Abstract
X-chromosome dosage compensation in Drosophila requires the male-specific lethal (MSL) complex, which up-regulates gene expression from the single male X chromosome. Here, we define X-chromosome-specific MSL binding at high resolution in two male cell lines and in late-stage embryos. We find that the MSL complex is highly enriched over most expressed genes, with binding biased toward the 3′ end of transcription units. The binding patterns are largely similar in the distinct cell types, with ∼600 genes clearly bound in all three cases. Genes identified as clearly bound in one cell type and not in another indicate that attraction of MSL complex correlates with expression state. Thus, sequence alone is not sufficient to explain MSL targeting. We propose that the MSL complex recognizes most X-linked genes, but only in the context of chromatin factors or modifications indicative of active transcription. Distinguishing expressed genes from the bulk of the genome is likely to be an important function common to many chromatin organizing and modifying activities.
Keywords: ChIP–chip, MSL complex, gene expression, chromatin modification, histone modification, dosage compensation
Organization of the genome into chromatin is crucial for proper cellular function and organismal development. Histone modifications mark chromatin domains for expression or silencing, but how the enzymes that catalyze these changes are initially targeted is poorly understood. Dosage compensation in Drosophila serves as an important model for the targeting and regulation of active chromatin domains (Gilfillan et al. 2004; Lucchesi et al. 2005).
Dosage compensation makes X-linked gene expression equivalent in males (XY) and females (XX). In Drosophila, this occurs primarily by increasing transcription of X-linked genes in males (Hamada et al. 2005; Straub et al. 2005). Five MSL (male-specific lethal) proteins and two noncoding roX (RNA on X) RNAs are known to associate specifically with the male X chromosome in a finely banded pattern along its length. The MSL complex is required for site-specific acetylation of histone H4 on Lys 16 (H4K16ac) on the X (Turner et al. 1992; Bone et al. 1994), which is likely to play a key role in up-regulation of transcription (Hilfiker et al. 1997). A complex with similar protein composition and H4K16 acetyltransferase activity is found in humans and plays an important role in the DNA damage response and cell cycle progression (Gupta et al. 2005; Smith et al. 2005; Taipale et al. 2005).
Understanding how chromatin modifying factors are specifically targeted to their sites of action is a key step toward understanding genome organization and function. Signature DNA sequences that could be responsible for the X-chromosome specificity of MSL complex binding have not been identified. There is evidence for spreading from roX genes in cis (Kelley et al. 1999; Park et al. 2002; Oh et al. 2003) and recognition of X segments in trans (Demakova et al. 2003; Fagegaltier and Baker 2004; Oh et al. 2004), but the rules for target recognition are not known. Insertion of a strong enhancer into some ectopic positions on the X chromosome can create new cytological sites of MSL binding, suggesting that transcription can activate MSL recognition (Sass et al. 2003). However, the actual identities of direct MSL targets and their key features are largely unknown. As a first step to defining the targeting mechanism, we have determined the genome-wide MSL-binding pattern in Drosophila SL2 cells (of embryonic origin), in Clone 8 cells (from larval wing imaginal disc), and in late embryos. We find that the MSL complex recognizes and binds to expressed genes and largely ignores intergenic regions. Our results support a model in which the MSL complex selectively identifies target genes in the context of active transcription.
Results
Establishment of a functional TAP-tagged MSL complex in transgenic flies and male tissue culture lines
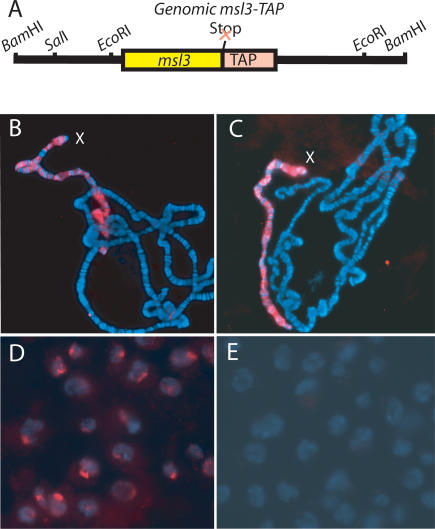
MSL complex was known to bind the X chromosome in a banded pattern along its length, but whether it associates with regulatory regions, genes, or elsewhere was not known. To precisely map MSL binding along the X chromosome at high resolution, we created a TAP-tagged MSL complex that could be isolated with high affinity (Puig et al. 2001). We TAP-tagged the MSL3 chromodomain protein at its C terminus, expressed from its native promoter without any loss of protein-coding information (Fig. 1A). To test our construct for MSL3 function, we created transgenic flies that contain a single copy of MSL3-TAP and found that this construct fully rescued msl3 mutant males (see Materials and Methods). Furthermore, the MSL3-TAP protein was detected along the length of the polytene male X chromosome in the normal MSL pattern (Fig. 1B). MSL3-TAP immunostaining of the X was not diminished in a wild-type background by the presence of endogenous MSL3 protein, suggesting that the epitope-tagged protein competes well with the native protein (Fig. 1C).
Figure 1.
MSL3-TAP protein targets the male X chromosome. (A) Genomic MSL3-TAP fusion construct. (B–E) Immunofluorescence in which the TAP epitope is recognized by the PAP antibody (red) and DNA is stained with DAPI (blue). (B) Immunostaining of male polytene chromosomes in the absence of endogenous MSL3 protein. (C) Immnostaining of male polytene chromosomes when both MSL3-TAP and wild-type MSL3 are present. (D) Immunostaining of MSL3-TAP SL2 cells, in which the fusion protein can be seen localized to a subnuclear region. (E) The MSL3-TAP signal is absent in untagged wild-type SL2 cells.
Given the full function of the MSL3-TAP fusion protein and its ability to compete with wild-type MSL3, we proceeded to create stably transfected cell lines to produce homogeneous tagged cell populations for analysis of MSL binding. We performed our initial studies in SL2 male tissue culture cells, originally derived from embryos. SL2 cells are male in character as demonstrated by subnuclear localization of the MSL complex (Copps et al. 1998) and specific decrease in X-linked transcription after RNA interference (RNAi) depletion of MSL2 (Hamada et al. 2005). We transfected SL2 cells and isolated several stable lines in which MSL3-TAP was expressed at levels comparable to native MSL3 (i.e., not overexpressed). We selected one line to utilize for subsequent experiments and demonstrated that MSL3-TAP copurified with other MSL proteins and displayed subnuclear localization consistent with X-chromosome association (Fig. 1D; data not shown).
