Abstract
The aim of this work was to investigate the behavior of thermophilic esterase EST2 from Alicyclobacillus acidocaldarius in milk and cheese models. The pure enzyme was used to compare the EST2 hydrolytic activity to the activity of endogenous esterase EstA from Lactococcus lactis. The results indicate that EST2 exhibits 30-fold-higher esterase activity than EstA. As EstA has thioesterase activity, EST2 was assayed for this activity under the optimal conditions determined for EstA (namely, 30°C and pH 7.5). Although it is a thermophilic enzyme, EST2 exhibited eightfold-higher thioesterase activity than EstA with S-methyl thiobutanoate. The abilities of EST2 and EstA to synthesize short-chain fatty acid esters were compared. Two methods were developed to do this. In the first method a spectrophotometric assay was used to monitor the synthesis of esters by the pure enzymes using p-nitrophenol as the alcohol substrate. The synthetic activities were also evaluated under conditions that mimicked those present in milk and/or cheese. The second method involved evaluation of the synthetic abilities of the enzymes when they were directly added to a model cheese matrix. Substantial ester synthesis by EST2 was observed under both conditions. Finally, esterase and thioesterase activities were evaluated in milk using the purified EST2 enzyme and in the model cheese matrix using a strain of L. lactis NZ9000 harboring the EST2 gene and thus overproducing EST2. Both the esterase and thioesterase activities measured in milk and in the cheese matrix were much greater than the activities of the controls.
In industrial cheese production pasteurization of raw milk has been reported to affect cheese aroma type and intensity, depending on the type of treatment used (35, 39). Besides destroying pathogenic bacteria, pasteurization also influences the type of lactic acid bacteria present during ripening (34) and therefore can adversely affect flavor development. To overcome losses in flavor and aroma produced by beneficial microorganisms destroyed by heating, alternative strategies have been suggested or tested. Either selected thermoresistant strains or strains (natural or mutant) with high levels of endogenous lipolytic and/or proteolytic activities have been used as starters or adjuncts for fermentation (1, 34, 35, 42, 43). Recently, enzymes with key activities, alone or in combination with starters, have been used to enhance cheese flavor (10, 21-23, 45). The aims of this study were to determine if thermophilic esterase EST2 (30) from Alicyclobacillus acidocaldarius, a bacterium first isolated from an acidic creek in Yellowstone National Park (6), exhibits high esterase activity in milk and in a standardized cheese model and to determine if this enzyme has thioesterase and ester-synthesizing activities like those of Lactococcus lactis EstA (38).
L. lactis is one of the most important lactic acid bacteria used in the dairy industry. By using esterase purification (4) and genome sequencing (3), it has been shown that this bacterium contains a single esterase (EstA) and has no lipase or alcohol acetyltransferase homologous to the enzymes found in yeast (14). EstA is probably a key enzyme in the development of food flavor through the degradation of esters and lipids and the synthesis of esters and thioesters (38). Studies using an EstA-negative mutant partially confirmed this (12). EstA has an optimal temperature of 30°C and exhibits maximal activity at pH 7.5 with p-nitrophenyl (pNP) esters having acyl chains that are four to six carbon atoms long. This enzyme exhibits sequence homology to S-formyl glutathionine hydrolase, which can explain its thioesterase activity (12). EstA belongs to a superfamily of phylogenetically related proteins with representatives in the domains Eukarya, Bacteria, and Archaea (18, 20, 24, 31). These proteins are divided into four groups based on sequence identity: the C group, which includes cholinesterases and fungal lipases; the L group, which includes lipoprotein lipases and bacterial lipases; the H group, named after the mammalian hormone-sensitive lipase (27), which comprises proteins exhibiting sequence similarity to hormone-sensitive lipase (18, 19); and the X group, which includes a wide variety of proteins that contain the α/β hydrolase motif and do not belong to any of the other groups. EstA has been classified in the X group (12). In contrast, A. acidocaldarius EST2 belongs to the H group (8, 18, 29). The functional protein was overexpressed in Escherichia coli, purified, and characterized. This enzyme has an optimal temperature of 70°C, exhibits remarkable temperature stability, has a half-life of 3 h at 75°C, and exhibits maximal activity at pH 7.0 with pNP esters with acyl chains that are six to eight carbon atoms long (30).
