSR are also required at a similar step, we depleted their respective contribution by RNAi and assessed the impact on RAFED activity using MAPK activation as readout. We conducted these experiments in the presence of RASV12 so that appropriate RAS signals were provided to endogenous CNK and KSR. In addition, we depleted endogenous RAF by RNAi and restored it with similar amounts of the two distinct RAF gain of functions tested above: RAFS346A and RAFED. As shown in Figure 2D, neither CNK nor KSR depletion reduced RAFED activity. In contrast, the activity of RAFS346A was affected by the removal of endogenous CNK and KSR. Together, these results are consistent with a model whereby CNK and KSR activities are both required for events leading to the activated state of the RAF activation loop occurring downstream of the dephosphorylation of the S259-like residue.
The kinase domain of KSR mediates CNK activity
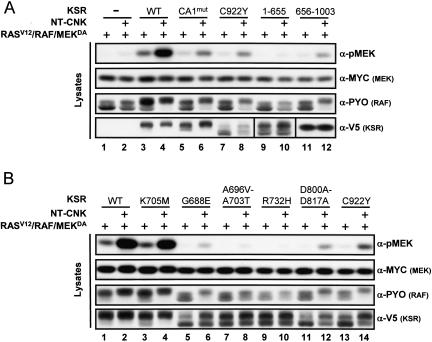
Using a strategy analogous to the one used for RAF, we next determined whether KSR is required for NT-CNK’s positive function and, if it is required, identified which structural feature of KSR is mediating the effect. As for the above experiments, endogenous KSR was depleted by RNAi (Supplementary Fig. S1). First, we conducted a control experiment in which RASV12, RAFWT, and MEKDA were cotransfected in the absence of KSR to show its strict requirement for MEK phosphorylation (Fig. 3A, lane 1). Strikingly, cotransfection of NT-CNK under those conditions did not significantly elevate MEK phosphorylation, nor did it increase RAF mobility shift (Fig. 3A, lane 2), thus demonstrating that KSR is essential for the positive effect of CNK on RAF. Indeed, introduction of KSR along with RASV12, RAFWT, and MEKDA restored MEK activation (Fig. 3A, lane 3), which could be further enhanced by NT-CNK (Fig. 3A, lane 4). We then tested the impact of KSR mutants on the ability of NT-CNK to stimulate MEK phosphorylation. We first evaluated two mutants that have either a decreased RAF-binding (KSRCA1mut) or a disrupted MEK-binding (KSRC922Y) activity. As previously shown (Roy et al. 2002), these mutations strongly reduced KSR activity (Fig. 3A, lanes 5,7). Interestingly, although the absolute output of pMEK was weaker than for KSRWT, they were nonetheless capable of supporting NT-CNK activity (Fig. 3A, lanes 6,8). These results are especially surprising for KSRC922Y, as this variant totally lost its capacity to interact with MEK (Roy et al. 2002; see below). Therefore, these findings suggest that NT-CNK depends on a functional aspect of KSR that is distinct from its scaffolding property. To roughly delineate the region of KSR that is integrating NT-CNK function, we tested the ability of the N-terminal portion (KSR1–655) (Fig. 1B) or the C-terminal portion (KSR656–1003) of KSR to mediate NT-CNK effect. Strikingly, the isolated kinase domain of KSR was capable, albeit weakly, of integrating NT-CNK function (Fig. 3A, cf. lanes 11 and 12), whereas the N-terminal domain was totally inert (Fig. 3A, lanes 9,10), thus suggesting that the NT-CNK effect is integrated by the kinase domain of KSR.
Figure 3.
CNK activity is KSR-dependent. (A,B) S2 cells were transfected using the indicated plasmid combinations. dsRNA (500 ng) targeting the 3′UTR of KSR was also included in each condition. The CA1mut amino acid change corresponds to L50S–R51G.
To further explore the relevance of the kinase domain of KSR in mediating CNK’s positive function, we compared the activity of a panel of kinase domain mutants. As shown above, replacement of the typically critical lysine residue (KSRK705M) located in subdomain II did not perturb KSR’s ability to integrate NT-CNK activity (Fig. 2B, lanes 1–4), which again indicated that the positive effect of KSR and CNK on RAF does not involve standard kinase activity. In support of this conclusion, we found that KSRD800A–D817A, another catalytically impaired mutant, retained the ability to respond to NT-CNK (Fig. 3A, lanes 11,12), even though it behaved as a strong loss of function given its disrupted MEK-binding activity (Roy et al. 2002; see below). In addition to the C922Y mutation, a number of point mutations located in the putative kinase domain have been recovered from genetic screens in Drosophila and C. elegans (Kornfeld et al. 1995; Sundaram and Han 1995; Therrien et al. 1995). We tested some of these mutations to determine whether any of them attenuated the response to NT-CNK. They included the G688E mutation and the A696V–A703T double point mutation that changed highly conserved residues within subdomain I (ATP-binding site) and subdomain II, respectively, as well as the R732H mutation that affects a relatively well-conserved residue located between subdomains III (αC helix) and IV. Interestingly, although KSRG688E retained some capacity to respond to NT-CNK (Fig. 3A, lanes 5,6), and thus behaved similarly as KSRC922Y and KSRD800A–D817A, the two other mutants (KSRA696V–A703T and KSRR732H) were totally inert (Fig. 3A, lanes 7–10). The failure of the A696V–A703T or R732H mutations to mediate NT-CNK effect suggests that they disabled an essential aspect of KSR function, thereby preventing any KSR-mediated activity.
