Abstract
Biomass-derived sugars, such as glucose, xylose, and other minor sugars, can be readily fermented to fuel ethanol and commodity chemicals by the appropriate microbes. Due to the differences in the optimum conditions for the activity of the fungal cellulases that are required for depolymerization of cellulose to fermentable sugars and the growth and fermentation characteristics of the current industrial microbes, simultaneous saccharification and fermentation (SSF) of cellulose is envisioned at conditions that are not optimal for the fungal cellulase activity, leading to a higher-than-required cost of cellulase in SSF. We have isolated bacterial strains that grew and fermented both glucose and xylose, major components of cellulose and hemicellulose, respectively, to l(+)-lactic acid at 50°C and pH 5.0, conditions that are also optimal for fungal cellulase activity. Xylose was metabolized by these new isolates through the pentose-phosphate pathway. As expected for the metabolism of xylose by the pentose-phosphate pathway, [13C]lactate accounted for more than 90% of the total 13C-labeled products from [13C]xylose. Based on fatty acid profile and 16S rRNA sequence, these isolates cluster with Bacillus coagulans, although the B. coagulans type strain, ATCC 7050, failed to utilize xylose as a carbon source. These new B. coagulans isolates have the potential to reduce the cost of SSF by minimizing the amount of fungal cellulases, a significant cost component in the use of biomass as a renewable resource, for the production of fuels and chemicals.
Rising costs and the finite nature of fossil fuels have led to renewed interest in lignocellulosic biomass as a renewable feedstock for the production of ethanol and other chemicals (5, 11, 17, 25). For this feedstock to be competitive, efficient technologies must be developed to extract and ferment the sugars from both the cellulosic and hemicellulosic portions of fibrous plants, comprising ca. 70% of the dry weight (1, 15). The most efficient process for the utilization of cellulose as a feedstock is yet to be defined but may represent either a two-step process with complete conversion to sugar prior to fermentation (sugar platform) or a one-step process in which saccharification of cellulose by cellulases occurs concurrently with fermentation (simultaneous saccharification and fermentation [SSF]) (24).
Yeast is the preferred organism for glucose conversion to ethanol and lactic acid bacteria serve as the primary organisms for the production of lactic acid (10, 26). Neither organism effectively ferments pentose (hemicellulose) sugars to high levels of products (1, 10). Although pentose-utilizing lactic acid bacteria are available, the phosphoketolase pathway used by these organisms converts two of the five carbons in pentoses to acetic acid, limiting yields and increasing the cost of product purification (7, 22). In addition, optimal growth and fermentation conditions for these traditional microbes are far removed from the optimum conditions for the fungal cellulases (50°C and pH 5.0) required for SSF of cellulose (24). Ethanologenic enteric bacteria have been developed that reduce the requirement for fungal cellulases by functionally expressing integrated cellulase genes from Erwinia chrysanthemi and ferment effectively at pH 5.0 (27). Although these strains efficiently ferment both hexose and pentose sugars, the lack of thermal tolerance precludes their use at temperatures above 43°C. The mismatch of optima for growth of and fermentation by microbes and activity of fungal cellulases increases the enzyme level required for an effective SSF process, arguably a major cost component in lignocellulose conversion to fuels and chemicals (24).
New gram-positive bacterial strains were recently reported that can grow and ferment at pH 5.0 and at temperatures up to 60°C, conditions that are optimal for cellulose hydrolysis by fungal enzymes (18, 19). These new isolates produced l(+)-lactic acid as the major fermentation product during SSF processes with low levels of cellulase (7.5 filter paper units [FPU]/g of glucan). Glucose and xylose were fermented efficiently with yields exceeding 90% by weight, an impossibly high yield for lactic acid bacteria which typically ferment pentoses using the phosphoketolase pathway (10).
We describe here the isolation and physiological characterization of three new gram-positive bacterial isolates with growth optima that match those for fungal cellulases. These strains, producing lactic acid from glucose and xylose, were identified as Bacillus coagulans based on a comparison of 16S rRNA sequences, fatty acid profiles, and physiological properties. Analyses of fermentation products revealed that these bacteria utilize the pentose-phosphate pathway for xylose fermentation, in contrast to the phosphoketolase pathway used by most other lactic acid bacteria (14).
MATERIALS AND METHODS
Culture media.
The mineral salts solution used for initial isolation of bacteria contained, per liter, 1.36 g of KH2PO4, 2.0 g of NaCl, 0.2 g of MgSO4 · 7H2O, 1.0 g of (NH4)2SO4, and 10 mg of FeSO4 · 7H2O. Initial pH of the mineral salts solution, as well as the complete media, was 5.0 and was adjusted with HCl. For Xylose-YE medium, this mineral solution was supplemented with 0.1% (wt/vol) yeast extract and 1% (wt/vol) xylose. In some experiments, xylose was replaced with other sugars at the same concentration. Yeast extract was replaced by corn steep liquor (CSL; 0.5% dry weight/vol) in pH-controlled fermentations. L broth (per liter, 10 g of Trypticase peptone, 5 g of yeast extract, and 5 g of NaCl; LB) supplemented with 1% sugar (wt/vol) served as rich medium (12). Agar was added (1.5% [wt/vol]) for the preparation of the solid medium. Aerobic cultures were grown in 10 ml of medium in 125-ml flasks in a shaker at 200 rpm at 50°C starting with an overnight-grown aerobic culture as inoculum at 1% (vol/vol). Cells from the aerobic cultures were used as needed either at 6 to 8 h of growth (early stationary phase) or after 16 to 18 h. Lactococcus lactis subsp. lactis strain NRRL B-4449 obtained from the U.S. Department of Agriculture was cultured in M17 broth (Difco) supplemented with either glucose or xylose (1% [wt/vol]).
