Abstract
The torpedo model of transcription termination by RNA polymerase II proposes that a 5′–3′ RNA exonuclease enters at the poly(A) cleavage site, degrades the nascent RNA, and eventually displaces polymerase from the DNA. Cotranscriptional degradation of nascent RNA has not been directly demonstrated, however. Here we report that two exonucleases, Rat1 and Xrn1, both contribute to cotranscriptional degradation of nascent RNA, but this degradation is not sufficient to cause polymerase release. Unexpectedly, Rat1 functions in both 3′-end processing and termination by enhancing recruitment of 3′-end processing factors, including Pcf11 and Rna15. In addition, the cleavage factor Pcf11 reciprocally aids in recruitment of Rat1 to the elongation complex. Our results suggest a unified allosteric/torpedo model in which Rat1 is not a dedicated termination factor, but is an integrated component of the cleavage/polyadenylation apparatus.
Keywords: RNA pol II, transcription termination, Rat1, 3′-end processing, Pcf11
The transcription cycle comprises initiation, elongation, and termination of the RNA chain. During elongation, RNA polymerase II (pol II) forms a remarkably stable complex with the template that can persist for many hours. Termination disrupts that association and releases polymerase so that it can be recycled. In budding yeast, a functional poly(A) site (Zaret and Sherman 1982) as well as several essential subunits of cleavage factor I (CF1) (Minvielle-Sebastia et al. 1997; Gross and Moore 2001), including Pcf11, Rna14, and Rna15, are necessary for termination (Birse et al. 1998), although the mechanism that links 3′-end processing to polymerase release remains unclear. Pol II termination, in cases where it occurs independent of a poly(A) site, requires helicases, TTF2 and Sen1, and sequence-specific RNA-binding proteins, Nab3 and Nrd1 (Steinmetz et al. 2001; Jiang et al. 2004). It is not known whether similar proteins are also necessary for poly(A) site-dependent termination. Two alternative models for coupling termination to mRNA 3′ processing have been proposed (for reviews, see Luo and Bentley 2004; Buratowski 2005): (1) The allosteric model proposes that polymerase is altered by a conformational change or loss of an anti-termination factor that depends on recognition of the poly(A) site, leading to loss of processivity and gradual termination (Logan et al. 1987; Greenblatt et al. 1993; Calvo and Manley 2001). (2) The torpedo model proposes that polymerase is targeted by a 5′–3′ exonuclease that attacks the 5′ PO4 exposed by endonucleolytic cleavage at the poly(A) site (Connelly and Manley 1988). In this model, the exonuclease catches up to the elongation complex and causes it to terminate. Whereas an allosteric change in polymerase could be effected by the cleavage factors themselves, the torpedo model requires that the exonuclease function as a dedicated termination factor that must act after cleavage has been completed.
There is considerable experimental evidence for and against both the allosteric and torpedo models, and a unified hypothesis for poly(A) site-dependent termination remains elusive. Some 3′-end processing factors contact pol II through its C-terminal domain (CTD) during transcription, and their binding or release could influence termination as predicted by the allosteric model (Licatalosi et al. 2002; Kim et al. 2004a; Calvo and Manley 2005). The CTD is a conserved domain comprising heptad repeats (YSPTSPS) that are phosphorylated and dephosphorylated at the Ser2 (S2) and Ser5 (S5) positions during the transcription cycle (Kobor and Greenblatt 2002; Buratowski 2003). Phosphorylation of S2 residues is required for Pcf11 binding to the CTD and has been implicated in recruitment of cleavage/polyadenylation factors to the elongation complex at the 3′ end of the gene (Licatalosi et al. 2002; Ahn et al. 2004). Protein:protein interaction with the phosphorylated CTD may provide an avenue through which a termination signal could be communicated to the polymerase active site. Furthermore, the CTD is required for optimal termination in both mammalian cells and yeast (McCracken et al. 1997; McNeil et al. 1998). Also consistent with the allosteric model, the CTD interaction domain (CID) of the yeast cleavage factor Pcf11 (Steinmetz and Brow 1996) can dismantle pol II elongation complexes in vitro, although this activity is not dependent on a poly(A) site (Z. Zhang et al. 2005). In a comparison of mutant Pcf11 proteins in yeast, binding to the CTD was found to correlate with the ability to support termination. In particular, the pcf11-2 mutant that has an intact CID was reported to support termination even though poly(A) site cleavage was disrupted (Sadowski et al. 2003).
The torpedo model, unlike the allosteric model, requires that termination be preceded by cleavage at the poly(A) site to expose an uncapped 5′ PO4 end that serves as substrate for a 5′–3′ exonuclease. This model has been challenged by reports that termination can occur without transcript cleavage. In yeast, mutants of Pcf11 and Ssu72 appear to disrupt cleavage without affecting termination (Dichtl et al. 2002; He et al. 2003; Sadowski et al. 2003), and, in metazoans, termination has been observed by EM of Miller spreads without evidence that it was preceded by RNA cleavage (Bauren et al. 1998; Osheim et al. 1999, 2002). The latter results would not rule out the torpedo mechanism, however, if the interval between cleavage and termination were very short. Indeed, it has been suggested that cleavage and termination might occur almost simultaneously, perhaps in a concerted reaction (Bauren et al. 1998). On the other hand, the torpedo model is strongly supported by the recent discovery that the RNA 5′–3′ exonucleases, yeast Rat1 and human hXrn2, are required for termination (Kim et al. 2004b; West et al. 2004). It is also of potential relevance that the human cleavage factor CPSF 73 has exonuclease activity (Dominski et al. 2005). Rat1 is a nuclear 5′–3′ riboexonuclease that together with its partner Rai1 (Xue et al. 2000) hydrolyzes single-stranded RNA with 5′-monophosphate end (Stevens and Poole 1995). Rat1 is homologous to Xrn1, an abundant exonuclease that is mainly, but not exclusively, cytoplasmic. Both enzymes contribute to maturation of rRNAs and snoRNAs in the nucleus (Petfalski et al. 1998; Xue et al. 2000; Fang et al. 2005), as well as mRNA turnover (Bousquet-Antonelli et al. 2000; He and Jacobson 2001). The rat1-1 allele was originally identified as a ts mutant that accumulates poly(A)+ RNA in the nucleus (Amberg et al. 1992), a phenotype that it shares with mutants in poly(A) cleavage factors, such as Pcf11 (Hammell et al. 2002). The temperature sensitivity of rat1-1 is complemented by nuclear-targeted Xrn1 (NLS-Xrn1) (Johnson 1997), indicating that Xrn1 can substitute for the essential functions of Rat1. It is not known whether nuclear Xrn1 complements the transcription termination defect of rat1-1.