Chromatin immunoprecipitation (ChIP) analysis of MSL complex on high-resolution genomic tiling arrays
To determine the precise locations of the MSL complex along the X chromosome, we designed genomic tiling arrays (NimbleGen Systems, Inc.) composed of 388,000× 50mers, spaced with 50-base-pair (bp) gaps along the entire nonrepetitive X chromosome (∼22 Mb), and most of chromosome 2L (∼19.6 Mb). We performed ChIPs with modifications designed to optimize our specificity. The inclusion of the TAP tag allowed us to use the same antibody for ChIP of both untagged control and tagged experimental samples (see Materials and Methods). This differs from ChIP procedures that rely on specific antibodies to endogenous proteins, in which the same extract is utilized for both control and experiment, but the antibody differs in each case (specific vs. nonspecific). In an additional variation from typical ChIP protocols, rather than removing all bound proteins and DNA from affinity beads by chemical methods, we cleaved off the tagged protein and cross-linked DNA with TEV protease that has a specific site engineered into the TAP tag.
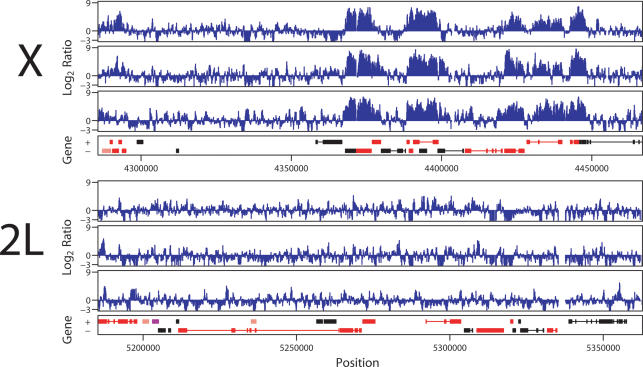
After ligation-mediated PCR amplification and random priming, DNA samples recovered from input and control IP, and input and experimental IP were hybridized to the genomic tiling arrays. Because cross-linked chromatin is sonicated into fragments of ∼250–800 bp in length, positive binding should always result in clusters of positive signals from sequential 50mers. We compared binding clusters identified on the X chromosome versus 2L and found strong enrichment for the X chromosome (Fig. 2). Biological replicates identified a strongly reproducible set of binding clusters. All subsequent analyses identifying genes as positive or negative for binding were dependent on a precise definition for a binding site, which can vary depending on the efficacy of specific experiments. Therefore, for each individual experiment we determined a threshold log2 signal and number of sequentially positive probes based on statistical analysis that minimizes the expected incidence of false positives (see Materials and Methods). It is important to note that even with perfect data, a statistical analysis cannot be expected to yield perfect correlations in large part due to the biology of the system. For example, overlapping genes or small genes embedded in gene clusters are relatively common in Drosophila, but clearly add ambiguity to precise correlations between specific binding clusters and the identification of specific targets. In multiple analyses of SL2 cells, 739 binding clusters were identified over X, while 11 were seen on chromosome 2L, confirming the chromosomal specificity of MSL binding. Representative data are shown in Figure 2, and the entire data set is available at http://www.chip.org/~ppark/Supplements/GD06.html.
Figure 2.
High-resolution ChIP–chip analysis of MSL3-TAP binding to chromosomes X and 2L. (Top) X chromosome. (Bottom) Chromosome 2L. A 180-Kb section is shown for each, representing the hybridization to ∼1800 50mers, or ∼0.5% of the total data set. For each chromosome, results from one experiment, a corresponding dye-swap, and a second independent experiment are aligned above the gene annotation for each segment. Genes expressed from left to right are shown above genes expressed from right to left. Rectangles represent exons, connected by lines that represent introns. Red genes are expressed while black genes are not expressed as determined by Affymetrix analysis of SL2 cells. MSL binding is enriched on the X chromosome over expressed genes.
Comparison with expression microarrays reveals that MSL complex prefers expressed genes, with stronger binding toward the 3′ end
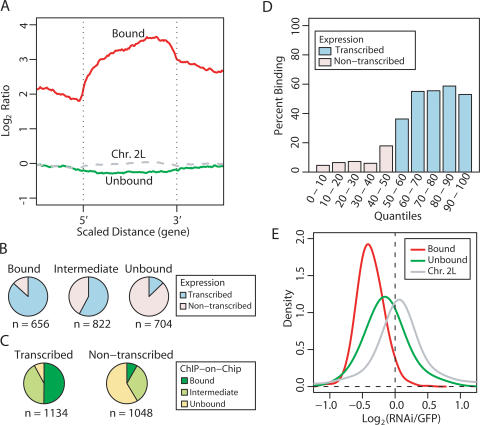
In parallel with our ChIPs, we purified RNA from MSL3-TAP tagged SL2 cells and performed expression analyses using Affymetrix Drosophila 2.0 microarrays. When the annotated genome was aligned with our expression and binding data, we saw a clear correlation of binding with expressed genes (e.g., red genes in Fig. 2) and not with nonexpressed genes (black in Fig. 2) or intergenic regions. When quantified, ∼90% of binding clusters were within expressed genes, while only 7% were within nonexpressed genes and <3% were in intergenic regions. Furthermore, binding was clearly not centered at 5′ regulatory regions, but often appeared to cover a large portion of each transcription unit. To analyze this objectively, we scaled all bound genes to the same relative length, and found that binding on average was enriched over the middle and 3′ end, and away from the 5′ end (Fig. 3A). This was seen in genes of all lengths, and was most evident in long genes (Supplementary Fig. S1). This pattern is clearly distinct from typical sequence-specific transcription factors, which bind to discrete target sequences generally in 5′ regulatory regions (Ren et al. 2000). The pattern is also distinct from general transcription factors thought to increase accessibility of promoter regions to RNA polymerase (Kim et al. 2005). The association of MSL complex to bodies of genes, with stronger binding toward the 3′ end is instead reminiscent of binding patterns for factors that regulate transcription elongation or termination (Simic et al. 2003; Carrozza et al. 2005; Keogh et al. 2005; Kizer et al. 2005; Rao et al. 2005).