KSR is more than a scaffold connecting MEK to RAF
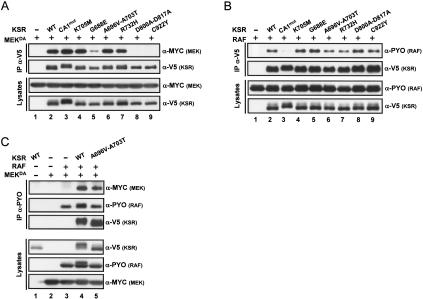
Mutations in KSR disrupting MEK or RAF binding, such as KSRCA1mut, KSRC922Y, or KSRD800A–D817A had been previously shown to impair KSR activity and were actually used as evidence to argue that KSR has a scaffolding role within the MAPK module (Roy et al. 2002). Compared with KSRWT, KSRG688E also displayed lowered MEK-binding activity (Fig. 4A, cf. lanes 2 and 5), but interacted normally with RAF (Fig. 4B, cf. lanes 2 and 5). The reduced association of KSRG688E to MEK is thus consistent with its loss-of-function behavior, although it is surprising that it is not more active than the two KSR mutants (KSRD800A–D817A and KSRC922Y) that entirely lost MEK binding (Fig. 4A, lanes 8,9). Unexpectedly, the two strongest mutants, KSRA696V–A703T and KSRR732H, showed normal association with either MEK (Fig. 4A, lanes 6,7) or RAF (Fig. 4B, lanes 6,7), and thus their inactivity could not be explained by a lack of MEK or RAF binding. Another property of KSR is its ability to bridge RAF and MEK. We verified this ability and found no evidence that these two mutants are less competent than KSRWT to induce a RAF/MEK complex (Fig. 4C; data not shown). Together, these findings demonstrated that the mere bridging of RAF and MEK by KSR is not sufficient for RAF to phosphorylate MEK, and thus KSR must control another event. A number of scenarios can be envisioned to explain these results. For example, in addition to link RAF and MEK together, KSR may be involved in properly orienting MEK with respect to RAF, so that phosphorylation could normally proceed. However, such a model would not suffice to explain why RAFED does not respond to KSR. A second possibility, which would be consistent with the RAFED results, would be that KSR also controls RAF catalytic function.
Figure 4.
Two inactive KSR mutants retain their ability to associate with RAF and MEK. S2 cells were transfected using the indicated plasmid combinations. (A,B) Cell lysates were immunoprecipitated using an α-V5 antibody to determine the amounts of MEK or RAF associated to KSR. Lysates were also directly probed to monitor protein levels. (C) RAF immunoprecipitation (α-PYO) was used to verify the capacity of KSRA696V–A703T, compared with wild-type KSR, to promote the formation of a RAF/MEK complex.
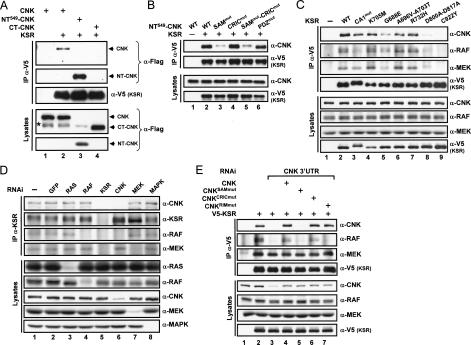
CNK links KSR to RAF
The results shown above suggest that CNK and KSR positively regulate RAF at a similar step and that CNK could not stimulate RAF activity without KSR. Based on this functional dependency, we investigated the possibility that CNK and KSR physically interacted. To verify this, we coexpressed Flag-tagged CNK and V5-tagged KSR in S2 cells, immunoprecipitated KSR from cell lysates, and looked for the presence of CNK in the immunoprecipitate. As shown in Figure 5A, we found that CNK and KSR indeed associated when coexpressed together (lane 2). Given that both proteins can also interact with RAF, we conducted a similar experiment under conditions whereby endogenous RAF had been depleted by RNAi to determine whether it mediates the CNK/KSR association, but found no evidence for this scenario (data not shown; see below). To narrow down the region of CNK involved in this association, we tested the ability of an N-terminal (NT549-CNK) and a C-terminal (CT-CNK) CNK construct (Fig. 1B) to coimmunoprecipitate with KSR. These experiments showed that only the N-terminal region of CNK associated with KSR (Fig. 5A, lanes 3,4), thus indicating that a KSR-binding site resides in this area. Similar results were obtained with the shorter NT-CNK variant used in the functional studies, although that variant migrated immediately below the immunoglobulin heavy chain, and thus was partly masked (data not shown). To determine whether any of the three conserved domains of the N-terminal region of CNK is required for the association with KSR, we tested the effect of individual point mutations inactivating each of these in the NT549-CNK context. Markedly, inactivation of the SAM domain resulted in a severe loss of KSR binding (Fig. 5B, lane 3), whereas the CRIC or PDZ domain mutations did not alter the association (Fig. 5B, lanes 4–6). We next sought to determine which region of KSR interacted with CNK. During the course of this work, we noticed that expression of V5-KSR in S2 cells associated with endogenous CNK, RAF, and MEK, as determined by coimmunoprecipitation and immunoblot analysis (Fig. 5C, lane 2; data not shown). We thus tested the isolated N- and C-terminal domains of KSR for their ability to associate with endogenous CNK. Consistent with the fact that NT-CNK activity is integrated by the kinase domain of KSR, we found that endogenous CNK specifically associated with the C-terminal KSR construct (Supplementary Fig. S3A). Similar results were obtained with Flag-tagged CNK or NT-CNK constructs (data not shown). Taken together, these results identify the SAM domain of CNK as a KSR-binding interface, and thus provide a molecular link to explain the functional dependency of NT-CNK toward KSR.
Figure 5.