While Rat1 is clearly essential for termination, its mechanism of achieving pol II displacement from the DNA template is not obvious. Rat1 is found with pol II at the 5′ ends of genes as well as at the 3′ end, where there is an apparent increase in recruitment close to the poly(A) site (Kim et al. 2004a). One potential mode of Rat1 interaction with pol II is through Rtt103, a protein that binds S2 phosphorylated CTD via a CID (Kim et al. 2004a). Although the involvement of 5′–3′ exonucleases in termination suggests a torpedo-like mechanism, it is unclear whether degradation of the nascent RNA would be sufficient to trigger termination since elongation complexes are stable after extensive RNase digestion in vitro (Gu et al. 1996; Kireeva et al. 2000). Furthermore, it has not been established that cotranscriptional degradation of nascent RNA downstream of the poly(A) site actually occurs and, if so, whether it correlates with termination. In Caenorhabditis elegans operons, there is evidence for a 5′–3′ exonuclease that degrades RNA downstream of the poly(A) site in the intergenic region, but it is not known whether it operates cotranscriptionally (Liu et al. 2003). In yeast and human cells, loss of Rat1 or hXrn2 stabilizes transcripts downstream of the poly(A) site in total cellular RNA (Kim et al. 2004b; West et al. 2004). This stabilization could be caused by diminished cotranscriptional degradation of nascent RNA, poor in vivo cleavage at the poly(A) site, or reduced post-transcriptional degradation. It remains possible, therefore, that Rat1 contributes to termination by a mechanism other than nascent RNA degradation, perhaps involving protein:protein interactions with the CTD and 3′-end processing factors.
In this report, we tested the torpedo model’s prediction that nascent RNA is cotranscriptionally degraded downstream of the poly(A) site, using the RNA IP technique (RIP) (Niranjanakumari et al. 2002; Gilbert et al. 2004). Consistent with the model, we found that nascent RNA is, in fact, degraded, and both Rat1 and Xrn1 contribute to this process. Unexpectedly, this degradation can occur without eliciting termination, showing that it is not sufficient to cause polymerase release. Surprisingly, we found that Rat1 is critical for recruitment of 3′-processing factors and for correct 3′-end formation. Conversely, 3′-processing factors are also required for normal Rat1 recruitment. In summary, our results argue that Rat1 is not a dedicated termination factor, but instead, like conventional 3′-end processing factors, it contributes to both poly(A) site cleavage and termination.
Results
GAL1-ADH4 assay for pol II transcription termination
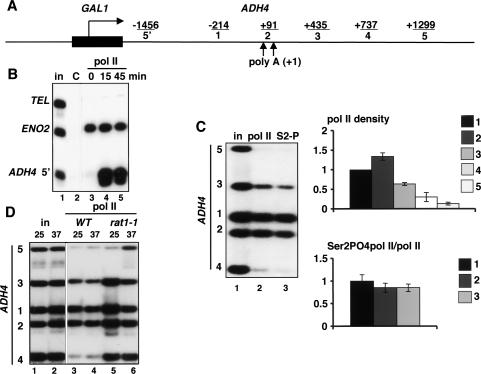
To study transcriptional termination in budding yeast, we measured pol II density within a gene, and in the 3′-flanking region, by anti-pol II chromatin immunoprecipitation (ChIP). ChIP has the advantage of being able to detect the presence of pol II on the DNA regardless of its elongation activity. By using an antibody that recognizes the phosphorylated and unphosphorylated forms of the CTD, a domain that is large and highly repetitive, we minimized the possibility of epitope masking as a result of conformational changes. Although budding yeast has the advantage of numerous informative mutants that affect termination, it has the disadvantage that most intergenic regions are small, making it difficult to resolve the termination region for a given gene from the terminator or promoter of an adjacent gene. To circumvent this problem, we adopted the chromosomal ADH4 gene as a reporter because it has an unusually long 3′-flanking region of ∼4.7 kb that is devoid of other transcription units (Fig. 1A). To drive high-level transcription, the ADH4 promoter was replaced with the GAL1 promoter. As expected, the pol II ChIP signal on GAL1-ADH4 was strongly induced by galactose (Fig. 1B lanes 3–5). Multiplex PCR analysis of an anti-pol II ChIP experiment on galactose-induced ADH4 is shown in Figure 1C (lane 2) with primer pairs located within the gene and in the 3′-flanking region. No signals were detected with these primers prior to galactose induction (data not shown). Multiplexing is a reliable measure of relative polymerase density at different positions because the signals for different segments of the gene are internally controlled. ChIP signals were normalized to input, and the average values for four experiments are plotted in Figure 1C (right graph, top). The results show a reproducible small increase in polymerase density at position 2 that overlaps the poly(A) sites, consistent with the pausing observed near poly(A) sites in other systems (Enriquez et al. 1991; Birse et al. 1997; Orozco et al. 2002). The poly(A) sites were mapped by an RNase protection assay (Supplementary Fig. 1A) to two positions: a major site 1 at approximately +112 and a minor site 2 at +177 bases relative to the termination codon. Downstream of the poly(A) sites, polymerase density steadily dropped to almost undetectable levels by position 5 located at +1299 relative to poly(A) site 1. We also measured pol II CTD phosphorylation at the S2 residue of the heptad repeats using a phospho-specific polyclonal antibody and normalized these values to total pol II (Fig. 1C, lane 3). Reproducible values were obtained at positions 1, 2, and 3 before the levels of polymerase became too low. We did not detect a major net dephosphorylation of the CTD at S2 over this region comprising sequences between 300 bases upstream and 350 bases downstream of the poly(A) cleavage site (Fig. 1C, right graph, bottom).
Figure 1.
Transcription termination at the chromosomal ADH4 gene. (A) Diagram of GAL1-ADH4. PCR products are indicated with the distances in base pairs from the center of each to the major poly(A) site (+1). Upward arrows indicate the two poly(A) sites. (B) Inducible transcription of GAL1-ADH4. Pol II ChIP of GAL1-ADH4 in wild-type cells (DBY548) induced with galactose for the indicated times. The ADH4 5′ primer pair (−1456 in A) was used. ENO2 is a positive control, and telomere VIR (TEL) is a nontranscribed negative control. IPs were with polyclonal anti-pan CTD that detects total pol II. (in) Input chromatin; (C) control IP with irrelevant antibody. (C) Transcription termination and CTD Ser2 phosphorylation. Anti-pol II ChIP with anti-pan CTD as in B, and anti-Ser2 phosphorylated CTD (S2) in the wild-type strain DBY548. Total pol II density relative to the value at position 1 in the ORF, normalized to input, is shown in the top graph. Ser2 phosphorylated pol II, normalized to total pol II, is shown in the bottom graph. (D) Termination at ADH4 is inhibited by inactivation of Rat1. Pol II ChIP of wild-type (DBY548, lanes 3,4) and rat1-1 (DBY745, lanes 5,6). Note the delayed termination with high pol II at positions 3 and 4 in rat1-1 at 25°C (lane 5) relative to wild type (lane 3).
To determine whether pol II ChIP of GAL1-ADH4 is an informative assay for termination defects, we analyzed the ts mutant rat1-1 that blocks termination at 37°C on other genes (Kim et al. 2004b). Termination downstream of ADH4 was indeed strongly inhibited by inactivation of Rat1 (Fig. 1D). Note that the distribution of pol II in the ADH4 3′-flanking region at 25°C in rat1-1 is unusual in that more polymerase extended to position 4, at +737 bases relative to poly(A) site 1, than in wild-type at 25°C (Fig. 1D, cf. lanes 3 and 5). This result suggests that although termination is still complete by position 5 in rat1-1 at 25°C, it is delayed relative to wild-type, as manifested by a shift in pol II density toward more distal positions downstream of the poly(A) site.