Isolation of thermotolerant, acidophilic bacteria.
Environmental samples were collected from various sources. Approximately 3.5 g of each was suspended in 50 ml of mineral salts solution with the aid of 2-g glass beads (125-ml flask, 200 rpm, 3 h, 50°C), and a sample of the suspension was plated on Xylose-YE medium. After 16 h of incubation at 50°C, representative colonies were selected and stored in 20% glycerol at −75°C for further physiological analysis.
For enrichment of xylose-utilizing bacteria, 44 ml of the suspension described above was supplemented with yeast extract (0.05%) and xylose (1%). After incubation (200 rpm) for 24 h at 50°C, a sample from this enrichment was plated on Xylose-YE medium and incubated at 50°C for 16 h. Individual colonies with distinct morphological characteristics were selected and stored for further analysis.
Fermentations.
Batch fermentations without pH control were conducted in 13-by-100-mm screw cap tubes filled to the top with appropriate medium. Inoculum (1% [vol/vol]) was an overnight aerobic culture grown in the same medium. Growth and fermentation profile of the isolates were determined after 24 h of growth at 50°C. In these fermentations, growth was restricted to the first 6 to 8 h, and the cell yield was limited by decreasing pH due to lactic acid accumulation. pH-controlled fermentations were performed in 500-ml vessels with 250 or 300 ml of medium in a custom-designed pH-stat (18, 19). The mineral salts solution described above supplemented with CSL (0.5% [wt/vol]) and glucose or xylose (3% [wt/vol]) served as the growth medium. The gas phase above the culture was air. The inoculum (1% [vol/vol]) was derived from a 3- to 4-h-old culture (mid-exponential phase of growth) grown aerobically at 50°C and pH 5.0 in L broth with glucose (1% [wt/vol]). The automatic addition of 2 N KOH maintained the pH at 5.0. Samples were removed at various times for determination of cell density and fermentation products. Sugars and fermentation products were determined by using a Hewlett-Packard HPLC chromatograph (HP1090) equipped with a filter photometric detector and a refractive index detector in series (23). An Aminex HPX-87H column (Bio-Rad laboratories, Hercules, CA) was used to separate fermentation products with 4 mM H2SO4 as the eluent.
SSF of cellulose.
SSF of crystalline cellulose was carried out in pH-controlled fermentations as described previously (19). The medium contained mineral salts, CSL (0.5% [wt/vol]), Solka Floc as crystalline cellulose (2% [wt/vol]; International Fiber Corp., North Tonawanda, NY), and different amounts of cellulase (GC220; Genencor International, Palo Alto, CA). The inoculum was derived from a pH-stat culture grown in mineral salts medium with glucose (1% [wt/vol]) and CSL (50°C, pH 5). Cells were collected by centrifugation at room temperature and resuspended in mineral salts solution at 1/10 of the original volume. These concentrated cells were used to inoculate the SSF medium at 1% (vol/vol). Fermentations were at 50°C and pH 5.0. Samples were removed at various times for determination of sugars and fermentation products by high-pressure liquid chromatography (HPLC).
13C-NMR experiments.
For evaluation of the metabolic pathway utilized by the three bacterial isolates during xylose fermentation, cultures were grown in LB plus xylose to mid-exponential phase in a pH-stat at pH 5.0 and 50°C. For the experiment with nongrowing cells, 40 ml of culture was centrifuged, and the cells were washed with 5.0 ml of LB at room temperature. The cells were resuspended in 4.75 ml of LB-xylose (1%) (pH 5.0). Sufficient d-[1-13C]xylose (99% enrichment; Cambridge Isotope Laboratories, Andover, MA) was added to the culture to bring the xylose concentration to 1.2%, a 13C enrichment of 20.8% at the xylose C-1 position. For the experiment with growing cells, cells from 0.5 ml of the mid-exponential-phase culture were removed from the pH-stat, washed with an equal volume of LB at room temperature, and resuspended in 4.75 ml of LB-xylose medium (pH 5.0). Both fermentations were carried out at 50°C with the manual addition of 1.0 N KOH to maintain the pH between 6.0 and 7.0. When acid production stopped, cells were removed by centrifugation, and the supernatant was analyzed by HPLC and 13C nuclear magnetic resonance (NMR). NMR spectra were obtained by using a modified Nicolet NT300 spectrometer (2) operating in the Fourier transform mode as follows: 75.46 MHz; excitation pulse width, 25 μs; pulse repetition delay, 9 s; spectral width, 18 kHz; and 1,000 scans. [1-13C]propionate (50 mM) served as an internal reference. Fermentation products were identified by comparison with known standards. L. lactis was grown in M17 broth with xylose (1% [wt/vol]) at pH 7.0 and 37°C in a pH-stat and fermentations were carried out in M17 broth. [1-13C]xylose fermentations by L. lactis were at 37°C and the pH was maintained between 6.0 and 7.0 by manual addition of 1.0 N KOH.