In this paper, we describe biochemical characterization of purified EST2, a comparison of this enzyme with EstA, and cloning and expression of the EST2 gene in L. lactis. EST2 may have potential for use in the dairy industry in the development and/or acceleration of the development of cheese flavor. Therefore, the esterase and thioesterase activities in milk and in a cheese model were investigated by using the purified enzyme and an EST2-overproducing strain, respectively. As the formation of esters is important in the development of cheese flavor components, analytical methods were developed in an aqueous solution and in a cheese matrix to investigate the ability of EST2 to synthesize esters of short-chain fatty acids.
MATERIALS AND METHODS
Chemicals.
pNP-hexanoate, S-methyl thiobutanoate, ethyl-hexanoate, 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB), 3,3′-diaminobenzidine, and chloramphenicol were purchased from Sigma Chemical Co. (St. Louis, MO). Nisin powder (nisin content, 2.5%) was purchased from ICN.
Bacterial strains, media, and growth conditions.
L. lactis cells were grown at 30°C in M17 medium (Merck, Darmstadt, Germany) supplemented with 0.5% (wt/vol) glucose. When needed, chloramphenicol (10 μg ml−1) was added to the medium.
Enzyme purification.
EST2 and EstA were purified as previously described by Manco et al. (30) and Fernandez et al. (12), respectively.
Cloning and overexpression of EST2 in L. lactis.
The expression vector pNZ7408 was obtained by cloning the EST2 gene in pNZ8148 (J. E. T. van Hylckama Vlieg, J. A. Wouters, R. van Kranenburg, M. Twigt, J. Muller-Beenhakkers, T. Jansen-van den Bosch, G. Rutten, G. Smit, and P. G. Gruinenberg, unpublished data). The EST2 gene was amplified by PCR with oligonucleotides est5′ (5′-GATATACCCATGGCGCTCGATCCCGTCATTCAG-3′) and est3′ (5′-CGTGAGTGCCTGCAGTCGCTTGCATCCGCCTTTTG-3′) using High Fidelity DNA polymerase. The pT7-SCII-AG vector (32) was used as the template. The amplification primer est5′ was designed to introduce an NcoI restriction site (underlined) upstream of the initiation site, whereas est3′ was designed to introduce a PstI restriction site (underlined) downstream of the stop codon of the EST2 gene. By insertion of the NcoI restriction site into the EST2 gene, the second amino acid was changed from Pro to Ala. This substitution did not alter the main characteristics of EST2. The PCR product, eluted from an agarose gel and digested with NcoI and PstI, was ligated into NcoI-PstI-digested expression vector pNZ8148 to create the pNZ7408 construct.
The cloned fragment was completely sequenced to verify that no undesired mutations were introduced during amplification. By using this plasmid the EST2 gene was expressed under direct control of the nisin-inducible promoter (7).
L. lactis NZ9000 cells containing pNZ7408 were cultured in 50 ml of M17 medium supplemented with 0.5% glucose and chloramphenicol (10 μg ml−1). Cells were induced at an optical density at 600 nm (OD600) of ∼0.5 with 1, 2, 5, and 10 ng/ml of nisin, and uninduced cells were used as a control.
Cells were harvested after 90 min of induction by centrifugation (5,000 × g, 4°C, 10 min) and were resuspended in 5 ml of 50 mM sodium phosphate buffer (pH 7.5).
Cells were disrupted with a French pressure cell (Aminco Co.), and debris was removed by centrifugation. The crude extracts obtained were used in spectrophotometric esterase assays and Western blot analyses.
Electrophoreses and Western blot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12.5%) was performed with a Bio-Rad Mini-Protean II cell unit as described by Laemmli (25). By using a transblot apparatus (Bio-Rad), proteins were electrotransferred (1 h at 100 V) onto a polyvinylidene difluoride (PVDF) immunoblot membrane (0.2 μm; Bio-Rad) in 0.375 M Tris-glycine buffer (pH 8.3) containing 10% methanol. Subsequently, the PVDF sheet was removed, soaked in blocking buffer (1× phosphate-buffered saline, 5% milk, 0.05% Tween 20) and incubated for 1 h with an anti-EST2 rabbit antiserum (30) diluted 1:1,000 in blocking buffer. The membrane was washed twice in blocking buffer (15 min each) and incubated for 1 h with horseradish peroxidase-linked whole antibody from goat diluted 1:3,000 in blocking buffer. The filter was washed again as described above and developed with 3,3′-diaminobenzidine and hydrogen peroxide.
Esterase activity.