Nuclear Xrn1 exonuclease does not rescue the termination defect in rat1-1
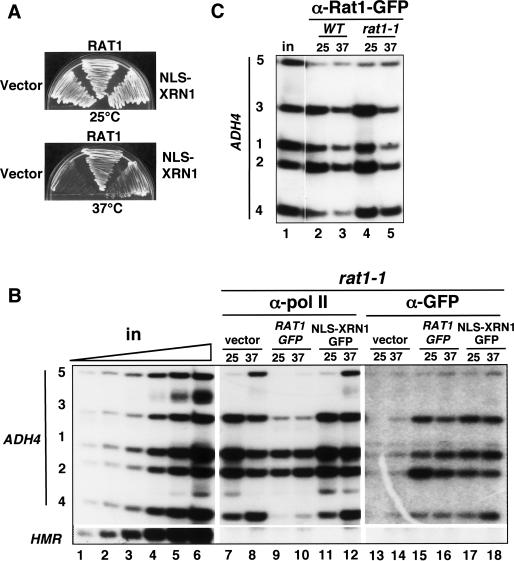
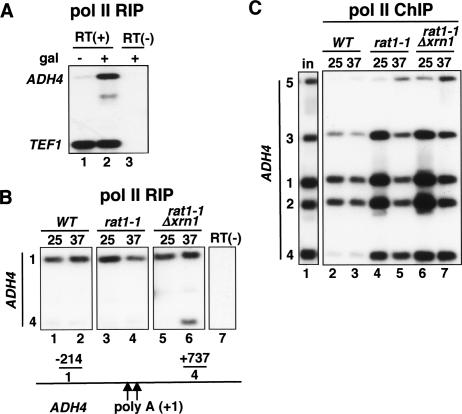
The ts growth phenotype of rat1-1 is complemented by overexpression of the related exonuclease Xrn1 with a nuclear localization signal (NLS-Xrn1) (Fig. 2A; Johnson 1997). We asked whether the termination defect caused by Rat1 inactivation is also complemented by NLS-Xrn1. The rat1-1 strain was transformed with expression plasmids for GFP-tagged wild-type Rat1 or NLS-Xrn1, or empty vector as a control. As expected, wild-type Rat1 completely restored termination at 37°C, whereas the empty vector did not (Fig. 2B, lanes 8,10). If an essential function of Rat1 is to cause transcription termination, then NLS-Xrn1 is expected to restore termination in rat1-1 at 37°C. Contrary to this expectation, NLS-Xrn1 did not restore termination in rat1-1 at 37°C (Fig. 2B, lane 12), although it permitted growth at this temperature. The essential function(s) of Rat1 that is complemented by NLS-Xrn1 is therefore in a process other than pol II termination, such as nuclear RNA turnover, transport, or processing.
Figure 2.
Nuclear Xrn1 does not rescue the termination defect in rat1-1. (A) NLS-Xrn1 complements the ts growth of rat1-1. The rat1-1 strain (DBY745) was transformed with control vector plasmid (pFL38), WTRat1-GFP (pAJ226), or NLS-Xrn1-GFP (pAJ237) and grown at 25°C or 37°C on SC-Ura plates. As shown previously, NLS-Xrn1 complements growth at 37°C (Johnson 1997). (B) NLS-Xrn1 is recruited to the gene, but it does not complement the termination defect in rat1-1. Anti-pol II (lanes 7–12) and anti-GFP (lanes 13–18) ChIP of galactose-induced GAL1-ADH4 in rat1-1 transformed with vector, RAT1-GFP, or NLS-XRN1-GFP plasmids (DBY768, DBY754, and DBY777). (Lanes 1–6) Titration of input demonstrates linearity of the PCR. HMR is a transcriptionally silent control that was run on a separate gel. Note the failure to terminate at 37°C with NLS-Xrn1 (lane 12) and the cross-linking of NLS-Xrn1 to the ADH4 gene (lanes 17,18). (C) Wild-type and ts mutant Rat1 proteins are recruited to the 3′ end at 25°C and 37°C. ChIP of GFP-tagged Rat1 and Rat1-1 on galactose-induced GAL1-ADH4 in DBY786 (WT) and DBY787 (rat1-1).
To determine whether NLS-Xrn1 can be recruited to transcribed genes, we performed ChIP on this protein using anti-GFP antibody. Surprisingly, NLS-Xrn1 was specifically recruited to the galactose-induced ADH4 transcription unit at 25°C and 37°C, and was localized similarly to pol II (Fig. 2B, cf. lanes 17,18 and the untagged control lanes 13,14). The molecular basis for the interaction of Xrn1 with pol II transcription units is not known but may involve binding to the RNA transcript. The localization of GFP-tagged NLS-Xrn1was compared with Rat1. Consistent with previous results (Kim et al. 2004b), at the permissive temperature we observed an increase in Rat1 cross-linking at the poly(A) site (Fig. 2B, lane 15, cf. positions 1 and 2) that closely paralleled the 3′-processing factors, Pcf11 and Rna15 (Figs. 3A, 4). In contrast, NLS-Xrn1 did not show a marked increase in cross-linking at the poly(A) site (Fig. 2B, cf. lanes 15 and 17, positions 1 and 2). These observations suggest that 3′-processing factors and Rat1, but not NLS-Xrn1, are specifically recruited to the pol II elongation complex at the poly(A) site.
Figure 3.
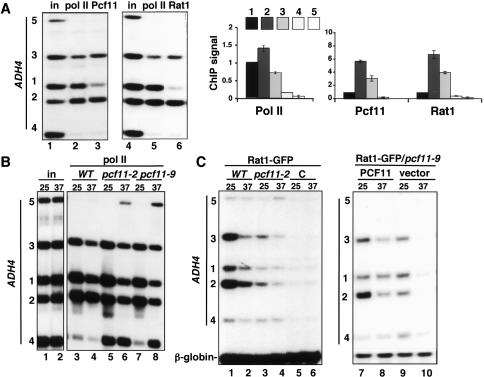
Coordinate recruitment of Rat1 and the 3′-processing factor Pcf11. (A) Highly coincident patterns of Pcf11 and Rat1 recruitment at the ADH4 3′ end. (Left) Pol II, Pcf11, and Rat1-GFP ChIP on galactose-induced GAL1-ADH4 in wild-type cells (DBY786) at 25°C. (Right) ChIP signals normalized to input were normalized relative to the value at position 1 in the ORF. Means and standard deviations from three experiments are shown in the graphs. (B) Termination defects in pcf11-2 and pcf11-9 mutants. Pol II ChIP of wild type (WT), pcf11-2, and pcf11-9 (DBY548, DBY585, DBY593) at 25°C and 37°C. Note delayed termination in pcf11-2 at 25°C (lane 5) and failure to terminate at 37°C (lane 6). (C) Rat1 recruitment to the ADH4 3′ end requires functional Pcf11. (Lanes 1–6) Rat1-GFP ChIP in wild-type (WT) and pcf11-2 strains (DBY786, DBY815) and the untagged control (C, DBY548). (Lanes 7–10) Rat1-GFP ChIP in pcf11-9 (DBY794) transformed with PCF11 plasmid (DBY796) or control vector (DBY795). β-globin is a loading control.
Figure 4.
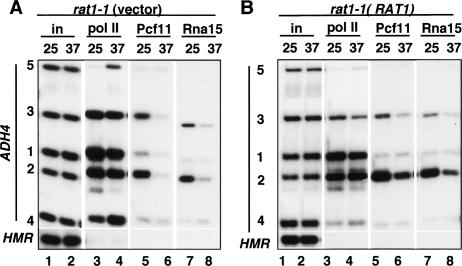
Normal recruitment of 3′-processing factors requires functional Rat1. (A) ChIP of pol II, Pcf11, and Rna15 on GAL1-ADH4 in rat1-1 transformed with control vector (DBY 768). (Lanes 6,8) Note recruitment of Pcf11 and Rna15 is reduced by inactivation of Rat1 at 37°C. (B) ChIP as in A of rat1-1 transformed with the wild-type RAT1 plasmid (DBY754). (Lanes 6,8) Note recruitment of Pcf11 and Rna15 at 37°C is partially restored relative to A. HMR-negative controls were run on separate gels.