Phylogenetic characterization.
Physiological characteristics of select isolates were determined by using API 20E and 50CH test kits (bioMérieux, Inc., Durham, NC) as described by the manufacturer. Inoculated tests were incubated at 50°C and read after 24 and 48 h. Fatty acid methyl ester profile and 16S rRNA (DNA) sequences of the isolates were determined by Midi Labs (Newark, DE) according to their standard protocol. The 16S rRNA (DNA) sequence of strain P4-102B was determined as described by Suzuki and Yamasato (21) using appropriate primers. The PCR amplified product was cloned into pUC19 and sequenced by using Licor automated DNA sequencer at the Department of Microbiology and Cell Science (University of Florida) DNA sequencing facility. The 16S rRNA sequences of the three isolates were compared to B. coagulans sequences from the GenBank and the database at the Ribosomal Database Project (3). A phylogram was drawn with the help of Treeview (16).
Materials.
Components for microbiological media were from either BBL or Difco and purchased through Fisher Scientific (Pittsburgh, PA). Analytical grade inorganic chemicals and organic chemicals were from Fisher Scientific or Sigma Chemical Co. (St. Louis, MO).
Nucleotide sequence accession numbers.
The 16S rRNA (DNA) sequences were deposited in GenBank under the accession numbers DQ297925 (strain 17C5), DQ297926 (strain 36D1), DQ297927 (strain P4-102B), and DQ297928 (B. coagulans strain ATCC 7050).
RESULTS AND DISCUSSION
Isolation of bacteria for optimum SSF and SSCF.
Using the procedures described in Materials and Methods, 380 bacterial isolates that grew in xylose mineral salts medium with 0.1% (wt/vol) yeast extract at pH 5.0 and 50°C were isolated from 77 environmental samples. A total of 270 of these isolates grew in filled tubes, suggesting anaerobic growth. This subsample of facultative anaerobes was tested for the following properties: (i) growth and cell yield in rich and mineral salts medium with either glucose or xylose at pH 5.0 and 6.8 under both aerobic and anaerobic conditions; (ii) fermentation profile; (iii) ability to grow and ferment the sugars in sugar cane bagasse hemicellulose acid hydrolysate (50% [vol/vol], with or without overliming with Ca[OH]2) with CSL; (iv) ethanol tolerance (≥4% [wt/wt]); cellulolytic activity (CM-cellulose hydrolysis); (v) fermentation of cellobiose; and (vi) xylanolytic activity (RBB-xylan hydrolysis). Based on the results of these experiments, pH-controlled fermentations of glucose and xylose in mineral salts medium with CSL, as well as fermentation of overlimed sugar cane bagasse hemicellulose acid hydrolysate (50%), two isolates, strains 17C5 and 36D1, that performed better than other isolates in lactate yield from both glucose and xylose, were selected for detailed study. Although the lactate yield of strain P4-102B in hemicellulose acid hydrolysate fermentations was only ca. 65% of the other two strains, this strain was found to be transformable with plasmid DNA by electroporation and was also selected in the present study. Strain 17C5 was isolated from a soil sample adjacent to Georgia Highway 121 about a mile north of the Florida border. Strain 36D1 was isolated from a mud sample in the effluent stream of Old Faithful Geyser 1, Calistoga, CA. Strain P4-102B was obtained from a mixed soil enrichment for isolates that can grow and ferment at pH 4.0 in xylose-yeast extract medium. Under aerobic conditions, the three isolates produced acetate as the primary product and, under anaerobic conditions, lactate was the primary fermentation product with small amounts of acetate, ethanol, formate, and succinate.
SSF of cellulose.