Figure 3.
Analysis of the link between MSL binding and transcription. (A) The average MSL-binding profile over bound and unbound genes. Genes of differing lengths were scaled to align 5′ and 3′ ends. The average bound profile covers the gene body, with stronger binding toward the 3′ end. (B) Relative fractions of bound, intermediate, and unbound genes that were transcribed (blue) or were not transcribed (pink) by Affymetrix analysis. (C) Relative fractions of transcribed genes and nontranscribed genes that were clearly bound (dark green), intermediate (light green), and clearly unbound (yellow) by MSL3-TAP ChIP–chip analysis. (D) Comparison of expression state and MSL binding. Genes were divided into quantiles by increasing Affymetrix expression values, and graphed to show the percent of genes in each quantile that were clearly bound by MSL complex in ChIP–chip analysis. Quantiles containing nonexpressed genes are labeled in pink, and quantiles with expressed genes are labeled in blue. Once genes reach a threshold expression level, the probability of robust MSL binding does not increase with increased expression. (E) Comparison of the effect of RNAi depletion for MSL2 (Hamada et al. 2005) on expression level of transcribed genes that are bound by MSL complex (red) versus transcribed genes that are clearly unbound (green). Genes on chromosome 2L show no effect of MSL2 depletion (gray).
To examine the potential link between transcription and MSL targeting, we divided all genes into three categories: bound, intermediate, and unbound. The bound category was identified as above a statistically determined threshold defined for each experiment, as discussed above. A second, lower threshold was used as an upper boundary for the unbound class, indicative of a strong lack of enrichment for MSL complex (see Materials and Methods). Lack of binding can be a difficult distinction to make in typical ChIP experiments, but here genes on autosomes such as chromosome 2L constituted a very useful control class. The remaining genes that did not fall into either of these defined groups were classified as intermediate. This distribution of genes into three categories allowed us to focus on characteristics of the two unambiguous classes (bound and unbound) while recognizing the ambiguous classification of genes that fell between the two groups.
We next asked what percentage of bound, intermediate, and unbound genes were transcribed versus nonexpressed as defined by microarray expression analysis of SL2 mRNA. (Fig. 3B). We found a clear enrichment for transcribed genes in the bound class (87%) as compared with the unbound group (13%). Conversely, we asked what percentage of transcribed genes fell into each of the three categories (Fig. 3C). Among transcribed genes, 50% were bound, 42% were intermediate, and 8% fell into the unbound category. Among nontranscribed genes, ∼8% were bound, 33% were intermediate, and 59% were unbound. These results suggest a correlation between transcription state and MSL binding.
We next asked whether the probability of being clearly bound by the MSL complex increased with absolute expression level (Fig. 3D). While MSL complex clearly prefers expressed genes, we found that beyond the threshold discriminating nonexpressed from expressed genes, there was not a direct correlation between expression level and probability of MSL binding. The probability of robust binding levels off at ∼60% regardless of expression level. Therefore, whether a gene is expressed or not is important, but the absolute expression level of a gene is not a key contributory factor for MSL binding.
Is there a direct consequence of MSL association with expressed genes on the X chromosome? We asked whether or not MSL binding correlates with up-regulation of transcription by comparing the consequence of RNAi depletion of MSL complexes in SL2 cells (Hamada et al. 2005) on expressed genes that are clearly bound by the complex versus genes that are expressed but devoid of complex (Fig. 3E). We found that bound genes significantly decreased expression after msl2 RNAi treatment (Fig. 3E, red curve), while unbound genes on average were less affected (Fig. 3E, green curve). However, the unbound genes on X showed more decrease on average than genes on chromosome 2L (Fig. 3E, gray curve) suggesting the interesting possibility that MSL action could affect at least some X-linked genes at a distance.
Previous analyses of X-chromosome specificity relied largely on comparing the whole X chromosome to autosomes for sequences that might specify regulation by dosage compensation. With our newly identified set of precise binding sites, we focused our search for sequences that were enriched in these specific segments, when compared with autosomes or X segments that were not bound by MSL complex. These searches once again failed to identify unique sequence signatures that might specify MSL recognition. Specificity could instead rely on combinations of degenerate sequences, a possibility that can only be addressed by rigorous experimentation on specific target sites.
Comparison of MSL binding in two male cell lines and in embryos reveals a large set of common MSL targets
Whether or not MSL complex is attracted to its targets by as yet unidentified DNA sequences or by some type of chromatin mark that changes according to cell type or transcription state is not known. To address this question, we created a second MSL3-TAP cell line, using Clone 8 cells (Currie et al. 1988). Clone 8 cells were originally derived from larval wing imaginal disc. Like SL2 cells, which are of embryonic origin, we found that Clone 8 cells are male in character as demonstrated by expression of MSL2 and subnuclear localization of the MSL complex (data not shown). ChIP analyses of MSL binding in Clone 8 cells showed strong parallels with SL2 cells; for example, in the relationship of transcription to binding (Supplementary Fig. S2). To expand our repertoire of cell types surveyed for MSL binding, we also grew staged collections of MSL3-TAP transgenic embryos at 25°C and collected 12- to 17-h embryos for analysis. Embryos differed from the two cell lines in several important aspects. First, the embryos had no endogenous MSL3 protein, so all MSL complexes in the cells contained MSL3-TAP. Second, the embryos were a mixture of males and females, but MSL3-TAP was only stabilized and bound to the X chromosome in the 50% of the population that was male. Most significantly, embryos are mixtures of many distinct cell types; therefore, if binding patterns differed in distinct cell populations, the embryonic pattern would be the average of those patterns rather than a homogeneous sample.