CNK mediates the formation of a KSR/RAF complex. (A–C) S2 cells were transfected using the indicated plasmid combinations and cell lysates were either directly probed or immunoprecipitated as indicated. The star in A denotes the position of a nonspecific protein revealed by the α-Flag antibody. (D) S2 cells were plated with the indicated dsRNAs (10 μg/mL). Four days later, cells were lysed and protein extracts were immunoprecipitated using an α-KSR monoclonal antibody. Two 100-mm dishes were pooled for each condition except for lane 7, where five dishes had to be pooled to obtain equal KSR protein levels, as depletion of endogenous MEK greatly destabilized KSR (not shown). (E) S2 cells were incubated with or without dsRNA (RNAi; 10 μg/mL) directed at CNK 3′UTR sequences. Cells were transfected 24 h later using the indicated combinations of plasmids (+500 ng of CNK 3′UTR dsRNA) and were processed as indicated.
Given the ability of V5-KSR to associate with endogenous CNK, RAF, and MEK (Fig. 5C, lane 2), we took advantage of our panel of KSR kinase domain mutants to determine which residues are critical for those associations. As expected, the CA1 mutant only affected the association with endogenous RAF (Fig. 5C, cf. lanes 2 and 3). Interestingly, the mutants that disturbed MEK binding (i.e., KSRG688E, KSRD800A–D817A, and KSRC922Y) also reduced the association with CNK (Fig. 5C, lanes 5,8,9), thus suggesting that MEK and CNK binding on KSR depend on a common region/architecture of the putative kinase domain. Nonetheless, we were intrigued by the fact that two of these mutations (D800A–D817A and C922Y) strongly affected endogenous CNK binding and yet responded to NT-CNK (Fig. 3B). We verified whether NT-CNK could still associate with these two mutants and found that although the interactions were decreased, there was significant binding, especially in a context of MEK overexpression (Supplementary Fig. S3B; data not shown), thus explaining the responsiveness of these mutants to NT-CNK. Another interesting observation was that the two most severe mutants, KSRA696V–A703T and KSRR732H, interacted normally with CNK as well as with MEK and RAF (Fig. 5C, lanes 6,7), therefore ruling out a binding defect with these proteins. Finally, and most surprisingly, the mutations affecting MEK and CNK binding also impeded endogenous RAF binding (Fig. 5C, lanes 5,8,9), which contrasts with the KSR/RAF co-overexpression results shown in Figure 4B. We thus suspected that there could be more than one site of interaction between KSR and RAF and that the one present on the kinase domain of KSR is not direct, but depends on an endogenous protein. Given their similar binding profiles, endogenous CNK and/or MEK could be acting as mediators of the KSR/RAF association. Before testing this hypothesis, we wanted to ensure that a KSR/CNK/RAF/MEK complex genuinely existed in vivo. To accomplish this, we immunoprecipitated endogenous KSR from plain S2 cells and probed the immunoprecipitates for the presence of endogenous CNK, RAF, and MEK. As shown in Figure 5D, the KSR antibody indeed brought down all four proteins (lane 1), whereas elimination of endogenous KSR by RNAi prevented their detection (lane 5), hence demonstrating the presence of such a complex in S2 cells. We then took advantage of this simple immunoprecipitation procedure and used RNAi-treated cells to ask whether endogenous RAS activity is required for complex formation and whether any of its known constituents mediates some of the associations. Depletion of endogenous RAS did not alter the ability of KSR to associate with CNK, RAF, and MEK (Fig. 5D, lane 3), thus indicating that complex formation pre-exists to RAS activation. Also, no effect was detected when MAPK was depleted (Fig. 5D, lane 8). Beside abrogating their own presence in the complex, depletion of endogenous RAF or MEK did not alter the association of the other members of the complex (Fig. 5D, lanes 4,7). In contrast, depletion of endogenous CNK eliminated the association of RAF to KSR. Taken together, these results strongly suggest that MEK and CNK interact independently with KSR, whereas the KSR/RAF association is mediated by CNK, which would explain why mutations in KSR that impede CNK binding also affect endogenous RAF binding.
We next sought to determine which domain(s)/element(s) in CNK are mediating the KSR/RAF association. To experimentally manipulate the CNK-dependent KSR/RAF association, we depleted endogenous CNK by RNAi and rescued its presence by transfecting nontargetable CNK constructs. As shown in Figure 5E, depletion of endogenous CNK prevented coimmunoprecipitation of endogenous RAF with V5-KSR (cf. lanes 2 and 3, panel α-RAF), whereas transfection of a wild-type CNK construct restored the interaction (lane 4). As predicted from the above binding studies, a rescue construct carrying a SAM domain mutation was incapable of restoring endogenous RAF binding, as it failed to associate with V5-KSR (Fig. 5E, lane 5, α-RAF and α-CNK panels, respectively), thereby identifying the SAM domain as one of the critical points of contact to bridge KSR and RAF. On the other hand, a loss-of-function mutation within the CRIC domain did not prevent the formation of the KSR/RAF complex, therefore suggesting that the positive action of the CRIC is distinct from KSR/RAF complex formation. We previously showed that the kinase domain of RAF binds a short sequence motif in CNK named the RIM (Douziech et al. 2003). We thus verified whether this element is involved in the KSR/RAF interaction. Strikingly, a CNK rescue construct carrying a mutation in this element completely failed to restore the KSR/RAF association, even though it interacted normally with KSR (Fig. 5E, lane 7). Together, these findings provide strong evidence that the SAM domain and the RIM element are, respectively, specific bridging sites for KSR and RAF, and thereby enable the assembly of a KSR/RAF complex.