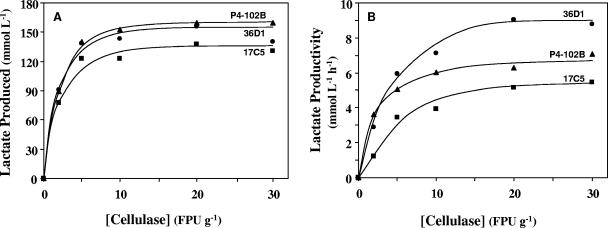
Since the main objective was to isolate bacterial strains that can ferment both glucose and xylose under conditions that are optimal for fungal cellulases, the three selected isolates were evaluated for a cellulase requirement in the SSF of the crystalline cellulose, Solka Floc (Fig. 1). In a typical SSF, all three strains fermented the glucose released by cellulases to lactic acid as the major product with small amounts of acetate and ethanol. Based on total lactic acid produced (Fig. 1A), the minimal amount of cellulase required for optimum SSF was about 10 FPU per g of cellulose. Strains 36D1 and P4-102B fermented ca. 85% of glucose equivalents in cellulose in about 72 h with 10 FPU per g of cellulose, whereas strain 17C5 required about 96 h to reach a similar level. Based on the volumetric productivity (Fig. 1B), the minimal amount of cellulase required was about 15 FPU per g of cellulose. This optimum for cellulase matches the minimal requirement for cost-effective cellulose conversion (24). Strain 36D1 was the most efficient strain in SSF, while strain 17C5 was the least efficient, probably due to its low rate of glucose fermentation (Fig. 2A). This lower lactic acid productivity of strain 17C5 can be accounted for by the higher-than-expected acetate and ethanol in the broth.
FIG. 1.
SSF of crystalline cellulose (Solka Floc) by selected isolates. Crystalline cellulose (20 g/liter) was saccharified by the addition of indicated amount of cellulase (GC220; Genencor) and simultaneously fermented to lactic acid by the specific bacterial isolate (strain 17C5, 36D1, or P4-102B) at 50°C and pH 5.0. Samples were withdrawn at different times, and the amount of lactate present in the broth was determined by HPLC. From these data, the total amount of lactate produced (A) and the volumetric productivity of lactate (B) were determined. For other details, see the text.
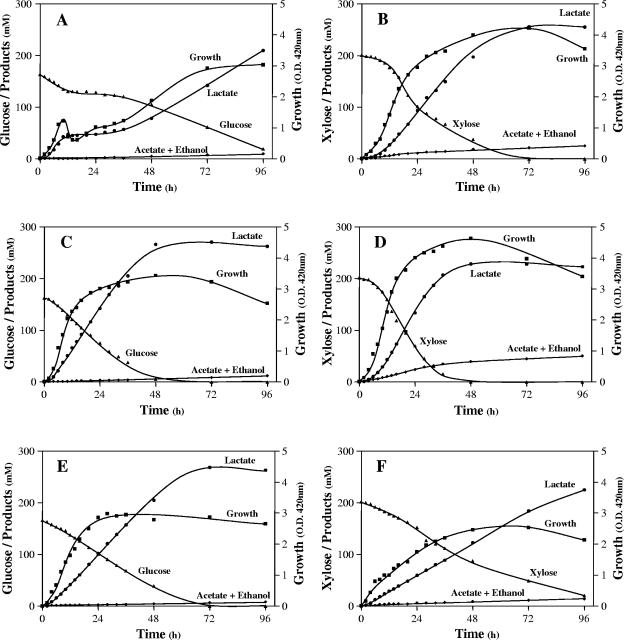
FIG. 2.
Glucose and xylose fermentation by selected isolates in mineral salts medium with 3% sugar (wt/vol) and 0.5% CSL (dry wt/vol) in a pH-stat at pH 5.0 and 50°C. (A and B) Strain 17C5; (C and D) strain 36D1; (E and F) strain P4-102B. A, C, and E, glucose; B, D, and F, xylose.
Fermentation characteristics.
The three selected isolates—strains 17C5, 36D1, and P4-102B—produced l(+)-lactic acid as the major fermentation product from both glucose and xylose (Fig. 2). Of these, strain 36D1 was the most efficient in converting glucose and xylose to lactic acid, with xylose being fermented more rapidly. Strain 17C5 fermented xylose at a higher rate than glucose, while the reverse was true for strain P4-102B. The primary product of glucose fermentation in all three strains was l(+)-lactic acid (ca. 95% of the fermentation products) with small amounts of acetate, ethanol, and succinate. With xylose as the carbon source, lactic acid represented 80 to 93% of the total fermentation products among the different strains. The presence of formate in the medium, as well as the higher fraction of lactic acid (>60%) in the products, suggests that the pentose phosphate pathway is the main pathway of xylose metabolism in these strains (Fig. 3) in contrast to the phosphoketolase pathway observed in other pentose utilizing lactic acid bacteria such as Lactobacillus pentosus and L. lactis (7, 22). If xylose was metabolized through the phosphoketolase pathway, the yield of lactate is not expected to be higher than 60% (Fig. 3).
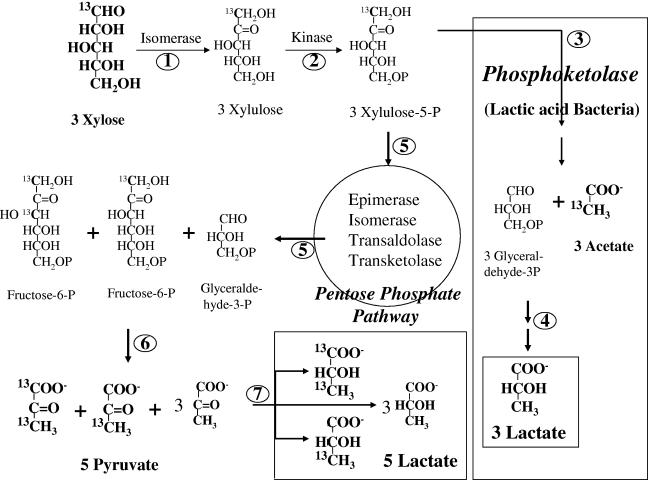
FIG. 3.