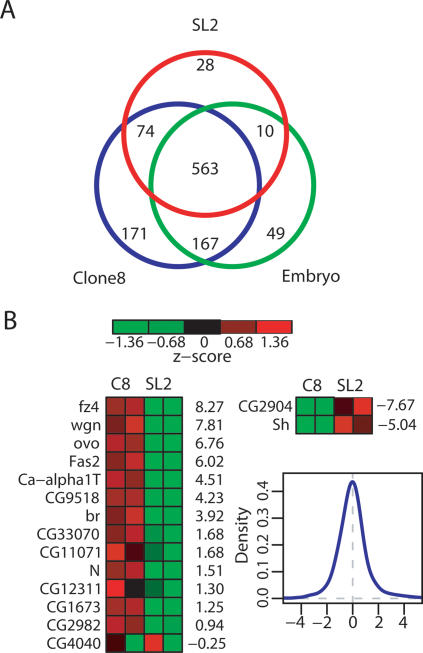
When we compared the lists of genes clearly bound by MSL complex in SL2 cells, Clone 8 cells, and embryos we found ∼600 genes that were common to all three data sets (Fig. 4A). This is a conservative estimate, as genes of the intermediate class are considered unbound in this comparison. Since MSL binding correlated with expressed genes in both SL2 cells and Clone 8 cells and the binding patterns were highly similar between experiments, our results suggested that the expression patterns in the different cell types might be largely common. Therefore, we analyzed our expression data, omitting analysis of embryonic transcription because the presence of multiple cell types within late embryos complicates the comparison of expression versus binding. We found that 1067 genes were commonly transcribed in SL2 and Clone 8 cells, while only 110 were differentially transcribed. The commonly transcribed genes were much more likely to be clearly bound in both cell types (50%) than clearly unbound (1.2%), whereas differentially transcribed genes were more likely to be unbound in both cell types (11% bound vs. 22% unbound) (Supplementary Table 1). These results suggest that commonly expressed genes account for the largely common MSL-binding pattern seen in distinct cell types.
Figure 4.
The MSL-binding pattern is largely common to late-stage embryos and two cell lines, but is not invariant. (A) Venn diagram showing the relationships between genes bound by SL2 cells (red), Clone 8 cells (blue), and embryos (green). This is a conservative number, as only the clearly bound genes are considered, while intermediate genes are not included in the totals. (B) Fourteen genes were identified as clearly bound in Clone 8 cells and clearly unbound in SL2 cells. Their normalized expression levels are shown on the left, ordered by the log fold ratios between the cell types, listed on the right of the heat map. The expression levels are higher in Clone 8 cells in all cases, except CG4040, which had low transcription resulting in an unreliable fold ratio (see Supplementary Table 2). For the two genes identified as clearly bound in SL2 and unbound in Clone 8, expression levels are much higher in SL2. The density plot shows that the distribution of log fold ratios for all genes is centered at zero and that the ratios observed here are not due to an overall shift.
The MSL-binding pattern is not invariant
Since SL2 cells, Clone 8 cells, and embryos display very similar patterns of MSL binding, it is possible that degenerate sequences have evolved on commonly expressed genes to identify them as MSL targets. To test whether sequence alone is sufficient for MSL recognition, we asked whether there were any genes that were bound in one cell type but unbound in the other cell type. Using our strict bound/unbound criteria, we identified 14 genes that were bound in Clone 8 cells and not in SL2 cells, and two genes that showed the opposite pattern (Fig. 4B). We then asked whether or not differential binding correlated with differential expression in the two cell types. By comparing the relative expression values for these genes in two microarray experiments for each cell type, we found that all of these genes are differentially expressed, with the exception of one ambiguous case in which expression levels were too low to obtain a meaningful fold change (CG4040). The absolute expression levels are listed in Supplementary Table 2, and the differences in gene expression level between the two cell lines are shown to the right of each row in Figure 4B. These differences are expressed as log2 ratios, which in most cases exceeded the amount of up-regulation (log2 ratio between 0 and 1) that would be indicative solely of MSL-mediated up-regulation. Overall, a clear correlation between differential expression of this set of genes and MSL binding is evident.
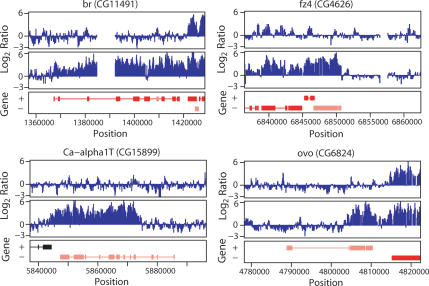
Figure 5 shows four examples of genes that are not bound by MSL complex in SL2 cells (top profiles) but are bound in Clone 8 cells (bottom profiles). In each case, the gene of interest is centered below the profiles. We validated the binding and transcription levels of these candidates by real time PCR analyses for differential MSL3-TAP binding (Fig. 6A), MSL1 binding (Fig. 6B), histone H4K16 acetylation (Fig. 6C), and transcript level (Fig. 6D). The enrichment of MSL1 and site-specific acetylation of histone H4 at Lys 16 both correlated well with differential binding of TAP-tagged MSL3 at these genes. Furthermore, real time PCR analysis of RNA levels validated the expression microarray differences seen in the two cell types. Our results strongly suggest that sequence alone is not sufficient for MSL binding because the same gene sequences can be clearly bound or clearly unbound depending on the cell type. Instead, our results suggest a model in which a majority of X-linked genes has evolved a mechanism to attract MSL complexes that is linked to gene activity or to the chromatin context of transcribed genes.
Figure 5.
Sequence alone is not sufficient to specify MSL binding. Examples of ChIP–chip tiling along four genes that were bound in Clone 8 cells but clearly unbound in SL2 cells. (Top profiles) SL2 cells. (Bottom profiles) Clone 8 cells. Gene annotation as in Figure 2. The central gene is the one of interest in each case. Note that in br and fz4 the whole genes appear covered by MSL binding, while the Ca-α1T and ovo genes show strong enrichment over the 3′ portions and no MSL complex detected over 5′ regions.
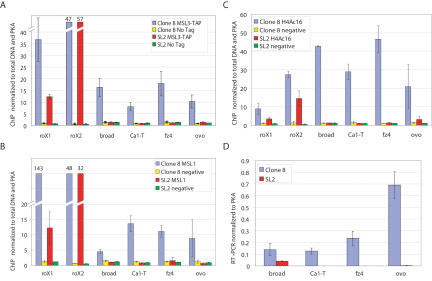
Figure 6.