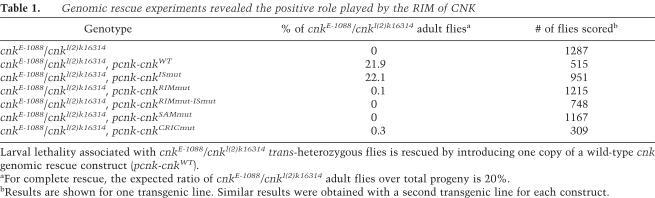
If the ability of CNK to bring together KSR and RAF is functionally relevant for the pathway, then one would predict that both the SAM domain and the RIM element are positively required for normal CNK function. While this appears to be the case for the SAM domain (Fig. 1A; Douziech et al. 2003), it is less clear for the RIM. Indeed, this element was initially defined as part of the so-called RAF inhibitory region (RIR) (Douziech et al. 2003), which, together with the IS element, is essential for CNK's inhibitory effect on RAF. For instance, unlike wild-type CNK, shorter versions of CNK that do not contain these two motifs or full-length RIR-inactivated mutants have been found to strongly collaborate with RAS when tested by overexpression in S2 cells or during Drosophila eye development (Fig. 1; Douziech et al. 2003). We recently conducted in vivo complementation assays using nonoverexpressed genomic rescue constructs to show that the IS element of CNK has a genuine negative role with respect to RAF and that the RTK-dependent binding of Src42 to a C-terminal binding site relieves its inhibitory effect (Laberge et al. 2005). Moreover, a construct in which only the IS element is disabled also fully restored the viability of cnk loss-of-function alleles, thus indicating that it does not fulfill a positive role.
To determine whether the RIM plays a positive role in a nonoverexpressed context, we tested a cnk genomic rescue construct containing a disrupted RIM element for its ability to prevent the lethality associated with cnk loss of function. We also tested mutations affecting either the SAM or the CRIC domains. In marked contrast to wild-type or IS-disrupted cnk constructs, a RIM-disrupted construct failed to complement cnk recessive lethality (Table 1). Similar findings were made with a double-mutant construct that affected both the RIM and the IS elements, thus indicating that inactivation of the RIM is disrupting a positive function that cannot be restored by deactivating the IS element. As expected, mutations targeting either the SAM or the CRIC domains were also unable to reverse cnk lethality. Together, these results strongly suggest that like the SAM and CRIC domains, the RIM is fulfilling an essential positive role in physiological conditions, which would be consistent with its ability to juxtapose RAF to CNK-bound KSR. Such a positive role for the RIM is not incompatible with its negative property. Indeed, the RIM can be viewed as a mere binding site for RAF which, on the one hand, allows the formation of a KSR/RAF complex and, on the other hand, enables the IS element to block RAF activity in the absence of proper RTK-induced signals. This scenario would also explain why several cnk loss-of-function alleles recovered in the KSR-dependent genetic screen have molecular lesions producing mutant proteins similar to NT-CNK and yet behaved as loss of functions (Therrien et al. 1998; Laberge et al. 2005). It is interesting, however, that when overexpressed, NT-CNK (or related CNK variants that lacked a functional RIM) is collaborating with RAS to mediate KSR activity even though it does not bridge KSR and RAF. Again, a number of models could explain this observation. The one we favor is that CNK does not only connect KSR to RAF, it is also controlling the RAF-activating property of KSR in a RAS-dependent manner. Therefore, when NT-CNK is overexpressed in the presence of activated RAS, a greater proportion of endogenous KSR become activated. As these activated KSR molecules still retain one physical contact to RAF through their CA1 region, the sum of their weaker effect results in nonnegligible RAF activity.
Table 1.
Genomic rescue experiments revealed the positive role played by the RIM of CNK
Larval lethality associated with cnkE-1088/cnkl(2)k16314 trans-heterozygous flies is rescued by introducing one copy of a wild-type cnk genomic rescue construct (pcnk-cnkWT).
aFor complete rescue, the expected ratio of cnkE-1088/cnkl(2)k16314 adult flies over total progeny is 20%.
bResults are shown for one transgenic line. Similar results were obtained with a second transgenic line for each construct.
The KSR/CNK interaction depends on a novel SAM domain-containing protein
One exciting finding of this study is the involvement of the SAM domain of CNK in the CNK/KSR interaction. Although initial studies on SAM domains revealed their ability to form homo- or hetero-oligomers with other SAM domains, mounting evidence suggests that they can also interact with unrelated proteins or even with RNA (Qiao and Bowie 2005). Hence, it is conceivable that the SAM domain of CNK directly contacts the kinase domain of KSR. However, it is also possible that the interaction is indirect and/or depends on additional components.
Our preliminary evidence suggests that the SAM domain of CNK does not homodimerize (data not shown). Yet, it is surprising that the L71K mutation used in this study and which is expected to preserve the structural integrity of the SAM domain while preventing it from oligomerizing (Stapleton et al. 1999), is as deleterious as a drastic mutation (W17S–I18S) predicted to unfold the domain (Douziech et al. 2003). This observation indicated that for proper function, the SAM domain of CNK possibly heterodimerizes with another SAM domain-containing protein. If this is the case, elimination of this hypothetical protein should impede RAS-mediated MAPK activation. To investigate this possibility, we targeted by RNAi a selected set of SAM domain proteins found in Drosophila and tested the effect on RAS-induced MAPK activation in S2 cells. In addition to CNK, Drosophila contains 35 independent SAM domain proteins according to the Single Modular Architecture Tool (SMART) database (http://smart.embl-heidelberg.de). Six of these, which are transcription factors or modulators of transcription factors, contain a variant form of the SAM domain also known as the Pointed (PNT) domain (Slupsky et al. 1998) and were not considered further. Another 14 were also not selected at this point because they corresponded either to chromatin-remodeling factors or RNA-binding proteins, or simply because available information did not suggest an involvement in RAS/MAPK signaling. Beside CNK, this left us with 15 candidate proteins to test (see Materials and Methods for corresponding CG numbers). Double-stranded RNA was thus produced for each of these and then tested separately. Interestingly, one of these dsRNAs, which targeted CG30476, significantly reduced MAPK activation induced by RASV12 (Fig. 6A, cf. lanes 2 and 3), while the others had no noticeable effect (data not shown). Moreover, in contrast to endogenous MEK depletion, elimination of CG30476 did not affect MAPK activity stimulated by RAFED (Fig. 6A, lane 6), thus positioning its requirement upstream or in parallel to RAF. Therefore, these findings not only identified a potential candidate regulating the function of CNK, they also unveiled a novel mediator of RAS/MAPK signals.