The initial rates of p-nitrophenoxide release from pNP-hexanoate were determined by measuring the absorbance at 410 nm in 1-cm-path-length cells with a Cary 100 spectrophotometer (Varian, Australia). The initial rates were calculated by linear least-squares analysis of time courses comprising less than 10% of the total substrate turnover. Assays were performed at 30°C in mixture containing 40 mM sodium phosphate buffer (pH 7.5), 4% acetonitrile, and 100 μM pNP-hexanoate. A stock solution of pNP-hexanoate was prepared by dissolving the substrate in pure acetonitrile. Assays were carried out in duplicate or triplicate, and the results were expressed as the means of two independent experiments.
One unit of enzymatic activity was defined as the amount of protein that released 1 μmol of p-nitrophenoxide/min from pNP-hexanoate (at the temperatures used) (30). The absorption coefficient used for p-nitrophenoxide was 14,000 M−1 cm−1.
Determination of the pH optimum for esterase activity.
The dependence of the initial velocity on pH was monitored at 348 nm (the pH-independent isosbestic point of p-nitrophenol and the p-nitrophenoxide ion) with a molar absorption coefficient of 5,000 M−1 cm−1 at 30°C. The following buffers were used: 50 mM sodium citrate in the pH range from 4.0 to 5.5, 50 mM sodium phosphate in the pH range from 5.5 to 7.0, and 50 mM Tris-HCl in the pH range from 7.0 to 8.0. Data were analyzed as described by Dixon and Webb (9).
Determination of esterase activity at different NaCl concentrations.
The dependence of the initial velocity on the NaCl concentration was monitored at 410 nm. Assays were performed at 30°C by using mixtures containing 40 mM sodium phosphate buffer (pH 7.5), 4% acetonitrile, 100 μM pNP-hexanoate, and different amounts of NaCl (0 to 10%, wt/vol). Assays were carried out in duplicate or triplicate, and the results were expressed as the means of two independent experiments. The absorption coefficients used for activity calculations were determined at each NaCl concentration used and ranged from 14,000 to 16,000 M−1 cm−1.
Thioesterase activity as determined by the DTNB assay.
Enzyme activity with thioesters was analyzed by the DTNB assay (44). The initial rates of formation of free thiol groups from S-methyl thiobutanoate were determined by measuring the absorbance at 410 nm of 5-thio(2-nitrobenzoic acid) in 1-cm-path-length cells with a Cary 100 spectrophotometer (Varian, Australia). The initial rates were calculated by linear least-square analysis of time courses comprising less than 10% of the total substrate turnover. The standard assay mixture contained 50 mM sodium phosphate buffer (pH 7.5), 2 mM DTNB, 20 mM S-methyl thiobutanoate, and 1 ng/ml of enzyme. A stock solution of S-methyl thiobutanoate was prepared by dissolving the substrate in dimethyl sulfoxide. One unit of enzymatic activity was defined as the amount of protein that released 1 μmol of 5-thio(2-nitrobenzoic acid) per min from DTNB. Assays were carried out in duplicate or triplicate, and the results were expressed as the means of two independent experiments. The absorption coefficient used for DTNB was 13,700 M−1 cm−1.
Determination of the pH optimum for thioesterase activity.
The dependence of the initial velocity on pH was monitored at 410 nm, and the assay was performed as described above for the esterase activity assay by using 2 mM DTNB and 20 mM S-methyl thiobutanoate as the substrate. The absorption coefficient for DTNB was calculated for each pH value.
Determination of thioesterase activity at different NaCl concentrations.
The dependence of the initial velocity on the NaCl concentration was monitored at 410 nm, and the assay was performed as described above for the esterase activity assay by using 2 mM DTNB and 20 mM S-methyl thiobutanoate as the substrate. The absorption coefficient used for the assay was determined at each NaCl concentration, and the values ranged from 13,700 to 14,100 M−1 cm−1.
Esterase and thioesterase activities in milk.
Ester degradation and thioester degradation were determined in highly pasteurized (HP) milk (full-fat milk heated at 102°C for 30 min and stored at 4°C until it was used) by using purified EST2 (3 ng/ml) and EstA (3 ng/ml). Ethyl-hexanoate and S-methyl thiobutanoate were added at concentrations of 5 mg/liter and 150 μg/liter, respectively. The samples were incubated for 16 h at 30°C, and at different times 3 ml of each sample was removed and boiled for 10 min to quench the reaction. Ester and thioester and their degradation products were analyzed by headspace gas chromatography (HS-GC), as described previously (1), by using flame photometric detection for the thioester and flame ionization detection for the ester. The analysis was standardized with exact quantities of ester and thioester in the HP milk matrix.
Esterase and thioesterase activities in cheese paste.