Differentially bound genes are differentially acetylated at H4Ac16 and differentially transcribed. (A) ChIP of MSL3-TAP binding to br, Ca1-T, fz4, and ovo confirms that these genes are differentially bound. Real-time PCR was used to quantify ChIP DNA, thereby validating the ChIP–chip studies. We show the average and standard deviations from three independent chromatin preparations, with roX genes assayed as positive controls. (B) ChIP using a polyclonal antibody against MSL1 shows that MSL3-TAP and MSL1 binding are similar at this subset of genes. (C) ChIP using an anti-H4K16ac antibody indicates that differentially bound genes are differentially acetylated, as expected for MSL targets. (D) RT–PCR analysis was used to validate the differential transcription data acquired from Affymetrix arrays. The absence of visible bars for SL2 cells (Ca1-T and fz4 genes) is due to low mRNA levels of these transcripts.
Discussion
The MSL complex performs a specialized function in flies, but its characterization is likely to have broad implications regarding the mechanism by which chromatin modification factors search for and identify active genes. We have identified the X-chromosome-specific binding pattern of MSL complex by ChIP microarray analysis in several distinct cell types. In all cases, we see strong enrichment for the X chromosome and not chromosome 2L, and strong enrichment over most active genes. When the profiles of all bound genes are scaled to align at the 5′ and 3′ ends, we see a marked preference for the middle and 3′ ends of genes rather than the 5′ end.
The MSL-binding profile correlates well with that of its targeted modification, H4K16ac, on selected X-linked genes (Smith et al. 2001). The skew toward the 3′ end of genes is unlike the profile of transcription initiation factors and instead reminiscent of factors that function in transcription elongation or termination. Together our results suggest that the MSL complex is unlikely to function directly at the promoter like a typical transcription factor. An appealing idea is that an improvement of transcription elongation might improve ultimate mRNA production, perhaps by local recycling of RNA polymerase or other components of the general transcriptional machinery (Smith et al. 2001). Recently, the genomic distribution of histone H3.3, a histone variant associated with transcription, showed increased enrichment on X-linked genes in Drosophila SL2 cells when compared with autosomal genes (Mito et al. 2005). This enrichment favors the 5′ ends of transcription units and so might reflect a stimulation of transcription initiation or elongation due to MSL action.
Our results suggest that the MSL complex targets genes predominantly in the context of active transcription. This is consistent with the predominance of MSL complex in interband regions of polytene chromosomes (Bone et al. 1994; Demakova et al. 2003), and with experiments in which enhancer sequences responsive to the Gal4 activator protein were able to create new MSL-binding sites that required the expression of Gal4 (Sass et al. 2003). At the same time, our results are also consistent with recent cytological comparisons of the elongating form of RNA polymerase II with the MSL pattern on the polytene X chromosome, in which colocalization was observed but was clearly incomplete (Kotlikova et al. 2006). For example, many genes that we identified as differentially transcribed between SL2 cells and Clone 8 cells were unbound in both (22% vs. 1.2% of commonly transcribed genes). This type of gene would show a lack of colocalization of RNA polymerase and MSL complex when transcribed. Consistent with the largely invariant pattern of MSL binding seen on polytene chromosomes by Kotlikova et al. (2006), we found that the majority of MSL targets are commonly expressed genes. Differentially regulated genes may have been less likely to evolve the ability to attract MSL complex and perhaps may have other mechanisms to compensate for dosage differences. Our results suggest intrinsic recognition of many, but not all, X-linked genes, within the context of transcription.
Recognition of expressed genes makes excellent biological sense for the MSL complex in two ways. The most obvious is that only expressed genes need to be up-regulated. In this regard, it is notable that binding is independent of the absolute transcription level of individual genes, as dosage compensation must be able to operate on genes with a wide range of intrinsic expression levels. Another important reason to link binding to transcription may be to prevent MSL complex from ectopically influencing genes that should not be expressed. When roX genes are inserted into P transposons and mislocalized at random positions in the genome, they attract MSL complexes that can spread from the site of insertion into flanking chromatin (Kelley et al. 1999). In several instances, such insertions have occurred in regions where the mini-white reporter gene is silenced in females, but activated in males through action of the MSL complex (Kelley and Kuroda 2003). MSL action appeared to have the capacity to overcome Polycomb, HP1, and unidentified modes of silencing. Clearly MSL complex must normally be limited in its targeting to avoid potentially catastrophic male-specific activation of silent genes.
How does MSL complex locate its target genes? Studies of roX genes suggest that spreading in cis can occur from high local concentrations of MSL complex. An interesting extension of this idea is that the covering of large segments of transcription units may occur by a very local spreading mechanism related to the much longer range spreading that can be seen from roX transgenes inserted on autosomes. Both long-range and local spreading could be the consequence of attraction of the MSL complex to chromatin modifications that mark RNA polymerase II transcription units, such as histone H3 methylated at Lys 36 (Carrozza et al. 2005; Keogh et al. 2005; Rao et al. 2005). Distinguishing expressed genes from the bulk of the genome is likely to be an important function common to many chromatin organizing and modifying activities.
Materials and methods
MSL3-TAP–pCaSpeR3 construct and cell transfection
A 5.5-kb BamHI genomic fragment containing the promoter and open reading frame of the msl3 gene (Gorman et al. 1995) was subcloned from cosmid msl3-5-1 (AE003560.1 position 60298–92793) into the pBluescript II SK(−) vector. A blunted NcoI/EcoRV fragment containing C-terminal TAP tag from the pBS1479 vector (Puig et al. 2001) was subcloned into the EspI blunted msl3 construct. The resulting MSL3-TAP BamHI fragment was subcloned into pCaSpeR3 to make the final MSL3-TAP–pCaSpeR3 construct, which was cotransfected with a hygromycin resistant vector (10:1 ratio) into SL2 and Clone 8 cells using a calcium phosphate transfection kit (Invitrogen). Stable transformants were selected by adding hygromycin B (final 0.33 mg/mL) into the medium.
Fly stocks and genetic crosses
For the MSL3-TAP–pCaSpeR3 construct, several independent lines were made by P-element-mediated transformation (Spradling and Rubin 1982). For the mutant rescue, the MSL3-TAP–pCaSpeR3–14 (2L insertion at 28D) stock was used. Males of the genotype yw; P{w+ MSL3-TAP–pCaSpeR3–14}/+; msl3/TM3 Sb were crossed to homozygous msl3 mutant females of the genotype yw; +/+; msl3/msl3. The resulting rescued males were identified by the absence of Sb and the presence of w+.