Figure 6.
KSR recruitment to CNK depends on Hyphen, a novel SAM domain-containing protein. (A) Depletion of endogenous HYP by RNAi reduces MAPK activation (pMAPK levels) induced by RASV12, but not by RAF-ALED. (B) HYP is required for MEK phosphorylation stimulated by NT-CNK. R-R-K-M denotes the cotransfected RASV12, RAF, KSR, and MEKDA plasmids. S2 cells in lanes 3–5 were plated with either dsGFP or dsHYP 3′UTR RNAs (6 μg/mL) for 24 h prior to transfection (500 ng of the same dsRNAs was included in the transfection mixture). (C) HYP is required for the CNK/KSR association. RNAi treatment was conducted as in B. (D,E) HYP associates with CNK independently of KSR and requires the integrity of the SAM domain of CNK. A PYO-tagged HYP (Δ3′UTR) variant was used for these experiments as the AU5-tagged variant did not immunoprecipitate quantitatively. PYO-HYP also fully rescued MEK activation and CNK/KSR interaction following depletion of endogenous HYP (not shown). (E) Endogenous HYP mediates KSR/CNK complex formation in vivo. The experiment was conducted as in Figure 5D.
CG30476 is an evolutionarily conserved low molecular weight protein of 106 amino acids that essentially consists of a single SAM domain followed by a short stretch of conserved residues (Supplementary Fig. S4). We renamed this protein Hyphen (HYP) based on its presumed ability to functionally link critical proteins within the RAS/MAPK pathway. If HYP is influencing the function of the SAM domain of CNK, then depletion of endogenous HYP should have an impact on NT-CNK-induced MEK activation. This is indeed what we observed as RNAi-mediated HYP knock-down potently counteracted RAF-dependent MEK phosphorylation stimulated by RASV12, KSR, and NT-CNK (Fig. 6B, cf. lanes 2 and 4). The specificity of the RNAi was demonstrated by rescuing the MEK activation defect using a nontargetable HYP construct (Fig. 6B, lane 5; see also Supplementary Fig. S4). We then tested whether HYP is relevant for the KSR/CNK association. We first addressed this point by verifying whether the interaction between V5-KSR and NT-CNK549 depended on endogenous HYP. Strikingly, depletion of endogenous HYP by RNAi severely reduced the KSR/CNK interaction (Fig. 6C, cf. lanes 2 and 3). Further, a HYP rescue protein that reversed the effect of HYP depletion (Fig. 6C, lane 4) could also be detected in the V5-KSR immunoprecipitate (Fig. 6C, lane 4, α-AU5 panel). Thus, these findings strongly suggest that HYP is essential for the formation of the KSR/CNK complex and that it is part of it.
Next, we asked whether HYP interacts independently with KSR and CNK or whether its association with one of these two proteins enables the recruitment of the other. To verify this, we coexpressed HYP either with NT-CNK or with KSR and determined by immunoprecipitation whether HYP forms a complex with either protein. As shown in Figure 6D, immunoprecipitation of PYO-tagged HYP brought down NT-CNK (lane 2) as well as KSR (lane 4). However, as endogenous KSR or CNK may participate in these binary interactions, we repeated these experiments in the presence of dsRNAs targeting endogenous KSR or CNK. Remarkably, while KSR RNAi only slightly decreased the HYP/CNK association (Fig. 6D, lane 5), removal of endogenous CNK totally eliminated the HYP/KSR interaction (Fig. 6D, lane 6). Therefore, these results suggest that the CNK/HYP association is a prerequisite for KSR recruitment, although the presence of KSR may stabilize the CNK/HYP interaction.
The results presented thus far are compatible with a model whereby HYP associates with the SAM domain of CNK, which in turn creates a binding interface for KSR. To verify the relevance of the SAM domain of CNK for the CNK/HYP association, we compared the ability of PYO-HYP to interact with wild-type and SAMmut (L71K) NT549-CNK by coimmunoprecipitation. As predicted, the SAMmut variant had a much reduced capacity to interact with HYP (Fig. 6E, cf. lanes 3 and 4), thus showing that the SAM domain of CNK is involved in the CNK/HYP association. A related observation also supported this conclusion. Indeed, we found that PYO-HYP also associated with endogenous CNK (Fig. 6E, lane 2) and that this interaction could be competed by the presence of wild-type NT549-CNK, but not by the SAMmut version (Fig. 6E, lanes 3,4).
Finally, we tested whether HYP is genuinely required for the formation of the endogenous KSR/CNK complex observed in S2 cells. We immunoprecipitated endogenous KSR from S2 cells treated or not with HYP dsRNA and look for the presence of associated CNK, RAF, and MEK. Consistent with the above findings, reduction of endogenous HYP lowered the natural KSR/CNK interaction, and as a result, decreased the amount of associated RAF, but did not perturb the KSR/MEK interaction (Fig. 6F).
Taken together, these results identify HYP as a novel component of the RAS/MAPK pathway, whose function through its association with the SAM domain of CNK, enables the recruitment of KSR to CNK. Although the CNK/HYP-binding mode and its structural consequence allowing KSR binding have yet to be determined, our findings are intriguingly similar to the recruitment of Rolled/MAPK to YAN, a Drosophila SAM/PNT domain-containing transcriptional repressor that is inactivated following its phosphorylation by MAPK (Rebay and Rubin 1995). In that particular case, the recruitment of MAPK to YAN depends on a short SAM domain protein, known as MAE, that heterodimerizes with the SAM domain of YAN (Baker et al. 2001).