To investigate further the interaction between Rat1 and the transcription elongation complex, we compared the cross-linking of pol II and Rat1 with the 3′ end of GAL1-ADH4. The results in Figure 2C showed that in wild-type and rat1-1 cells at 25°C, cross-linking of the Rat1 protein was elevated relative to pol II at the poly(A) site and immediately downstream (positions 2, 3) compared with the ORF (position 1) (see also graphs in Fig. 3A). In rat1-1, delayed termination at 25°C was associated with elevated cross-linking of Rat1 at position 4 (Fig. 2C, lane 4). At 37°C in the rat1-1 mutant, although Rat1 was inactivated, the protein still cross-linked to the gene downstream of the poly(A) site, in a distribution that closely paralleled pol II (Fig. 2C, lane 5). These results demonstrate association of both active and inactive Rat1 with pol II elongation complexes in the 3′-flanking region.
Pcf11 is required for Rat1 recruitment to the 3′ end
We investigated the possibility that Rat1 may function in termination by interacting with 3′-end processing factors. This idea is suggested by the finding that the Rat1-associated CTD-binding protein Rtt103 copurified with Pcf11 and other subunits of the CF1A 3′-end processing complex (Kim et al. 2004b). To test whether Rat1 and Pcf11 might also interact at a poly(A) site, we compared the cross-linking of these two proteins at the 3′ end of ADH4 (Fig. 3A). Pcf11 and Rat1 were present in low amounts within the gene (position 1), and the cross-linking of both proteins increased in parallel over the poly(A) site (positions 2 and 3) (Fig. 3A, lanes 3,6). Further downstream at positions 4 and 5, Rat1 and Pcf11 levels declined in parallel with pol II (Fig. 3A). As expected, Rna15 had an identical distribution to Pcf11 (Fig. 4A, lanes 5,7). We conclude that there is a striking coincidence in the cross-linking patterns of the exonuclease Rat1 and subunits of the 3′-end processing factor CF1A. Furthermore, we found no evidence that release of 3′-end processing factors from the elongation complex preceded release of the polymerase itself.
The interactions responsible for Rat1 recruitment to the elongation complex are not well understood. The observation that Rat1, Rna15, and Pcf11 have such similar patterns of recruitment (Figs. 3A, 4) suggests that a functional interaction between these proteins might contribute to Rat1 association with the pol II transcription complex. We therefore investigated how Pcf11 inactivation affected pol II termination and Rat1 recruitment. Inactivation of Pcf11 in two mutant strains pcf11-2 and pcf11-9 that are defective for 3′-end cleavage at 37°C (Amrani et al. 1997; Birse et al. 1998; Sadowski et al. 2003) strongly inhibited termination at ADH4 (Fig. 3B, lanes 5–8). The termination defect in pcf11-2 was unexpected as it differs from a previous report where normal termination was detected in this mutant by nuclear run-on analysis of a gene on a high-copy plasmid (Sadowski et al. 2003). This discrepancy might be explained by the fact that unlike ChIP, the run-on assay would not detect polymerases that failed to terminate if they did not resume elongation under the in vitro conditions used. In summary, our results do not support the conclusion that inactivation of Pcf11 in the pcf11-2 mutant uncouples termination from 3′-end processing. The Pcf11-2 mutant protein, which has a wild-type CID (confirmed by sequencing of the gene from our strain), was recruited normally to the ADH4 poly(A) site at 25°C, but at 37°C recruitment was very poor, as determined by anti-Pcf11 ChIP (Supplementary Fig. 2B). Remarkably, at 25°C pcf11-2 demonstrated delayed termination, like that seen in rat1-1 at 25°C (Fig. 1D, lane 4), characterized by a high pol II density at position 4 in the ADH4 3′-flanking region (Fig. 3B, lanes 3,5,7).
We next tested whether recruitment of Rat1 to the 3′ end of ADH4 is affected by interaction with Pcf11. ChIP analysis of GFP-tagged Rat1 showed that its recruitment to the ADH4 3′ end was markedly reduced by Pcf11 inactivation in both pcf11-2 and pcf11-9 as shown by the low signals relative to the β-globin loading control (Fig. 3C, lanes 3,4,9,10). Overall Rat1 recruitment was reduced in the wild-type strain at 37°C in this experiment, but it still increased at the poly(A) site (position 2) relative to the ORF (position 1) (Fig. 3C, lanes 1,2). In contrast, discretely localized Rat1 recruitment at the poly(A) site was eliminated in pcf11-9 even at the permissive temperature (25°C) (Fig. 3C, lanes 7,9, cf. positions 1 and 2). Hence, even under conditions when there is no detectable defect in 3′-end formation, the pcf11-9 mutation prevented normal recruitment of Rat1. Complementation of pcf11-9 with the wild-type PCF11 gene on a plasmid restored Rat1 recruitment to the poly(A) site at 25°C (Fig. 3C, lanes 7,9). Although complementation of pcf11-9 with wild-type PCF11 increased overall Rat1 recruitment to positions 1–3 at 37°C, (Fig. 3C, lanes 8,10), it did not restore the specific increase in cross-linking that occurs normally over the poly(A) site. In summary, these results show that (1) pcf11-2 and rat1-1 have similar delayed termination defects under permissive conditions; (2) the recruitment of Rat1 and CF1A subunits is highly coincident; and (3) association of Rat1 with the elongation complex is very sensitive to mutation of the CF1A subunit, Pcf11, even under conditions where poly(A) site cleavage is not affected.
Rat1 is required for the recruitment of 3′-end processing factors at the 3′ end
If the exonuclease functions as a dedicated termination factor after cleavage at the poly(A) site, as suggested by the torpedo model, then recruitment of 3′-end processing factors should be independent of Rat1. We tested this prediction by asking if Pcf11 and Rna15 cross-linking to the GAL1-ADH4 3′ end was affected by inactivating Rat1. In fact, loss of Rat1 function by shifting rat1-1 cells to 37°C markedly inhibited recruitment of Pcf11 and Rna15 to the poly(A) site (Fig. 4A, lanes 5–8, cf. positions 1 and 2). Both the overall levels of Pcf11 and Rna15 and the specific increase in cross-linking over the poly(A) site were diminished by Rat1 inactivation (Fig. 4A, lanes 5–8, cf. positions 1 and 2). Complementation with the wild-type RAT1 gene restored both termination and recruitment of Pcf11 and Rna15 at 37°C (Fig. 4B, lanes 4,6,8). In summary, the results in Figure 4 show that Rat1 is required for the normal recruitment of 3′-end processing factors that are essential for termination.