Immunostaining
Polytene chromosome immunostaining was performed as described in Kelley et al. (1999). Slides were treated with PAP antibody (1:100 dilution, Sigma), and then incubated with anti-rabbit Texas Red secondary antibody (Jackson Laboratories). Immunolocalization of MSL proteins for SL2 and Clone 8 cells was performed as described in Copps et al. (1998) with minor modifications. The cells were centrifuged onto slides at 2000 rpm for 5 min in a Shandon Cytospin 3 cytocentrifuge, fixed by incubation in PBS plus 2% formaldehyde for 30 min. Fixed cells were washed twice in PBS, then dehydrated in acetone for 3 min at −20°C. After two more washes in PBS, slides were blocked by PBS plus 10% donkey serum for 30 min. For MSL3-TAP transfected cells slides were treated with PAP antibody (Sigma, 1:500), washed two times in PBST (0.1% Tween20) and then incubated for 1 h with an appropriate Alexa Fluor 555 secondary antibody (Invitrogen). The stained samples were washed two times in PBST and mounted in Prolong Gold (Invitrogen).
Embryo chromatin preparation
For a single experiment, 0.5 g of 12- to 17-h-old mixed sex embryos from MSL3-TAP and wild-type stocks were collected. Embryos were dechorionated and dounced on ice in 40 mL of NE-EM buffer (10 mM Hepes at pH 7.6, 1.5 mM MgCl2, 0.1 mM EGTA, 10 mM KCl, 0.3 M sucrose, protease inhibitors). Immediately, formaldehyde was added to a final concentration of 1% and the extract was incubated for 15 min at room temperature. Glycine was added to a final concentration of 0.25 M. Cells were collected and washed one time in ice-cold PBS, 0.5 mM EDTA and resuspended in 5 mL of RIPA buffer (50 mM Tris-HCl at pH 8.0, 2 mM EDTA, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP-40, protease inhibitors).
Chromatin preparation for cultured cells
MSL3-TAP and wild-type SL2 and Clone 8 (Cl.8+) cell lines were grown to a density of 2 ×106 to 4 × 106 cells/mL. Cells (108) were fixed with 1% formaldehyde for 10 min at room temperature. Glycine was added to a final concentration of 0.25 M and the incubation was continued for an additional 5 min. Cells were collected and washed three times in ice-cold PBS, 0.5 mM EDTA, two times in NE buffer (15 mM Hepes at pH 7.6, 5 mM MgCl2, 0.2 mM EDTA, 0.5 mM EGTA, 10 mM KCl, 350 mM sucrose, 0.1% Tween 20, protease inhibitors), and resuspended in RIPA buffer.
The cells were disrupted by sonication on ice as follows: four intervals of 15 sec (cultured cells) or four intervals of 20 sec (embryo) (2-min pauses between intervals), on power setting “60%” using a Fisher Scientific Sonic Dismembrator Model 500. Chromatin was sheared into lengths of 250–800 bp. The chromatin solution was clarified by centrifugation at 15,000g at 4°C for 10 min. Clarified sample was incubated with 30 μL Rabbit-IgG agarose beads (Sigma) for 10–14 h. The beads were washed once with RIPA buffer, twice with 50 mM Tris-HCl (pH 8.0), 2 mM EDTA, 800 mM NaCl, 1% NP-40; four times with 50 mM Tris-HCl (pH 8.0), 2 mM EDTA, 500 mM NaCl, 0.1% SDS, 1% NP-40; twice with 100 mM Tris-Cl (pH 8), 2 mM EDTA, 0.5M LiCl, 1% deoxycholate acid, 1% NP-40; and two times with TEV buffer (10 mM Tris-HCl at pH 8.0, 0.5 mM EDTA, 150 mM NaCl, 2 mM DTT, 0.1%, Tween 20).
DNA isolation, amplification, and hybridization
The immunoprecipitated material was eluted from the beads by adding 10 μL (100 U) of AcTEV protease into 450 μL of TEV buffer followed by incubation at 25°C for 1 h. To reverse the cross-links, samples were brought to 0.3M NaCl and 1% SDS and incubated at 65°C for 12 h. The samples were then extracted with phenol/chloroform/isoamyl alcohol followed by chloroform, and precipitated by ethanol in the presence of glycogen. The resulting precipitated DNA was amplified using a DNA linker as described by Strutt et al. (1997). LM-PCR (21–24 cycles) was performed using Platinum HiF Tag (Invitrogen). The resulting DNA was labeled and hybridized to arrays by NimbleGen Systems, Inc.
Tiling array data analysis
Experimental design: Tiling arrays containing 388,000 probes were designed based on FlyBase 3.2. Chromosome X and 19.6 Mb of chromosome 2L were tiled; chromosome 2L was useful as a control to verify that the features identified on the X are specific to it. For SL2 cells, two independent experiments were performed. Regions identified as bound in initial dye-swap replicates for the two SL2 experiments were very similar, and no further dye-swaps were performed. Two experiments and a dye-swap for one of them were used in the SL2 analysis. For Clone 8 cells and embryos, two experiments were performed for each cell type. Each experiment involved two arrays, one serving as a mock control in which the same protocol was followed in the absence of the TAP-tagged MSL complex. In total, seven log-ratio signals derived from 14 arrays were used: three for SL2 cells, two for Clone 8 cells, and two for embryos. The reproducibility between independent experiments within a cell type was excellent and care was taken to insure that the analysis is not biased by the uneven number of samples. In the embryo data, for instance, 776 and 766 clusters were identified in the two independent experiments and 757 were shared between the two (also see Fig. 2).