Discussion
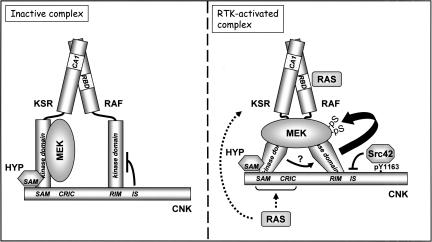
We previously demonstrated the ability of KSR to promote the formation of RAF/MEK complexes independently of RAS signals and have proposed that this scaffolding effect is a key functional aspect of KSR (Roy et al. 2002). Here, we showed that KSR does not act alone to bring RAF and MEK together, but requires at least two other proteins, namely, CNK and HYP (Fig. 7). Importantly, our data suggest that CNK/HYP-bound KSR activates RAF in a RAS-dependent manner and that this function occurs at a step regulating the activation loop of RAF. Given that Drosophila KSR does not appear to have intrinsic kinase activity, as mutagenesis of an essential residue for catalysis (i.e., K705M) still displays strong activity, it suggests that KSR does not phosphorylate the activation loop residues of RAF, and thus either another kinase is recruited to accomplish this task or RAF itself is executing it.
Figure 7.
Model summarizing the scaffolding and the RTK-dependent activating property of the KSR/HYP/CNK complex with respect to RAF and MEK. For simplicity, the model (detailed in the text) does not include the presumed effects of 14–3–3 protein binding on RAF and KSR conformation and localization. Dotted arrows represent possible entry points for RAS activity that is integrated by the complex independently of the RAF RBD. The arrow with a question mark between KSR and RAF kinase domains illustrates the putative RAS-dependent RAF-activating effect of KSR in association with CNK/HYP.
Interestingly, CNK and HYP do not exhibit any positive activity unless KSR is present (Fig. 3; data not shown), while KSR overexpression can induce RAF-mediated MEK phosphorylation independently of RAS and CNK (Fig. 1; Roy et al. 2002; data not shown). It thus appears that KSR mediates the effect of RAS and CNK and that it can even bypass their requirement when expressed at sufficiently high levels as if it carries an intrinsic RAF-activating property that is unveiled when overexpressed along with wild-type RAF and MEK. Various models can be envisioned to explain the RAF-activating property of KSR. The one we favor is based on the position in which KSR operates during this event and on the strong architectural and amino acid sequence homology between KSR and RAF members. A recent crystallographic study of the inactive B-RAF catalytic domain has uncovered an inhibitory interaction that takes place between the P loop and the DFG motif/activation loop (Wan et al. 2004). Structural analysis of this interaction strongly suggests that phosphorylation of the activation loop interferes with the interaction and thereby helps in switching and/or locking the DFG motif/activation loop into the active conformation. The importance of disrupting the inhibitory configuration is also strikingly suggested by the finding that up to 90% of a large number of B-RAF oncogenic mutations found in human melanomas affect a valine residue (V600) that stabilizes the inactive conformation (Davies et al. 2002; Wan et al. 2004). In fact, most of the other oncogenic B-RAF mutations recovered in melanoma cells could also be understood by their ability to disturb the inhibitory configuration. Surprisingly, some affected residues participating in catalysis, and hence, decreased intrinsic kinase activity. As these mutations were capable of elevating endogenous ERK activity by their ability to stimulate endogenous wild-type RAF proteins, it has been proposed that a catalytically impaired but conformationally derepressed RAF kinase domain transduces its effect to inactive RAF proteins, possibly via an allosteric process, and as a result promotes their catalytic activation. KSR may act through a similar mechanism. Its overexpression along with MEK and RAF may allow it to adopt a conformation that in turn disrupts the inhibited configuration of the RAF catalytic domain. This event would then position the activation loop of RAF in a suitable configuration for phosphorylation, which ultimately stabilizes the catalytically activated state. In physiological conditions, KSR may also operate via this process, but presumably in a regulated manner (Fig. 7). For example, the conformation of the kinase domain of KSR might be controlled allosterically by the CNK/HYP complex in a RAS-dependent manner, which in turn induces an activating conformational change in the kinase domain of RAF. This scenario might explain why the RAF-ALAA mutant still responded to NT-CNK (Fig. 2A), as even if its activation state could not be stabilized by phosphorylation, its conformation might still be controllable allosterically, thereby resulting in detectable catalytic activity. Although not mutually exclusive, KSR may also work by bringing other RAF-activating proteins or sequestering inhibitory proteins from RAF. The identification of two mutations (KSRA696V–A703T and KSRR732H) that completely eliminate the RAF-activating property of KSR, but that do not affect its RAF/MEK scaffolding function, should prove valuable to ascertain this novel function biochemically and structurally.
Collectively, our characterization of CNK’s functional elements/domains is providing novel insights as to how scaffold proteins can dynamically influence signaling within a given pathway. Indeed, it appears that prior to signal activation, the CNK/HYP pair juxtaposes a KSR/MEK complex to RAF and, owing to the IS of CNK, maintains this higher-order complex in an inactive state by selectively repressing RAF catalytic function (Douziech et al. 2003; Fig. 7, left). Then, upon signal activation, CNK integrates two RTK-elicited signals that together leads to RAF activation (Fig. 7, right). First, RTK-induced phosphorylation of the Y1163 residue of CNK allows the binding of Src42, which in turn releases the inhibitory effect of the IS motif (Laberge et al. 2005). Second, RTK-induced RAS activity not only acts through the RBD of RAF, but also via the SAM–CRIC region of CNK (Douziech et al. 2003; this study), thereby enabling KSR to activate RAF. How the N-terminal domains of CNK integrate RAS activity is currently unknown. One possibility is that the SAM domain, in association with HYP, merely acts as a binding interface for KSR, while the CRIC region is the one that perceives RAS activity and communicates it to KSR. It is also conceivable that RAS sends signals to KSR independently of CNK, and as a result, allows KSR to respond to NT-CNK.