Predicted flow of [1-13C]xylose tracer into the metabolic products of xylose fermentation either by the pentose-phosphate pathway or phosphoketolase pathway. Numbers represent the corresponding enzymes that catalyze the particular step as follows: 1, xylose isomerase; 2, xylulose kinase; 3, phosphoketolase (phosphoketolase produces glyceraldehyde-3-phosphate and acetyl phosphate that is converted to acetate and ATP by acetate kinase with ADP); 4, enzymes of glycolysis (glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerokinase, enolase, pyruvate kinase, and lactate dehydrogenase); 5, enzymes of the pentose phosphate pathway (phosphopentose epimerase, phosphopentose isomerase, transketolase, and transaldolase); 6, same enzymes as step 4 with phosphofructokinase, aldolase, and triose phosphate isomerase; 7, lactate dehydrogenase.
The small amount of acetate and ethanol produced by the three isolates during xylose fermentation is in agreement with the presence of pyruvate formate-lyase activity.
Xylose is utilized by the pentose-phosphate pathway.
In pentose-fermenting lactic acid bacteria, phosphoketolase cleaves xylulose-5-phosphate to glyceraldehyde-3-phosphate and acetyl-phosphate (7, 10, 22). These two intermediates are converted to 1 mol each of lactic acid and acetic acid as end products of fermentation (Fig. 3). In other organisms, such as Escherichia coli, the xylulose-5-phosphate enters the pentose-phosphate pathway, yielding fructose-6-phosphate and glyceraldehyde-3-phosphate (13). These compounds can be further metabolized to homo-lactic acid by appropriate bacteria. To establish the primary pathway for xylose metabolism in these new bacterial isolates, [13C]xylose labeled at the C-1 position was used as a substrate (8, 20). [1-13C]xylose metabolism by the phosphoketolase pathway is expected to yield [2-13C]acetate and unlabeled lactate (Fig. 3) (8, 14). In contrast, [13C]xylose metabolism by the pentose phosphate pathway would label both the C-1 and C-3 of lactate. Lactate labeled at both the C-1 and C-3 positions is expected to account for one-fifth of the total lactate, with C-3-labeled lactate accounting for another one-fifth of total lactate. The remaining three-fifths of the lactate is not expected to carry the 1-13C label of xylose except for a small amount produced by the cycling of carbon through the pentose phosphate pathway (Fig. 3). Carbon cycling through the pentose-phosphate pathway would also account for any label in the C-2 of lactate. Enrichment of the 13C label in lactate and acetate would differentiate between the two pentose utilization pathways and provide information on the primary pathway from xylose to lactic acid in this group of lactic acid bacteria.
Pyruvate formate-lyase is expected to yield [2-13C]acetate from both 3-13C- and 1,3-13C-labeled pyruvate. Although this acetate is indistinguishable from the [2-13C]acetate produced by the phosphoketolase pathway, the concentration of labeled acetate is expected to be very low in comparison with [13C] lactate in an organism with pentose phosphate pathway.
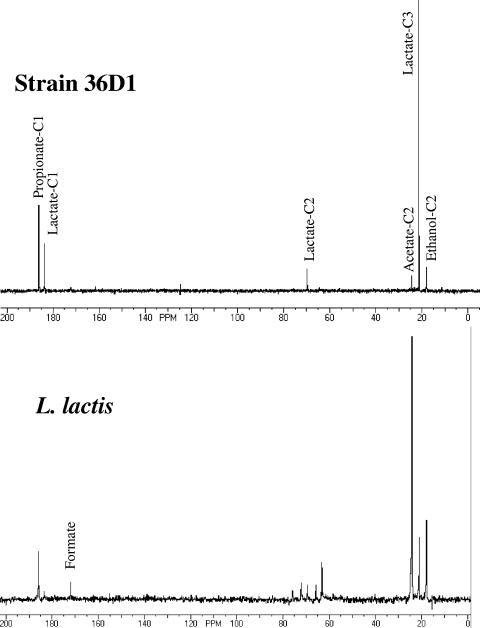
Integration of the peaks in the 13C-NMR spectrum of the spent medium from [1-13C]xylose fermentation by strain 36D1 and comparison of the peaks to known standards revealed that 91% of the 13C label from [1-13C]xylose was present in lactate (Fig. 4). Acetate and ethanol accounted for the remainder. This distribution of label among the products was comparable to the product ratio as determined by HPLC. In the lactate fraction, 74% of the label was in the C-3 position. Label at the C-1 and C-2 positions of lactate was about 21 and 5%, respectively. The unexpected label at the C-2 position is apparently a result of a small amount of carbon cycling through the pentose phosphate pathway that randomized the 1-13C label of xylose. In both acetate and ethanol, almost all of the label was found at the C-2 position, as expected. Although the presence of label in acetate may suggest that a fraction of the xylose is metabolized by the phosphoketolase pathway, it is unlikely due to the presence of formate and ethanol among the fermentation products.