Rat1 affects poly(A) site choice of yeast ACT1 and ADH4 mRNA
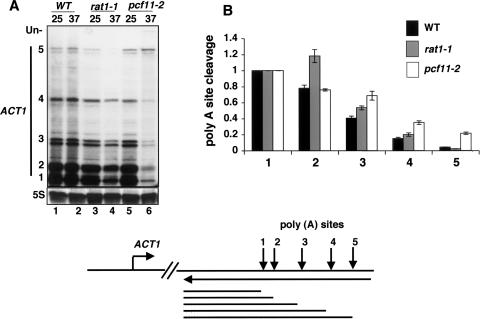
To confirm and extend the evidence for interaction between Rat1 and the CF1A 3′-end processing complex, we asked whether the rat1-1 and pcf11 ts mutants might have similar 3′-end processing defects. A characteristic phenotype of mutants that inhibit cleavage/polyadenylation in vivo, including pcf11, rna14, and rna15, is that they cause a shift in poly(A) site choice at the ACT1 gene, which has five alternative cleavage sites. At the nonpermissive temperature, transcripts that have used more distal poly(A) cleavage sites accumulate at the expense of those with more proximal poly(A) sites (Mandart and Parker 1995). This phenomenon may reflect slower kinetics of the cleavage reaction in the mutants.
ACT1 poly(A) cleavage sites were mapped by RNase protection analysis of total RNA from wild-type, rat1-1, and pcf11-2 cells grown at 25°C or 37°C. As seen previously for rna14 and rna15 (Mandart and Parker 1995), inactivation of Pcf11 shifted the poly(A) site choice to the more distal sites, 4 and 5 (Fig. 5A, lanes 5,6). The rat1-1 mutant also demonstrated a shift in ACT1 poly(A) site choice at 37°C, with reduced usage of the proximal site 1 relative to the more distal sites 2, 3, and 4 (Fig. 5A [lanes 3,4], B), though the effect was not as great as in pcf11-2. Furthermore, at ADH4, the distal poly(A) site 2 was favored relative to site 1 in rat1-1 compared with the wild type; however, unlike at ACT1, this effect was not enhanced by shifting the cells to 37°C (Supplementary Fig. 1). In conclusion, the experiment in Figure 5 shows that Rat1 affects poly(A) site choice in vivo in a similar way to mutants affecting subunits of CF1A.
Figure 5.
Rat1 is necessary for normal ACT1 poly(A) site selection. (A) ACT1 poly(A) site choice is shifted in favor of more 3′ sites when Rat1 is inactivated. Total RNA from wild-type (WT), rat1-1, or pcf11-2 strains (DBY548, DBY745, and DBY593) grown at 25°C or 37°C for 90 min was analyzed by RPA with 3′ ACT1 and 5S rRNA probes. (Un) Uncleaved RNA. The diagram shows poly(A) cleavage sites 1–5, the ACT1 anti-sense riboprobe, and RNase protection products. (B) Quantification of ACT1 mRNA cleavage at sites 1–5 in wild-type (WT), rat1-1, and pcf11-2 cells at 37°C. Cleavage relative to site 1 is shown after normalization for the 32P-U content of the protection products 1–5. Values are means and standard deviations from two experiments.
Cotranscriptional degradation of nascent RNA by Rat1 and Xrn1 is not sufficient to cause termination
Rat1-mediated cotranscriptional degradation of nascent RNA 3′ of the poly(A) cleavage site is the central premise of the torpedo model. To determine whether nascent RNA downstream of the poly(A) site is in fact degraded, we used anti-pol II RIP (Niranjanakumari et al. 2002; Gilbert et al. 2004). Cross-linked extracts were immunoprecipitated with anti-pol II antibody, DNA was removed with DNase I, and coprecipitated RNA was analyzed by RT–PCR to quantify nascent transcripts upstream and downstream of the ADH4 poly(A) sites. In parallel, the extracts were analyzed by anti-pol II ChIP to map polymerase density. As a control for the anti-pol II RIP assay, GAL1-ADH4 was analyzed before and after galactose induction (Fig. 6A). A specific RT–PCR signal corresponding to ADH4 nascent RNA upstream of the poly(A) site at position 1 was strongly induced by galactose relative to the constitutively transcribed TEF1 gene (Fig. 6A, lanes 1,2). No signal was detected in a control without RT (Fig. 6A, lane 3). The stability of nascent RNA upstream and downstream of the poly(A) cleavage sites was compared in wild-type and rat1-1 strains. In wild-type cells induced with galactose at 25°C or 37°C, a significant amount of nascent RNA was detected within the ORF at position 1, as expected. In contrast, little or no nascent RNA was detected downstream of the poly(A) site at position 4 (Fig. 6B, lanes 1,2), consistent with the fact that most polymerase molecules have terminated before this position (Fig. 6C, lanes 2,3). We next examined ADH4 nascent RNA in the rat1-1 strain at 25°C when transcription terminates, and at 37°C when it does not (Fig. 6C, lanes 4,5). As expected, in rat1-1 at both temperatures nascent RNA was associated with pol II within the gene at position 1. At 25°C, although there was a substantial amount of pol II at position 4 (Fig. 6C, lane 4) downstream of the poly(A) site, no nascent RNA was detectable at this position. We confirmed that the efficiency of RT–PCR amplification of regions 1 and 4 is approximately equal using a synthetic T7 RNA transcript spanning regions 1–4 (data not shown). Most importantly, at 37°C, when termination had been abrogated (Fig. 6C, lane 5), nascent RNA was still undetectable in association with pol II at position 4 (Fig. 6B, lane 4). Failure to terminate in rat1-1 at 37°C, therefore, was not associated with stabilization of a large fraction of nascent RNA.
Figure 6.
Degradation of nascent RNA by Rat1 and Xrn1 is not sufficient to cause pol II termination. (A) Anti-pol II RIP detects nascent transcripts. The wild-type strain (DBY548) was analyzed without (−) and with (+) galactose (gal) induction. Immunoprecipitated RNA was analyzed by RT–PCR with primers for GAL1-ADH4 (position 1) and constitutively expressed TEF1. RT(+) and RT(−) signify plus and minus RT, respectively. (B) Nascent RNA is degraded downstream of the poly(A) site. Anti-pol II RIP of GAL1-ADH4 in galactose-induced wild-type (WT), rat1-1, and rat1-1Δxrn1 strains (DBY548, DBY745, and DBY772) at 25°C or 37°C. Lane 7 is a control − RT. Note the absence of nascent RNA associated with pol II that has failed to terminate at position 4 in lanes 3–5 and stabilization of nascent RNA when both Rat1 and Xrn1 were eliminated (lane 6). This result is representative of four independent experiments. PCR product 4 has 65% as many 32P-dC residues as product 1. (C) Anti-pol II ChIP of the samples in B. (Lanes 4–6) Note pol II that failed to terminate is present at high density at position 4, although nascent RNA was not detectable in B.
It is possible that another exonuclease might be responsible for degrading nascent RNA downstream of the poly(A) site in rat1-1 at 37°C. The only other known 5′–3′ RNA exonuclease in yeast is Xrn1, and, therefore, we analyzed a rat 1-1, Δxrn1 double mutant. In the double mutant, ADH4 termination was indistinguishable from the single rat1-1 mutant at 25°C and 37°C, indicating that XRN1 deletion did not exacerbate the termination defect caused by rat1-1 (Fig. 6C, cf. lanes 4,5 and 6,7). Deletion of XRN1 alone had no detectable effect on termination at 25°C or 37°C (Supplementary Fig. 2A). In rat 1-1, Δxrn1 at 25°C, nascent RNA was present within the ORF (position 1) but not in the 3′-flanking region (position 4), consistent with degradation of most nascent RNA 3′ of the poly(A) site. In contrast, when both Rat1 and Xrn1 were eliminated by shifting the double mutant to 37°C, a substantial amount of nascent RNA could be detected at position 4, consistent with its stabilization (Fig. 6B, lane 6). This result, therefore, shows that nascent RNA downstream of the poly(A) site is, in fact, specifically degraded cotranscriptionally as predicted by the torpedo model, and that it can be stabilized if both Rat1 and Xrn1 are eliminated. After compensating for the number of labeled residues in the RT–PCR products corresponding to positions 1 and 4, we determined that on average 55% (n = 3) of nascent ADH4 RNAs were stabilized at position 4 relative to position 1 in the rat 1-1, Δxrn1 double mutant (Fig. 6B, lane 6) at 37°C. Stabilization may not be 100% effective, either because other nucleases are also involved or because Rat1-1 retains some exonuclease activity at 37°C. The latter possibility is consistent with our identification of the rat1-1 mutation as a single substitution, Y657C, in a residue that lies outside the N-terminal exonuclease domain (1–346). In summary, the results in Figure 6 show that although nascent RNA is degraded downstream of the poly(A) site, there is no clear correlation between this process and termination. Specifically, termination can be abrogated even when the nascent RNA has been degraded.