In summary, this study has identified CNK as a molecular platform coordinating the assembly and activity of a RAF-activating complex and has unexpectedly found that KSR, which is recruited to CNK-bound RAF by the novel protein HYP, is a central component of the RAF activation process. Regardless of the exact mechanism used by KSR to drive RAF activation, it is likely that a similar functional interaction between the kinase domains of KSR and RAF has been conserved during evolution and, in fact, might be a basic feature governing RAF activation across metazoans.
Materials and methods
Plasmids
Plasmids used in transfection experiments were all derived from the copper-inducible pMet vector (Therrien et al. 1998). Point mutations and deletions were generated by standard procedures and modified inserts were completely sequenced. Specific residues that were changed are indicated in the text or figure legends. CNK, KSR, RAF, and HYP cDNA inserts ended at their respective Stop codon followed by the adh 3′UTR sequences so that they could be used in RNAi-based rescue experiments as previously described (Douziech et al. 2003).
NT549-CNK construct was made by introducing an L550Stop mutation in pMet-full length CNK. Other CNK constructs used in transfection experiments have been previously described (Douziech et al. 2003). For in vivo rescue experiments, assembly of a pBS-based (Stratagene) construct containing cnk genomic sequences was described in Laberge et al. (2005). Point mutations were separately introduced in this template (SAMmut [L71K]; CRICmut [del A162-H163-R164]; RIMmut [T1068A-L1069A-K1070A]; ISmut [G1092A-V1093A-E1094A]; or double RIMmut–ISmut) and mutant inserts were then transferred to a pCaSpeR P-element vector (Pirrotta 1988).
KSR constructs contained a V5 epitope (GKPIPNPLLGLDST) at their N terminus, while RAF constructs contain two PYO (Glu) epitopes (MEYMPME). Hyphen full-length cDNA was amplified by PCR from the LD cDNA library (Berkeley Drosophila Genome Project). Two AU5 epitopes (TDFYLK) or two PYO epitopes were introduced at its N terminus. pMet-haRASV12 and pMet-mycMEKDA were described in Roy et al. (2002), while pMet-haMAPK was presented in Douziech et al. (2003).
Cell transfection, RNAi, cell lysates, and immunoprecipitations
S2 cells were maintained in FBS-containing Schneider (Invitrogen) or serum-free insect cell medium (Sigma). For transfection experiments, 7 × 106 cells were plated per 60-mm dish and transfected the next day with various plasmid combinations with or without specific dsRNAs (1 μg total of nucleic acids) using the Effectene transfection reagent (Qiagen). Protein expression was induced by adding CuSO4 (0.7 mM) to the medium at 18–24 h post-transfection. dsRNA production for RNAi experiments were produced and used in transfection experiments essentially as described in Roy et al. (2002). The 15 SAM domain-containing proteins selected for RNAi were CG4719, CG9098, CG9126, CG10743, CG11199, CG11206, CG12424, CG13320, CG13859, CG13996, CG15625, CG16757, CG30476, CG31163, and CG31187.
To prepare protein lysates, cells were harvested 36 h post-induction in ice-cold PBS and then incubated for 15 min on ice in 500 μL of lysis buffer (20 mM Tris at pH 8.0, 137 mM NaCl, 10% glycerol, 1% Igepal CA-630, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 0.15U/mL aprotinin, 20 μM leupeptin). Cell debris were removed by centrifugation at 12,000g for 15 min (4°C).
For immunoprecipitations, antibodies and protein A/G Plus-agarose beads (Santa Cruz Biotechnology, Inc.) were added together to cell lysates (∼2 mg total proteins) and gently rocked for 4 h at 4°C. Immunoprecipitated proteins were then washed three times with cold lysis buffer before analysis.
Western blot analysis and antibodies
For immunoblot analysis, cell lysates and immunoprecipitated proteins were resolved on 8%–10% SDS-PAGE and transferred to nitrocellulose membranes. Sources for α-CNK, α-MAPK, α-pMAPK, α-MEK, α-pMEK, α-RAF, α-HA, α-MYC, and α-PYO antibodies were mentioned in Douziech et al. (2003). Monoclonal α-V5 and polyclonal α-AU5 antibodies were obtained from Invitrogen and Bethyl Laboratories, respectively, whereas the α-KSR monoclonal antibody was described in Therrien et al. (1996).
Genetics
Fly husbandry, crosses, and P-element-mediated germline transformations were conducted according to standard procedures.