FIG. 4.
13C-NMR spectrum of spent medium of strain 36D1 and L. lactis NRRL B-4449. Strain 36D1 was grown in LB plus [1-13C]xylose at 50°C. L. lactis was grown in M17 broth with [1-13C]xylose at 37°C. [1-13C]propionate (50 mM) served as a standard and reference. See Materials and Methods for details.
Taking the amount of label in the C-2 of lactate as 1 to account for the randomization of carbon due to cycling, an enrichment at the other two positions was calculated (Table 1). The enrichment ratio at C-1 of lactate was about 5.0 for all three strains (17C5, 36D1, and P4-102B). Enrichment at the C-3 position was about 15 for the three strains.
TABLE 1.
13C enrichment ratios of fermentation products produced from [1-13C]xylosea
| Product | Carbon position | Isotope enrichment ratio
|
||||
|---|---|---|---|---|---|---|
| 17C5 | 36D1 | P4-102B | L. lactis | E. coli W3110 | ||
| Lactate | C-1 | 5.5 | 4.6 | 5.7 | 2.5 | 11.0 |
| C-2 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| C-3 | 15.1 | 15.8 | 12.6 | 7.7 | 11.1 | |
| Acetate | C-1 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| C-2 | 15.0* | 10.7* | 5.5* | 234.7* | 4.6 | |
| Ethanol | C-1 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| C-2 | 32.3* | 36.7* | 13.6 | 17.0 | 5.8 | |
Distribution of [1-13C]xylose label in fermentation products of various bacteria was determined as described in the text. All enrichment ratios were based on the abundance of 13C at the indicated positions with C-2 of lactate or C-1 of acetate and ethanol as reference. An asterisk indicates that the C-1 carbons of acetic acid and ethanol were not labeled or the amount of label at the C-1 position was below the detection limit. The presented values were computed using 0.5 mM at these positions as the minimal sensitivity of the instrument for 13C.
L. lactis subsp. lactis NRRL B-4449 is a xylose-utilizing lactic acid bacterium and, as with other lactic acid bacteria, utilizes the phosphoketolase pathway for xylose metabolism (6). In the [1-13C]xylose fermentation, 13C label enrichment at the C-2 position of acetate was about 230 (compared to C-1 of acetate) in contrast to less than 15 at C-2 of acetate in the three Bacillus strains (Fig. 4 and Table 1). The E. coli W3110 xylose fermentation profile is also included in this table for comparison since this bacterium, lacking phosphoketolase, ferments xylose through the pentose phosphate pathway.
These results show that the pentose phosphate pathway is the main metabolic pathway for xylose utilization in these new isolates. This is in agreement with the xylose fermentation profile of these bacteria (Fig. 2). Since these bacteria are effective in SSF of cellulose to lactate in mineral salts medium with 0.5% (wt/vol) of CSL (Fig. 1), as well as in hemicellulose acid hydrolysate medium (18, 19), the taxonomic positions of these isolates were determined by using the fatty acid profile and 16S rRNA (DNA) sequence.
Taxonomic characterization.
Strains 36D1 and P4-102B had similar fatty acid methyl ester (FAME) profiles that were comparable, with minor variations, to that of Bacillus coagulans strain 7050 (4), an ATCC type strain. Strain 17C5 differed from the other two by the presence of higher iso-C15:0 and lower anteiso-C15:0, iso-C16:0, C16:0, and anteios-C17:0. Fatty acid levels of strain 17C5 also differed from the observed composition of strains 36D1, P4-102B, and B. coagulans 7050 (data not presented). Based on the fatty acid profiles, isolates 36D1 and P4-102B can be grouped with B. coagulans. Since B. coagulans is a very diverse group (4, 21), strain 17C5 is also included in this group.
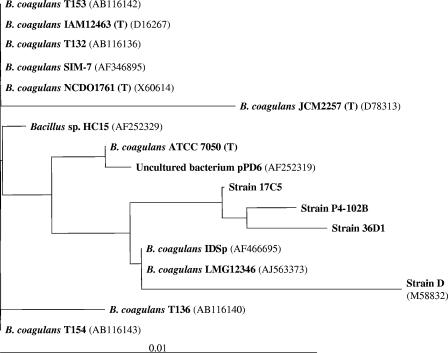
Identification of the three isolates as B. coagulans is also supported by the 16S rRNA (DNA) sequence-based phylogenetic analysis. In the phylogram (Fig. 5), the three strains (17C5, 36D1, and P4-102B) cluster together, indicating the closeness of the isolates to each other. The three isolates also comprise a larger group that includes a B. coagulans ATCC type strain, 7050, and two other strains identified as B. coagulans strains IDSp and LMG12346. Based on the comparison of the 16S rRNA (DNA) sequence, B. coagulans type strain ATCC 7050 from three different culture collections—IAM12463, JCM2257, and NCDO1761—clusters away from the present isolates, as well as from the sequence of strain ATCC 7050 obtained recently. Although strains 17C5, 36D1, and P4-102B are separated from other type strains and other bacilli identified as B. coagulans, a continuum exists among the various B. coagulans strains analyzed in the present study. This diversity within B. coagulans has been recognized by others (4, 21). Due to the heterogeneity of this species, as well as in the same isolate from different culture collections, we have identified all of the three strains isolated in the present study as B. coagulans.