Ester degradation and thioester degradation by EST2 were monitored in a cheese model (Ch-easy model) (42). L. lactis NZ9000 or L. lactis NZ9000 containing pNZ7408 was incubated in cheese paste for 2 weeks at 17°C in the presence of ethyl-hexanoate (5 mg/kg) or S-methyl thiobutanoate (150 μg/kg). The control did not contain added any lactic acid bacteria. After incubation, samples were analyzed by HS-GC to determine the extent of (thio)ester degradation.
In vitro ester synthesis assays.
Different parameters, such as pH, salt concentration, and the presence of sugars or protein, and combinations of these parameters were evaluated. The assays were carried out in buffered solutions without organic solvents. The enzyme was used at a concentration of 10 ng/ml. The reaction buffer contained 50 mM sodium phosphate buffer (pH 5.5 or 7.5), 50 μM p-nitrophenol, and 500 μM hexanoic acid. In some cases the buffer contained 5% NaCl, 4% lactose, or 20 mg/ml bovine serum albumin (BSA) (see Table 3), and several combinations of these compounds were tested in the assays. Ester synthesis was measured spectrophotometrically by monitoring the decrease in absorbance at 410 nm (Cary 100; Varian). All the assays were performed at 20°C. The initial rate was calculated by linear least-squares analysis of time courses comprising less than 20% of the total substrate turnover. Assays were performed in duplicate or triplicate, and the results were expressed as the means of two independent experiments. The absorption coefficients were determined for each different condition used. One unit of synthetic activity was defined as the amount of protein that synthesized 1 μmol of pNP-hexanoate/min.
TABLE 3.
Ester synthesis and degradation in milk- and cheese-like conditions in a buffered systema
| Conditions | Synthesis (U/mg) | Degradation (U/mg) |
|---|---|---|
| pH 7.5 | 11.0 ± 2.0 | 1,850.0 ± 90.0 |
| pH 5.5 | 48.0 ± 2.5 | 62.0 ± 2.2 |
| pH 7.5, 5% NaCl | 105.0 ± 6.0 | 805 ± 7.4 |
| pH 5.5, 5% NaCl | 635.0 ± 20.0 | 0.24 ± 0.01 |
| pH 7.5, 4% lactose | 15.0 ± 0.5 | 1,105.0 ± 75.0 |
| pH 5.5, 4% lactose | 32.5 ± 0.5 | 18.4 ± 0.8 |
| pH 7.5, 20 mg/ml BSA | 3.6 ± 0.2 | 1,015.0 ± 20.0 |
| pH 5.5, 20 mg/ml BSA | 12.0 ± 1.2 | 122.0 ± 1.3 |
| pH 7.5, 5% NaCl, 20 mg/ml BSA | 264.0 ± 11.0 | 3.7 ± 0.2 |
| pH 5.5, 5% NaCl, 20 mg/ml BSA | 330.0 ± 40.0 | 0.45 ± 0.01 |
| pH 7.5, 4% lactose, 20 mg/ml BSA | 95.0 ± 1.0 | 1,070.0 ± 50.0 |
| pH 5.5, 4% lactose, 20 mg/ml BSA | 261.0 ± 18.0 | 735.0 ± 10.0 |
The following assay conditions were used: 50 mM sodium phosphate buffer, 50 μM p-nitrophenol, 500 μM hexanoic acid (C6), and 20°C. All the components added to the reaction mixtures were obtained from stock solutions in sodium phosphate buffer (pH 5.5 or 7.5). One unit of enzymatic activity was defined as the amount of protein that synthesized 1 μM pNP-ester/min or released 1 μM pNP/min. For each set of conditions, the absorption coefficient of p-nitrophenol was calculated. Assays were done in duplicate or triplicate, and the data are the means ± standard deviations of two independent experiments.
Ester synthesis in cheese paste.
An analysis of ester synthesis in cheese paste was performed in order to confirm the ester synthesis in a cheese paste model (42). We added 1.5 ml of 50 mM sodium phosphate buffer (pH 5.5) containing ethanol (final concentration in the paste, 20 mg/kg), hexanoic acid (final concentrations in the paste, 200 mg/kg), 1% (wt/wt) NaCl, and the enzyme (8 ng) to 25 g of cheese paste. The same mixture without enzyme was used as a control. The samples were incubated for 24 h at 17°C. After incubation, the samples were analyzed by HS-GC with flame ionization detection to determine the amount of ethyl-hexanoate formed. Exact amounts of ethyl-hexanoate in the cheese paste matrix were used to standardize the analysis.