Acknowledgments
We are grateful to Sylvain Meloche, Frank Sicheri, and David Wassarman, as well as members of the Therrien laboratory for helpful discussion and critical reading of the manuscript. M.T. is recipient of a Canadian Research Chair. This work was supported in part by grants from the Canadian Institutes of Health Research and the National Cancer Institute of Canada with funds from the Canadian Cancer Society.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1390406
References
- Anselmo A.N., Bumeister R., Thomas J.M., White M.A. Critical contribution of linker proteins to Raf kinase activation. J. Biol. Chem. 2002;277:5940–5943. doi: 10.1074/jbc.M110498200. [DOI] [PubMed] [Google Scholar]
- Baker D.A., Mille-Baker B., Wainwright S.M., Ish-Horowicz D., Dibb N.J. Mae mediates MAP kinase phosphorylation of Ets transcription factors in Drosophila. Nature. 2001;411:330–334. doi: 10.1038/35077122. [DOI] [PubMed] [Google Scholar]
- Bumeister R., Rosse C., Anselmo A., Camonis J., White M.A. CNK2 couples NGF signal propagation to multiple regulatory cascades driving cell differentiation. Curr. Biol. 2004;14:439–445. doi: 10.1016/j.cub.2004.02.037. [DOI] [PubMed] [Google Scholar]
- Chong H., Vikis H.G., Guan K.L. Mechanisms of regulating the Raf kinase family. Cell. Signal. 2003;15:463–469. doi: 10.1016/s0898-6568(02)00139-0. [DOI] [PubMed] [Google Scholar]
- Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Dhillon A.S., Kolch W. Untying the regulation of the Raf-1 kinase. Arch. Biochem. Biophys. 2002;404:3–9. doi: 10.1016/s0003-9861(02)00244-8. [DOI] [PubMed] [Google Scholar]
- Dhillon A.S., Meikle S., Yazici Z., Eulitz M., Kolch W. Regulation of Raf-1 activation and signaling by dephosphorylation. EMBO J. 2002;21:64–71. doi: 10.1093/emboj/21.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douziech M., Roy F., Laberge G., Lefrancois M., Armengod A.V., Therrien M. Bimodal regulation of RAF by CNK in Drosophila. EMBO J. 2003;22:5068–5078. doi: 10.1093/emboj/cdg506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaumot M., Hancock J.F. Protein phosphatases 1 and 2A promote Raf-1 activation by regulating 14–3–3 interactions. Oncogene. 2001;20:3949–3958. doi: 10.1038/sj.onc.1204526. [DOI] [PubMed] [Google Scholar]
- Kolch W. Meaningful relationships: The regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 2000;351:289–305. [PMC free article] [PubMed] [Google Scholar]
- Kornfeld K., Hom D.B., Horvitz H.R. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- Laberge G., Douziech M., Therrien M. Src42 binding activity regulates Drosophila RAF by a novel CNK-dependent derepression mechanism. EMBO J. 2005;24:487–498. doi: 10.1038/sj.emboj.7600558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanigan T.M., Liu A., Huang Y.Z., Mei L., Margolis B., Guan K.L. Human homologue of Drosophila CNK interacts with Ras effector proteins Raf and Rlf. FASEB J. 2003;17:2048–2060. doi: 10.1096/fj.02-1096com. [DOI] [PubMed] [Google Scholar]
- Light Y., Paterson H., Marais R. 14–3–3 antagonizes Ras-mediated Raf-1 recruitment to the plasma membrane to maintain signaling fidelity. Mol. Cell. Biol. 2002;22:4984–4996. doi: 10.1128/MCB.22.14.4984-4996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M., Barbacid M. RAS oncogenes: The first 30 years. Nat. Rev. Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- Morrison D.K., Cutler R.E. The complexity of Raf-1 regulation. Curr. Opin. Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- Morrison D.K., Davis R.J. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- Pearson G., Robinson F., Beers Gibson T., Xu B.E., Karandikar M., Berman K., Cobb M.H. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- Qiao F., Bowie J.U. The many faces of SAM. Sci. STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-element transformation in Drosophila. In: Rodriguez R.L., Denhardt D.T., editors. Vectors: A survey of molecular cloning vectors and their uses. Butterworths; Boston, MA: 1988. pp. 437–456. [Google Scholar]
- Rebay I., Rubin G.M. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–866. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Rocheleau C.E., Ronnlund A., Tuck S., Sundaram M.V. Caenorhabditis elegans CNK-1 promotes Raf activation but is not essential for Ras/Raf signaling. Proc. Natl. Acad. Sci. 2005;102:11757–11762. doi: 10.1073/pnas.0500937102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy F., Laberge G., Douziech M., Ferland-McCollough D., Therrien M. KSR is a scaffold required for activation of the ERK/MAPK module. Genes & Dev. 2002;16:427–438. doi: 10.1101/gad.962902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupsky C.M., Gentile L.N., Donaldson L.W., Mackereth C.D., Seidel J.J., Graves B.J., McIntosh L.P. Structure of the Ets-1 pointed domain and mitogen-activated protein kinase phosphorylation site. Proc. Natl. Acad. Sci. 1998;95:12129–12134. doi: 10.1073/pnas.95.21.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton D., Balan I., Pawson T., Sicheri F. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol. 1999;6:44–49. doi: 10.1038/4917. [DOI] [PubMed] [Google Scholar]
- Sundaram M., Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- Therrien M., Chang H.C., Solomon N.M., Karim F.D., Wassarman D.A., Rubin G.M. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- Therrien M., Michaud N.R., Rubin G.M., Morrison D.K. KSR modulates signal propagation within the MAPK cascade. Genes & Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- Therrien M., Wong A.M., Rubin G.M. CNK, a RAF-binding multidomain protein required for RAS signaling. Cell. 1998;95:343–353. doi: 10.1016/s0092-8674(00)81766-3. [DOI] [PubMed] [Google Scholar]
- Wan P.T., Garnett M.J., Roe S.M., Lee S., Niculescu-Duvaz D., Good V.M., Jones C.M., Marshall C.J., Springer C.J., Barford D., et al. Mechanism of activation of the RAF–ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Wellbrock C., Karasarides M., Marais R. The RAF proteins take centre stage. Nat. Rev. Mol. Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- Zhang B.H., Guan K.L. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. EMBO J. 2000;19:5429–5439. doi: 10.1093/emboj/19.20.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziogas A., Moelling K., Radziwill G. CNK1 is a scaffold protein that regulates Src-mediated Raf-1 activation. J. Biol. Chem. 2005;280:24205–24211. doi: 10.1074/jbc.M413327200. [DOI] [PubMed] [Google Scholar]