FIG. 5.
Phylogenetic relationship of various isolates from the present study and other known B. coagulans strains based on their 16S rRNA sequence. Numbers in parentheses represent the GenBank accession numbers for the respective sequences. Strains marked with “(T)” represent a type strain in that specific collection, and all are equivalent to ATCC 7050 originally isolated by Hammer (9). Strain collections: IAM, Institute of Applied Microbiology, Japan; NCDO, National Collection of Dairy Organisms, now incorporated into National Collection of Industrial Food and Marine Bacteria, Scotland, United Kingdom; JCM, Japan Collection of Microorganisms, Japan; ATCC, American Type Culture Collection. The scale represents 1% divergence between sequences.
B. coagulans is a facultative, slightly acidophilic, moderately thermophilic bacterium originally isolated from spoiled milk (9), and the members of this species constitute a very heterogeneous group (4). Although strains 17C5, 36D1, and P4-102B are closely related to B. coagulans strain 7050 based on their FAME profiles and 16S rRNA (DNA) sequence analyses, significant differences in the physiological properties of the three isolates and that of the ATCC type strain 7050 can be detected. The most striking and relevant difference between strain 7050 and the three present isolates is the inability of strain 7050 to utilize xylose either aerobically or anaerobically. Although De Clerck et al. (4) reported that xylose utilization by strain 7050 is variable, in several attempts in our laboratory xylose failed to support growth of or fermentation by strain 7050. Since xylose is one of two major components of biomass, its utilization by the three new isolates elevates them to potential use as organisms for biomass conversion to products. Minor differences among the three isolates and the emended description of B. coagulans (4) are also readily identifiable. For example, De Clerck et al. described that the optimum pH for growth of B. coagulans as 7.0 (4). The three isolates described in here had a very broad optimum pH for growth, between 4.5 and 6.0, and growth at pH 7.0 was poor (19).
Summary.
Three unique members of B. coagulans have been described that can ferment both hexoses and pentoses to lactic acid at pH 5.0 and 50°C, conditions that are also optimal for fungal cellulases during SSF processes. These strains utilize the pentose-phosphate pathway for the fermentation of pentose sugars, facilitating the near-complete conversion of the pentose sugars such as xylose and arabinose (in addition to glucose) into optically pure l(+)-lactic acid.
Acknowledgments
We thank A. P. Rooney, USDA-ARS, for providing the L. lactis strain and M. Moniruzzaman, Genencor, for the cellulase preparation used in this study.
This study was supported by funds from the U.S. Department of Energy (DE-FC36-01GO11073, DE-FG36-04GO14019, and DEFG02-96ER20222), the U.S. Department of Agriculture (00-52104-9704 and 01-35504-10669), and the State of Florida, University of Florida Agricultural Experiment Station.
REFERENCES
- 1.Aden, A., M. Ruth, K. Ibsen, J. Jechura, K. Neeves, J. Sheehan, B. Wallace, L. Montague, A. Slayton, and J. Lukas. 2002. Lignocellulosic biomass to ethanol process design and economics utilizing co-current dilute acid prehydrolysis and enzymatic hydrolysis for corn stover. NREL/TP-510-32438. National Renewable Energy Laboratory, Golden, Colo. [Online.] http://www.nrel.gov/docs/fy02osti/32438.pdf.