Discussion
Termination and cotranscriptional degradation of nascent RNA
Two models for coupling 3′-end processing to termination have dominated thinking about this process for almost 20 years: (1) the torpedo model (Connelly and Manley 1988), and (2) the anti-terminator or allosteric model (Logan et al. 1987; Greenblatt et al. 1993; Calvo and Manley 2001). We investigated this coupling mechanism by studying the role of the yeast 5′–3′ RNA exonuclease, Rat1, in both 3′-end processing and transcription termination. For these experiments we exploited the ADH4 gene, which has an exceptionally long 3′-flanking region devoid of other transcription units. At ADH4, most pol II is released from the template within 750 bases 3′ of the major poly(A) site 1 (Fig. 1C), and termination is apparently preceded by pausing (Fig. 1C), as in other genes (Enriquez et al. 1991; Chedin et al. 1998; Yonaha and Proudfoot 1999; Orozco et al. 2002). Within the resolution of our analysis, neither release of 3′-end processing factors nor loss of CTD Ser2 phosphate (Fig. 1C) clearly preceded release of pol II itself. The complex that is ultimately released from the DNA template may, therefore, have phosphorylated Ser2 residues in the CTD and 3′-end processing factors associated with it.
Termination downstream of ADH4 was virtually abolished in the ts rat1-1 mutant at 37°C, consistent with previous observations showing that this exonuclease and its human homolog, hXrn2, are essential termination factors (Kim et al. 2004b; West et al. 2004). The torpedo model suggests that when Rat1 is inactivated, failure to terminate is caused by a defect in degradation of nascent RNA downstream of the poly(A) cleavage site. We tested the prediction that Rat1-1 inactivation would stabilize nascent transcripts that coimmunoprecipitate with pol II in the 3′-flanking region. In rat1-1 at 37°C, polymerases that had failed to terminate were abundant in the 3′-flanking region, but there was little or no nascent RNA associated with them (Fig. 6B). A lack of nascent RNA could be due to release of the RNA transcript without release of polymerase from the template; however, this is unlikely because the RNA–DNA hybrid is necessary for stable association of polymerase with DNA (Kireeva et al. 2000). Nascent RNA downstream of the poly(A) site was substantially stabilized by loss of both Xrn1 and Rat1 in the rat 1-1, Δxrn1 double mutant at 37°C, but not when either one alone was inactivated (Fig. 6B). A role for Xrn1, which is predominantly cytoplasmic, in degradation of nascent pol II transcripts is unexpected, but consistent with the fact that this exonuclease functions in the nucleus in rRNA and snoRNA processing (Petfalski et al. 1998; Xue et al. 2000; Fang et al. 2005). While Xrn1 participates in degradation of nascent RNA, its deletion had no detectable effect on termination (Fig. 6C; Supplementary Fig. 2A). Although it complemented ts growth and localized to the ADH4 gene, NLS-Xrn1 failed to correct the termination defect in rat1-1 (Fig. 2B, lanes 17,18). In contrast, the termination defect in rat 1-1, Δxrn1 at 37°C was fully corrected by a single-copy RAT1 plasmid (data not shown). Our results, therefore, did not confirm the hypothesis that the termination defect in rat1-1 is necessarily correlated with stabilization of RNA associated with polymerases downstream of the poly(A) site. Kim et al. (2004b) showed that Rat1 with a point mutant in a conserved residue (D235A) of the exonuclease domain failed to complement the termination defect of rat1-1, arguing for the importance of exonuclease activity in termination. It remains possible, however, that this negative result could be caused by other factors such as an inability to localize correctly at transcribed genes. In summary, our results demonstrate that degradation of nascent RNA downstream of the poly(A) site is not sufficient to trigger polymerase release.
Rat1 is not a dedicated termination factor; it enhances both 3′-end processing and termination
If nascent RNA degradation is not sufficient to cause termination, then why is Rat1 an essential termination factor? Our results suggest that, in fact, Rat1 enhances termination by promoting recruitment of 3′-end processing factors including Pcf11 and Rna15, which are necessary for termination (Birse et al. 1998). Rat1, Pcf11, and Rna15 turn out to be recruited to the poly(A) site in a highly coordinated fashion. In support of this conclusion, Rat1 recruitment was severely inhibited by mutation of Pcf11 (Fig. 3C), and, conversely, Pcf11 and Rna15 recruitment was inhibited by mutation of Rat1 (Fig. 4A). The close integration of Rat1 and Pcf11 functions is also suggested by the fact that the rat1-1 and pcf11-2 mutants have the same delayed termination phenotype at 25°C (Figs. 1D, 3B). Furthermore, inactivation of Rat1 and Pcf11 caused a similar shift in favor of more downstream poly(A) sites at ACT1 (Fig. 5). The latter result shows that Rat1 function is not limited to events after cleavage at the poly(A) site. It will be of interest to determine which domains of Rat1 participate in recruitment of 3′-end processing factors. One possibility is that its interactions with Rai1 and Rtt103 are important in this regard. It is not known how these interactions are affected by the rat1-1 mutation. Rtt103 binds Ser2 phosphorylated CTD heptad repeats and copurifies with CF1 and CPF (Kim et al. 2004b); however, in agreement with Kim et al. (2004b), deletion of Rtt103 did not detectably inhibit termination in our hands (data not shown). Together, the experiments show the interdependent recruitment of Rat1 and 3′-end processing factors, suggesting that these proteins are partners in both cleavage/polyadenylation and termination.
A unified model for coupled 3′-end processing and termination
We confirmed one important prediction of the torpedo model—namely, that nascent RNA downstream of the poly(A) site is degraded cotranscriptionally—however, we did not find a correlation between nascent RNA degradation and termination. Our results do not exclude the possibility that RNA degradation, specifically by Rat1, is a necessary step in termination. Whether or not nascent RNA degradation by Rat1 plays a role, our experiments implicate a second function of this protein in termination—namely, its ability to interact with 3′-end processing factors and promote their recruitment to the pol II transcription complex at the poly(A) site. This finding suggests that rather than acting as a dedicated termination factor after the RNA has been cleaved, Rat1 actually participates in both processing and termination like the 3′-end cleavage factors do (Birse et al. 1998; Z. Zhang et al. 2005).