Normalization: Due to both experimental and technical variability, the signal must be standardized for proper comparison among experiments. This can be done using the stochastic part of log-ratio distributions corresponding to the regions in which there is no binding. Because the distribution of log-ratios may be asymmetric with a heavy tail on the right corresponding to binding, the standard variance estimates are biased by the amount of binding. In order to estimate the variance of only the noise part, we employ a robust measure of deviation based only on the difference between successive probes along the chromosome (σ* = s × median |di − median(d)|, where di is xi + 1 − xi and xi is the value of the ith probe after smoothing, and the scaling factor is s ≈ 1.4826 √ 2) Because this difference should not be affected by bound regions, we can estimate the standard deviation in each experiment more accurately. It is possible to use only the left side of the distribution to estimate the variance (Gibbons et al. 2005) but we found that the distribution of the noise may not be symmetrical. We have noticed that there is a strong dye bias in our data at low intensities, but the effect is the same in each pair of experimental and mock data. Because our analysis is based on the difference between the two, normalization for this effect was not necessary. We have verified that a lowess-type correction gives essentially the same result.
Identification of bound regions: There are two parameters to consider in determining whether a region is bound: the threshold for significant log-ratio value and the number of minimum probes needed to define a cluster of probes. For log-ratio value, we used >2σ* to define “bound” region and <σ* unbound, similar to standard approaches (e.g., Kim et al. 2005). For cluster size, we required a minimum of eight probes, corresponding to 750 bp. This was chosen based on the false discovery rate threshold of 10−5 for SL2 cells, calculated by permuting the location of the probes. Briefly, the steps in the analysis are the following: (1) Smooth the data using median smoothing, with a window size of 7; (2) identify clusters in each experiment separately using the 2σ* threshold, as this was more robust than using the averaged profile; (3) compute the overlapping regions between replicate experiments (minimum overlap of three probes); (4) for “bound” genes, overlapping clusters must appear in at least two separate biological replicates in SL2 cells or in both experiments for Clone 8 cell and embryo data; (5) for “unbound” genes, no cluster can be found using the lower threshold σ* or, if an isolated cluster is found, it must be very small (<10% of the gene length); and (6) the rest are classified as “intermediate.” The gene length requirement in step 5 was necessary because there was a much greater chance of observing a cluster by chance for long genes.
ChIP protocol for quantitative real-time PCR
Chromatin samples were prepared as described above. Three independent chromatin samples were used that were distinct from the original ChIP–chip samples. Antibodies against MSL1 (10 μL/IP), H4 acetylated at Lys 16, AHP417 (5 μL/IP) (Serotec) and control rabbit IgG antibodies, SC2027 (5 μL/IP) (Santa Cruz Biotechnology) were used for immunoprecipitation. Fifty microliters of protein A agarose beads (Upstate) were used for each immunoprecipitation. Washing and cross-link reversing steps were performed as previously described (Kageyama et al. 2001). Real-time PCR was performed using the ABI PRISM 7000 Sequence Detection System (Applied Biosystems). The pka gene was used as the internal reference for normalizing the variations in the quality and the amount of Total and IP DNAs. The primer sequences were as follows: for pka: forward, 5′-TTC TCGGAGCCGCACTCGCGCTTCTAC-3′, and reverse, 5′-CAATCAGCAGATTCTCCGGCT-3′; for Br-C (CG11491): forward, 5′-AATAGACTCCTCCTGCCCCTGC-3′, and reverse, 5′-TGCTGCTCCTTGTGGCGTTTCA-3′; for Ca-α1T (CG15899): forward, 5′-TCGCACCACCATCATAACAA-3′, and reverse, 5′-AACAGCACGCATAGTCCTCC-3′; for fz4 (CG4626): forward, 5′-AAATCTGGTGGGTAACGAAATG -3′, and reverse, 5′-GGAACAGCTTCAACTGGGAAC-3′; for ovo (CG6824): forward, 5′-CCATCAGCACCACAGTAACAGC-3′, and reverse, 5′- GCAGACGCGGCAGACGAACT-3′; for roX1: forward, 5′-ATGCGAGCGAGACAATGATACT-3′, and reverse, 5′-GACTTGCAGTCCGCCCTATG-3′; for roX2: forward, 5′-AGCTCGGATGGCCATCGA-3′, and reverse, 5′-CGT TACTCTTGCTTGATTTTGC-3′. PCR amplification was performed in duplicate in a 25 μL final volume using Platinum SYBR Green qPCR SuperMix UDG with ROX (Invitrogen). The PCR protocol used an initial denaturing step at 94°C for 10 min followed by 40 cycles of 94°C for 30 sec, 52°C for 30 sec, and 72°C for 1 min. Dissociation curve analysis was run at the end of 40 cycles to verify PCR product identity. Relative quantification for each probe was performed by comparative CT method based on manufacturer’s instruction (ABI Prism 7700 sequence detection system User Bulletin #2, Applied Biosystems). Standard curves for each set of primers were constructed using serial dilutions of total DNA to verify equal amplification efficiency of all primer sets.
Preparation of RNA for Affymetrix array and quantitative real-time RT–PCR
Total RNA was isolated from 2 × 106 to 4 × 106 cells using the RNeasy Mini Kit (Qiagen). cDNA preparation and hybridization to Affymetrix Drosophila Genome 2.0 arrays were performed using a protocol suggested by Affymetrix, Inc. The Affymetrix Drosophila 2.0 array was designed based on FlyBase 3.1 and contained probes for 18,500 transcripts. For each cell type, two independent experiments were performed and data were processed using the Affymetrix GeneChip Operating Software 1.1 (Affymetrix, Inc). Transcripts receiving the “Present” call by the software in both samples in a cell type were deemed “transcribed,” and those receiving the “Absent” calls in both were deemed “nontranscribed.” Where multiple probes were present for a single gene, the probe with the highest average expression was used. Real time RT–PCR was performed as described in (Bai et al. 2004) using the same sets of primers as for the ChIP protocol. To investigate the effect of MSL protein on transcription, expression data were from a previous experiment utilizing MSL2 RNAi (Hamada et al. 2005).