- 2.Buszko, M. L., D. Buszko, and D. C. Wang. 1998. Internet technology in magnetic resonance: a common gateway interface program for the worldwide web NMR spectrometer. J. Magn. Reson. 131:362-366. [DOI] [PubMed] [Google Scholar]
- 3.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Clerck, E., M. Rodriguez-Diaz, G. Forsyth, L. Lebbe, N. A. Logan, and P. DeVos. 2004. Polyphasic characterization of Bacillus coagulans strains, illustrating heterogeneity within this species, and emended description of the species. Syst. Appl. Microbiol. 27:50-60. [DOI] [PubMed] [Google Scholar]
- 5.Duff, S. J. B., and W. D. Murray. 1996. Bioconversion of forest products industry waste cellulosics to fuel ethanol: a review. Bioresource Technol. 55:1-33. [Google Scholar]
- 6.Erlandson, K. A., J. H. Park, W. E. Khal, H. H. Kao, P. Basaran, S. Brydges, and C. A. Batt. 2002. Dissolution of xylose metabolism in Lactococcus lactis. Appl. Environ. Microbiol. 66:3974-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garde, A., G. Jonsson, A. S. Schmidt, and B. K. Ahring. 2002. Lactic acid production from wheat straw hemicellulose hydrolysate by Lactobacillus pentosus and Lactobacillus brevis. Bioresource Technol. 81:217-223. [DOI] [PubMed] [Google Scholar]
- 8.Gunsalus, I. C., B. L. Horecker, and W. A. Wood. 1955. Pathways of carbohydrate metabolism in microorganisms. Bacteriol. Rev. 19:79-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammer, B. W. 1915. Bacteriological studies on the coagulation of evaporated milk. Iowa Agric. Exp. Station Res. Bull. 19:119-131. [Google Scholar]
- 10.Hofvendahl, K., and B. Hans-Hagerdal. 2000. Factors affecting the fermentative lactic acid production from renewable resources. Enz. Microb. Technol. 26:87-107. [DOI] [PubMed] [Google Scholar]
- 11.Kheshgi, H. S., R. C. Prince, and G. Marland. 2000. The potential of biomass fuels in the context of global climate change: focus on transportation fuels. Annu. Rev. Energy Environ. 25:199-244. [Google Scholar]
- 12.Lee, J. H., P. Patel, P. Sankar, and K. T. Shanmugam. 1985. Isolation and characterization of mutant strains of Escherichia coli altered in H2 metabolism. J. Bacteriol. 162:344-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, E. C. C. 1996. Dissimilatory pathways for sugars, polyols, and carboxylates, p. 307-342. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 14.Lokman, B. C., P. van Santen, J. C. Verdoes, J. Kruse, R. J. Leer, M. Posno, and P. H. Pouwels. 1991. Organization and characterization of three genes involved in d-xylose catabolism in Lactobacillus pentosus. Mol. Gen. Genet. 230:161-169. [DOI] [PubMed] [Google Scholar]
- 15.Lynd, L. R., C. E. Wyman, and T. U. Gerngross. 1999. Biocommodity engineering. Biotechnol. Prog. 15:777-793. [DOI] [PubMed] [Google Scholar]
- 16.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 17.Parajo, J. C., J. L. Alonso, and V. Santos. 1996. Lactic acid from wood. Process Biochem. 31:271-280. [Google Scholar]
- 18.Patel, M., M. Ou, L. O. Ingram, and K. T. Shanmugam. 2004. Fermentation of sugar cane bagasse hemicellulose hydrolysate to l(+)-lactic acid by a thermotolerant acidophilic Bacillus sp. Biotechnol. Lett. 26:865-868. [DOI] [PubMed] [Google Scholar]
- 19.Patel, M. A., M. Ou, L. O. Ingram, and K. T. Shanmugam. 2005. Simultaneous saccharification and co-fermentation of crystalline cellulose and sugar cane bagasse hemicellulose hydrolysate to lactate by a thermotolerant acidophilic Bacillus sp. Biotechnol. Prog. 21:1453-1460. [DOI] [PubMed] [Google Scholar]
- 20.Scott, A. I., and R. L. Baxter. 1981. Applications of 13C NMR to metabolic studies. Annu. Rev. Biophys. Bioeng. 10:151-174. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki, T., and K. Yamasato. 1994. Phylogeny of spore-forming lactic acid bacteria based on 16S rRNA gene sequences. FEMS Microbiol. Lett. 115:13-18. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka, K., A. Komiyama, K. Sonomoto, A. Ishizaki, S. J. Hall, and P. E. Stanbury. 2002. Two different pathways for d-xylose metabolism and the effect of xylose concentration on the yield coefficient of l-lactate in mixed-acid fermentation by the lactic acid bacterium Lactococcus lactis IO-1. Appl. Microbiol. Biotechnol. 60:160-167. [DOI] [PubMed] [Google Scholar]
- 23.Underwood, S. A., S. Zhou, T. B. Causey, L. P. Yomano, K. T. Shanmugam, and L. O. Ingram. 2002. Genetic changes to optimize carbon partitioning between ethanol and biosynthesis in ethanologenic Escherichia coli. Appl. Environ. Microbiol. 68:6263-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wooley, R., M. Ruth, D. Glassner, and J. Sheehan. 1999. Process design and costing of bioethanol technology: a tool for determining the status and direction of research and development. Biotechnol. Prog. 15:794-803. [DOI] [PubMed] [Google Scholar]
- 25.Wyman, C. E. 2003. Potential synergies and challenges in refining cellulosic biomass to fuels, chemicals, and power. Biotechnol. Prog. 19:254-262. [DOI] [PubMed] [Google Scholar]
- 26.Zaldivar, J., J. Nielsen, and L. Olsson. 2001. Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Appl. Microbiol. Biotechnol. 56:17-34. [DOI] [PubMed] [Google Scholar]
- 27.Zhou, S., F. C. Davis, and L. O. Ingram. 2001. Gene integration and expression and extracellular secretion of Erwinia chrysanthemi endoglucanase CelY (celY) and CelZ (celZ) in ethanologenic Klebsiella oxytoca P2. Appl. Environ. Microbiol. 67:6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]