Pcf11 has been suggested as an allosteric effector that destabilizes the transcription complex via binding of its CID to the S2 phosphorylated pol II CTD (Sadowski et al. 2003; Z. Zhang et al. 2005). Pcf11 is essential for both 3′-end processing and termination. Previous analysis of the pcf11-2 allele suggested that termination was supported independently of 3′-end processing (Sadowski et al. 2003). Reinvestigation of termination in pcf11-2 showed that, in fact, termination defects are evident at both 25°C and 37°C (Fig. 3B). We conclude that the Pcf11-2 mutant protein, although it has a wild-type CID, is not able to support termination uncoupled from 3′-end processing in vivo.
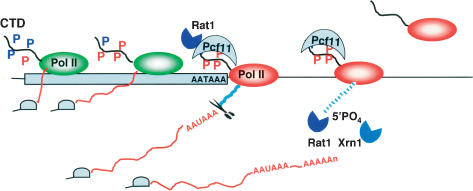
Our results indicate that, by themselves, neither allosteric modification of pol II by Pcf11 CID binding nor nascent RNA degradation by the torpedo exonucleases, Rat1 and Xrn1, is sufficient to cause poly(A)-site-dependent pol II termination in vivo. Instead we suggest a hybrid of the allosteric and torpedo models (Fig. 7) in which cleavage is stimulated by both Rat1 and conventional 3′-end processing factors like Pcf11. Cleavage may be a prerequisite for termination; however, there is currently no direct demonstration of this point. In this unified model, we propose that the complex that carries out poly(A) site cleavage comprises pol II, cleavage factors bound to the phosphorylated CTD, and Rat1. This complex undergoes one or more transitions that achieve cleavage at the poly(A) site, nascent RNA degradation 3′ of the cleavage site, and an allosteric change that is transmitted to the pol II catalytic site, causing it to release from the template DNA.
Figure 7.
Hybrid model for coupling termination with 3′-end processing. This model incorporates aspects of both the antiterminator/allosteric and torpedo models. Cooperative association of Rat1 and cleavage/polyadenylation factors exemplified by Pcf11 with the CTD Ser2 phosphorylated pol II elongation complex is shown at the poly(A) site (AATAAA). This interaction is proposed to result in an allosteric change in pol II (designated by a change from green to red) that favors termination. (Blue Ps) Ser5-PO4; (red Ps) Ser2-PO4. Nascent RNA downstream of the poly(A) cleavage site (blue line) is degraded by both Xrn1 and Rat1. This degradation could facilitate termination; however, it is not sufficient to cause termination.
Materials and methods
Yeast strains and growth conditions
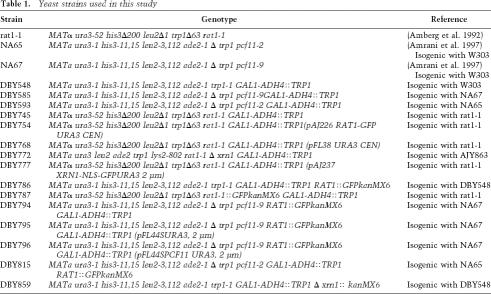
Strains are listed in Table 1.
Table 1.
Yeast strains used in this study
Unless otherwise stated, all experiments were performed on cultures induced with galactose for 1 h as follows: Cultures were grown at 25°C in YEP or SC-Ura3 with 2% raffinose to OD600 nm = 0.8–1.0 and induced by addition of galactose to 2%. After galactose induction for 15 min, the cultures were divided in half, pelleted, and resuspended in YEP with 2% raffinose and 2% galactose for 60 min at either 25°C or 37°C.
Plasmids
The RAT1-GFP (pAJ226), NLS-XRN1-GFP (pAJ237) (Johnson 1997), and pFL44S PCF11 (Amrani et al. 1997) have been described. pBSAct1 poly(A) and pBS ADH4 poly(A) used to make riboprobe contain a fragment spanning bases −100 to +250 relative to the termination codon cloned in Bluescripts KS−.
ChIP and RIP
ChIP was as described (Schroeder et al. 2000). Multiplex PCR was for 21 cycles of 40 sec at 94°C, 40 sec at 48°C, and 1 min at 72°C, with 32P-dCTP added for the last two cycles. MgCl2 concentration was optimized for each primer pair combination. Gels were quantified by a PhosphorImager. Primer pairs 1–5 correspond to positions +1201 to +1390, +1513 to +1689, +1840 to +2050, +2199 to +2349, and +2671 to +2947, relative to the ATG start codon. In some experiments, a human β-globin plasmid (5 pg) was added before elution from the protein A beads to serve as a control for recovery and gel loading. The RIP procedure is based on previously described methods (Niranjanakumari et al. 2002; Gilbert et al. 2004). Cultures were cross-linked with 1% formaldehyde for 20 min at room temperature, treated with glycine (0.2 M, 5 min), washed in PBS, and lysed with glass beads in 0.5 mL of FA lysis buffer with proteinase inhibitors and RNA guard (40 U, Amersham) plus 5 mM DTT. Sonicated extracts (0.5 mL) were digested with DNase I (20 U) plus 25 mM MgCl2 and 5 mM CaCl2 for 30 min at 37°C. Digestion was terminated by addition of EDTA to 20 mM. Extracts were supplemented with BSA (1 mg/mL), sonicated salmon sperm DNA (100 μg/mL), and poly(U) (250 μg/mL), and precleared with magnetic protein G beads (Dynal) for 1 h at 4°C. Precleared extracts were incubated with anti-panCTD antibody overnight at 4°C plus additional RNA guard (40 U), then bound to blocked protein G beads for 2 h. Beads were washed for 10 min at room temperature each with FA lysis buffer twice, FA lysis + 500 mM NaCl, 250 mM LiCl buffer, and TE + 100 mM NaCl as for ChIP. Immunoprecipitated RNA was eluted and reverse-cross-linked, then purified with Trizol (Invitrogen) and redigested with DNase I. cDNA was made with Superscript RT III (Invitrogen). cDNA was PCR amplified for 27 cycles, with 32P-dCTP added for the last two cycles. PCR products were analyzed on 6% denaturing polyacrylamide gels.
RNase protection assay
Total RNA was prepared by hot phenol extraction, and RPA was performed as described (Licatalosi et al. 2002). ADH4 riboprobes were made from wild-type (W303) and rat1-1 genomic DNA sequences, which differ by a single C–T substitution at position +201 relative to the termination codon. PCR-generated fragments containing sequences +1301 to +1633 relative to the ATG were inserted into the Hind III–BamHI site of pBSKS−. Antisense ACT1 probe comprising bases +1404 to +1787 relative to the ATG was made from pVZyAct1.
Antibodies
Rabbit anti-panCTD (Schroeder et al. 2000), anti-Ser2-PO4 (L. Zhang et al. 2005), anti-Pcf11 (Licatalosi et al. 2002), and anti-Rna15 (Bonneaud et al. 1994) have been described. Anti-GFP was made by immunizing rabbits with His-tagged GFP.