Acknowledgments
We are grateful to R. Richman, J. Racine, and A. Sarovschii for excellent technical assistance, and to H. Oh for the msl3 genomic clone. We thank members of the Kuroda laboratory for critical reading of the manuscript; V. Solovyev, S. Peng, and M. Gelbart for many helpful discussions; and P. Becker and colleagues for sharing unpublished results. This work was supported by the National Institutes of Health (GM45744 to M.I.K and GM67825 to P.J.P), a grant to E.L., a Leukemia and Lymphoma Society Fellow (5198-05), and the Howard Hughes Medical Institute. M.I.K. is an HHMI Investigator.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1400206
References
- Bai X., Alekseyenko A.A., Kuroda M.I. Sequence-specific targeting of MSL complex regulates transcription of the roX RNA genes. EMBO J. 2004;23:2853–2861. doi: 10.1038/sj.emboj.7600299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone J.R., Lavender J., Richman R., Palmer M.J., Turner B.M., Kuroda M.I. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes & Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Li B., Florens L., Suganuma T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Copps K., Richman R., Lyman L.M., Chang K.A., Rampersad-Ammons J., Kuroda M.I. Complex formation by the Drosophila MSL proteins: Role of the MSL2 RING finger in protein complex assembly. EMBO J. 1998;17:5409–5417. doi: 10.1093/emboj/17.18.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie D., Milner M., Evans C. The growth and differentiation in vitro of leg and wing imaginal disc cells from Drosophila melanogaster. Development. 1988;102:805–814. [Google Scholar]
- Demakova O.V., Kotlikova I.V., Gordadze P.R., Alekseyenko A.A., Kuroda M.I., Zhimulev I.F. The MSL complex levels are critical for its correct targeting to the chromosomes in Drosophila melanogaster. Chromosoma. 2003;112:103–115. doi: 10.1007/s00412-003-0249-1. [DOI] [PubMed] [Google Scholar]
- Fagegaltier D., Baker B.S. X chromosome sites autonomously recruit the dosage compensation complex in Drosophila males. PLoS Biol. 2004;2:1854–1861. doi: 10.1371/journal.pbio.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons F.D., Proft M., Struhl K., Roth F.P. Chipper: Discovering transcription-factor targets from chromatin immunoprecipitation microarrays using variance stabilization. Genome Biol. 2005;6:R96. doi: 10.1186/gb-2005-6-11-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan G.D., Dahlsveen I.K., Becker P.B. Lifting a chromosome: Dosage compensation in Drosophila melanogaster. FEBS Lett. 2004;567:8–14. doi: 10.1016/j.febslet.2004.03.110. [DOI] [PubMed] [Google Scholar]
- Gorman M., Franke A., Baker B.S. Molecular characterization of the male-specific lethal-3 gene and investigation of the regulation of dosage compensation in Drosophila. Development. 1995;121:463–475. doi: 10.1242/dev.121.2.463. [DOI] [PubMed] [Google Scholar]
- Gupta A., Sharma G.G., Young C.S., Agarwal M., Smith E.R., Paull T.T., Lucchesi J.C., Khanna K.K., Ludwig T., Pandita T.K. Involvement of human MOF in ATM function. Mol. Cell. Biol. 2005;25:5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada F.N., Park P.J., Gordadze P.R., Kuroda M.I. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes & Dev. 2005;19:2289–2294. doi: 10.1101/gad.1343705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfiker A., Hilfiker-Kleiner D., Pannuti A., Lucchesi J.C. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y., Mengus G., Gilfillan G., Kennedy H.G., Stuckenholz C., Kelley R.L., Becker P.B., Kuroda M.I. Association and spreading of the Drosophila dosage compensation complex from a discrete roX1 chromatin entry site. EMBO J. 2001;20:2236–2245. doi: 10.1093/emboj/20.9.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R.L., Kuroda M.I. The Drosophila roX1 RNA gene can overcome silent chromatin by recruiting the male-specific lethal dosage compensation complex. Genetics. 2003;164:565–574. doi: 10.1093/genetics/164.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R.L., Meller V.H., Gordadze P.R., Roman G., Davis R.L., Kuroda M.I. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Keogh M.C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., et al. Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizer K.O., Phatnani H.P., Sibata Y., Hall H., Greenleaf A.L., Strahl B.D. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 methylation with transcript elongation. Mol. Cell. Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlikova I.V., Demakova O.V., Semeshin V.F., Shloma V.V., Boldyreva L.V., Kuroda M.I., Zhimulev I.F. The Drosophila dosage compensation complex binds to polytene chromosomes independently of developmental changes in transcription. Genetics. 2006;172:963–974. doi: 10.1534/genetics.105.045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi J.C., Kelly W.G., Panning B. Chromatin remodeling in dosage compensation. Annu. Rev. Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Mito Y., Henikoff J.G., Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Oh H., Park Y., Kuroda M.I. Local spreading of MSL complexes from roX genes on the Drosophila male X chromosome. Genes & Dev. 2003;17:1334–1339. doi: 10.1101/gad.1082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Bone J.R., Kuroda M.I. Multiple classes of MSL binding sites target dosage compensation to the X chromosome of Drosophila. Curr. Biol. 2004;14:481–487. doi: 10.1016/j.cub.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Park Y., Kelley R.L., Oh H., Kuroda M.I., Meller V.H. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science. 2002;298:1620–1623. doi: 10.1126/science.1076686. [DOI] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rao B., Shibata Y., Strahl B.D., Lieb J.D. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell. Biol. 2005;25:9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Sass G.L., Pannuti A., Lucchesi J.C. Male-specific lethal complex of Drosophila targets activated regions of the X chromosome for chromatin remodeling. Proc. Natl. Acad. Sci. 2003;100:8287–8291. doi: 10.1073/pnas.1332749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R., Lindstrom D.L., Tran H.G., Roinick K.L., Costa P.J., Johnson A.D., Hartzog G.A., Arndt K.M. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.R., Allis C.D., Lucchesi J.C. Linking global histone acetylation to the transcription enhancement of X-chromosomal genes in Drosophila males. J. Biol. Chem. 2001;276:31483–31486. doi: 10.1074/jbc.C100351200. [DOI] [PubMed] [Google Scholar]
- Smith E.R., Cayrou C., Huang R., Lane W.S., Cote J., Lucchesi J.C. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol. Cell. Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A.C., Rubin G.M. Transposition of cloned P-elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Straub T., Gilfillan G.D., Maier V.K., Becker P.B. The Drosophila MSL complex activates the transcription of target genes. Genes & Dev. 2005;19:2284–2288. doi: 10.1101/gad.1343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H., Cavalli G., Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M., Rea S., Richter K., Vilar A., Lichter P., Imhof A., Akhtar A. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol. Cell. Biol. 2005;25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M., Birley A.J., Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]