Acknowledgments
We thank F. Wyers for pcf11 mutants, plasmids, and anti-Rna15 antibody; C. Cole for the rat1-1 mutant; J. Svejstrup for advice on RIP; G. Geiger for anti-GFP; and the UCHSC Cancer Center sequencing facility. We are grateful to T. Blumenthal, J. Jaehning, S. Kim, and S. Johnson for helpful suggestions. D.B. thanks D. Fox and P. Tjaden for scenic accommodation. This work was funded by NIH grants GM063873 to D.B. and GM53655 to A.J.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1409106
References
- Ahn S.H., Kim M., Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- Amberg D.C., Goldstein A.L., Cole C.N. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Amrani N., Minet M., Wyers F., Dufour M.E., Aggerbeck L.P., Lacroute F. Pcf11 encodes a 3rd protein-component of yeast cleavage and polyadenylation factor I. Mol. Cell. Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauren G., Belikov S., Wieslander L. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes & Dev. 1998;12:2759–2769. doi: 10.1101/gad.12.17.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse C.E., Lee B.A., Hansen K., Proudfoot N.J. Transcriptional termination signals for RNA polymerase II in fission yeast. EMBO J. 1997;16:3633–3643. doi: 10.1093/emboj/16.12.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse C.E., Minvielle-Sebastia L., Lee B.A., Keller W., Proudfoot N.J. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- Bonneaud N., Minvielle S.-L., Cullin C., Lacroute F. Cellular localization of RNA14p and RNA15p, two yeast proteins involved in mRNA stability. J. Cell Sci. 1994;107:913–921. doi: 10.1242/jcs.107.4.913. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Presutti C., Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- Buratowski S. The CTD code. Nat. Struct. Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Connections between mRNA 3′ end processing and transcription termination. Curr. Opin. Cell Biol. 2005;17:257–261. doi: 10.1016/j.ceb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Calvo O., Manley J.L. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- Calvo O., Manley J.L. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 2005;24:1009–1020. doi: 10.1038/sj.emboj.7600575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin S., Riva M., Schultz P., Sentenac A., Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes & Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S., Manley J.L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes & Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- Dichtl B., Blank D., Ohnacker M., Friedlein A., Roeder D., Langen H., Keller W. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell. 2002;10:1139–1150. doi: 10.1016/s1097-2765(02)00707-4. [DOI] [PubMed] [Google Scholar]
- Dominski Z., Yang X.C., Marzluff W.F. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Enriquez H.P., Levitt N., Briggs D., Proudfoot N.J. A pause site for RNA polymerase II is associated with termination of transcription. EMBO J. 1991;10:1833–1842. doi: 10.1002/j.1460-2075.1991.tb07709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F., Phillips S., Butler J.S. Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA. 2005;11:1571–1578. doi: 10.1261/rna.2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C., Kristjuhan A., Winkler G.S., Svejstrup J.Q. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell. 2004;14:457–464. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Nodwell J., Mason S. Transcriptional antitermination. Nature. 1993;364:401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- Gross S., Moore C. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc. Natl. Acad. Sci. 2001;98:6080–6085. doi: 10.1073/pnas.101046598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Wind M., Reines D. Increased accommodation of nascent RNA in a product site on RNA polymerase II during arrest. Proc. Natl. Acad. Sci. 1996;93:6935–6940. doi: 10.1073/pnas.93.14.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell C.M., Gross S., Zenklusen D., Heath C.V., Stutz F., Moore C., Cole C.N. Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 2002;22:6441–6457. doi: 10.1128/MCB.22.18.6441-6457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Jacobson A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol. 2001;21:1515–1530. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Khan A.U., Cheng H., Pappas D.L., Jr., Hampsey M., Moore C.L. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes & Dev. 2003;17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Liu M., Spencer C.A., Price D.H. Involvement of transcription termination factor 2 in mitotic repression of transcription elongation. Mol. Cell. 2004;14:375–385. doi: 10.1016/s1097-2765(04)00234-5. [DOI] [PubMed] [Google Scholar]
- Johnson A.W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Ahn S.H., Krogan N.J., Greenblatt J., Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004a;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Krogan N.J., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004b;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kireeva M.L., Komissarova N., Waugh D.S., Kashlev M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- Kobor M., Greenblatt J. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta. 2002;1577:261–275. doi: 10.1016/s0167-4781(02)00457-8. [DOI] [PubMed] [Google Scholar]
- Licatalosi D.D., Geiger G., Minet M., Schroeder S., Cilli K., McNeil J.B., Bentley D.L. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Liu Y., Kuersten S., Huang T., Larsen A., MacMorris M., Blumenthal T. An uncapped RNA suggests a model for Caenorhabditis elegans polycistronic pre-mRNA processing. RNA. 2003;9:677–687. doi: 10.1261/rna.2128903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan J., Falck P.E., Darnell J.E.J., Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse β maj-globin gene. Proc. Natl. Acad. Sci. 1987;84:8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Bentley D. A ribonucleolytic rat torpedoes RNA polymerase II. Cell. 2004;119:911–914. doi: 10.1016/j.cell.2004.11.041. [DOI] [PubMed] [Google Scholar]
- Mandart E., Parker R. Effects of mutations in the Saccharomyces cerevisiae RNA14, RNA15, and PAP1 genes on polyadenylation in vivo. Mol. Cell. Biol. 1995;15:6979–6986. doi: 10.1128/mcb.15.12.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G.H., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L. The C-terminal domain of RNA polymerase II couples messenger RNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- McNeil J.B., Agah H., Bentley D. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes & Dev. 1998;12:2510–2521. doi: 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker P.J., Wiederkehr T., Strahm Y., Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor Ia and functions in premessenger RNA 3′-end formation. Proc. Natl. Acad. Sci. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S., Lasda E., Brazas R., Garcia-Blanco M.A. Reversible cross-linking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- Orozco I.J., Kim S.J., Martinson H.G. The poly(A) signal, without the assistance of any downstream element, directs RNA polymerase II to pause in vivo and then to release stochastically from the template. J. Biol. Chem. 2002;277:42899–42911. doi: 10.1074/jbc.M207415200. [DOI] [PubMed] [Google Scholar]
- Osheim Y.N., Proudfoot N.J., Beyer A.L. EM visualization of transcription by RNA polymerase II: Downstream termination requires a poly(A) signal but not transcript cleavage. Mol. Cell. 1999;3:379–387. doi: 10.1016/s1097-2765(00)80465-7. [DOI] [PubMed] [Google Scholar]
- Osheim Y.N., Sikes M.L., Beyer A.L. EM visualization of pol II genes in Drosophila: Most genes terminate without prior 3′ end cleavage of nascent transcripts. Chromosoma. 2002;111:1–12. doi: 10.1007/s00412-002-0183-7. [DOI] [PubMed] [Google Scholar]
- Petfalski E., Dandekar T., Henry Y., Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M., Dichtl B., Hubner W., Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder S., Schwer B., Shuman S., Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes & Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E.J., Brow D.A. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E.J., Conrad N.K., Brow D.A., Corden J.L. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- Stevens A., Poole T.L. 5′-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′-exonuclease-1. J. Biol. Chem. 1995;270:16063–16069. doi: 10.1074/jbc.270.27.16063. [DOI] [PubMed] [Google Scholar]
- West S., Gromak N., Proudfoot N.J. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- Xue Y., Bai X., Lee I., Kallstrom G., Ho J., Brown J., Stevens A., Johnson A.W. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to Rat1p. Mol. Cell. Biol. 2000;20:4006–4015. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonaha M., Proudfoot N.J. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell. 1999;3:593–600. doi: 10.1016/s1097-2765(00)80352-4. [DOI] [PubMed] [Google Scholar]
- Zaret K.S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982;28:563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- Zhang L., Schroeder S., Fong N., Bentley D.L. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress.’. EMBO J. 2005;24:2379–2390. doi: 10.1038/sj.emboj.7600711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Fu J., Gilmour D.S. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes & Dev. 2005;19:1572–1580. doi: 10.1101/gad.1296305. [DOI] [PMC free article] [PubMed] [Google Scholar]