RESULTS AND DISCUSSION
In this study we compared thermophilic esterase EST2 from A. acidocaldarius and EstA from L. lactis in light of the potential use of EST2 in the dairy industry for development of cheese flavors and/or for acceleration of cheese ripening. EST2 exhibits activity with esters of glycerol (30, 33), which are the main lipids in milk and are partially responsible for the development of characteristic cheese flavors (17, 22, 23) during maturation. Lipids are degraded by esterases and lipases and are the primary sources of free fatty acids, particularly short-chain free fatty acids, which confer characteristic flavors to some specific aged cheeses. However, through ester resynthesis reactions, new esters conferring other characteristic flavors to cheeses are produced. The formation of ethyl esters, such as ethyl-butanoate and ethyl-hexanoate, is thought to play a major role in the development of fruity flavors in, for example, Italian cheeses (2, 11, 36, 41). It is well known (13, 17) that esterase-containing tissues of ruminants are used in standardized procedures for cheese production, while esterases and lipases from starter and nonstarter lactic acid bacteria are also thought to play an important role (15).
In vitro analysis of esterase and thioesterase activities.
In the first part of this work we compared EST2 and EstA esterase and thioesterase activities under the optimal assay conditions previously reported for EstA (namely, 30°C and pH 7.5) (12).
The kinetic constants were measured by using pNP-hexanoate and S-methyl thiobutanoate as substrates for esterase and thioesterase activities, respectively (Table 1). Both the catalytic rate (kcat) and the efficiency of esterase activity were higher for EST2 than for EstA (∼6- and 150-fold higher, respectively). The lower Michaelis constant (Km) for EST2 suggests that the EST2 substrate affinity is higher than the EstA substrate affinity. The catalytic rate of thiosterase activity was approximately eight times higher for EST2 (235 s−1) than for EstA (30.5 s−1), but as the Km was 10 times lower for EstA, the catalytic efficiencies were similar. These results suggest that both EST2 activities are superior or, at worst, similar to the activities of EstA under conditions optimal for EstA activity. It has been reported previously that EST2 also acts on triglycerides, on esters of pheromones, and on sugar derivatives (30, 33). Thioesterase activity in the temperature range from 50 to 70°C has been reported recently for EST2 (33). This study demonstrated that EST2 has very high thioesterase activity at 30°C, even though EST2 is a thermophilic enzyme.
TABLE 1.
Comparison of EST2 and EstA kinetic parameters at 30°C and pH 7.5, determined by using pNP-hexanoate (0-150 μM) and S-methyl thiobutanoate (0-40 mM) as the substrates in the esterase and thioesterase assays, respectivelya
| Activity | Enzyme | Sp act (U/mg) | kcat (s−1) | Km | Efficiency |
|---|---|---|---|---|---|
| Esterase | EST2 | 2,750 ± 150 | 1,580 ± 85 | 20 ± 1 μM | 79 ± 4 μM−1 s−1 |
| EstA | 90 ± 3.5 | 45 ± 0.8 | 90 ± 4 μM | 0.50 ± 0.01 μM−1 s−1 | |
| Thioesterase | EST2 | 410 ± 70 | 235 ± 40 | 37 ± 2 mM | 6.3 ± 0.7 mM−1 s−1 |
| EstA | 53 ± 1 | 30.5 ± 0.6 | 3.5 ± 0.3 mM | 8.7 ± 0.4 mM−1 s−1 |
Assays were done in duplicate or triplicate, and the data are the means ± standard deviations of two independent experiments.
Since EST2 may be used to enhance flavor and/or accelerate cheese ripening and since cheese has well-defined pH and salt concentration values (pH 5.5 and 5% NaCl), the EST2 esterase and thioesterase activities as a function of pH and NaCl concentration were evaluated.
Figure 1A shows the pH dependence of EST2 esterase and thioesterase activities at 30°C. In this experiment three different buffers were used; the pH values ranged from 4.0 to 8.0, and there was overlap in the pH values of the buffers. The profiles of esterase and thioesterase activities appeared to be very similar, and EST2 exhibited maximum activity at pH 7.0 to 8.0. At pH 4.0 esterase activity was barely detectable, and at pH 5.5 both activities were present, although the levels were substantially reduced. Figure 1B shows the dependence of the activities on the NaCl concentration at pH 7.5. An interesting observation is that the two activities had different salt sensitivities. At an NaCl concentration of 5% (wt/vol) both activities were about 50% of the maximum activity, while at an NaCl concentration of 10% the decreases in the esterase and thioesterase activities were 90% and 60%, respectively.
FIG. 1.
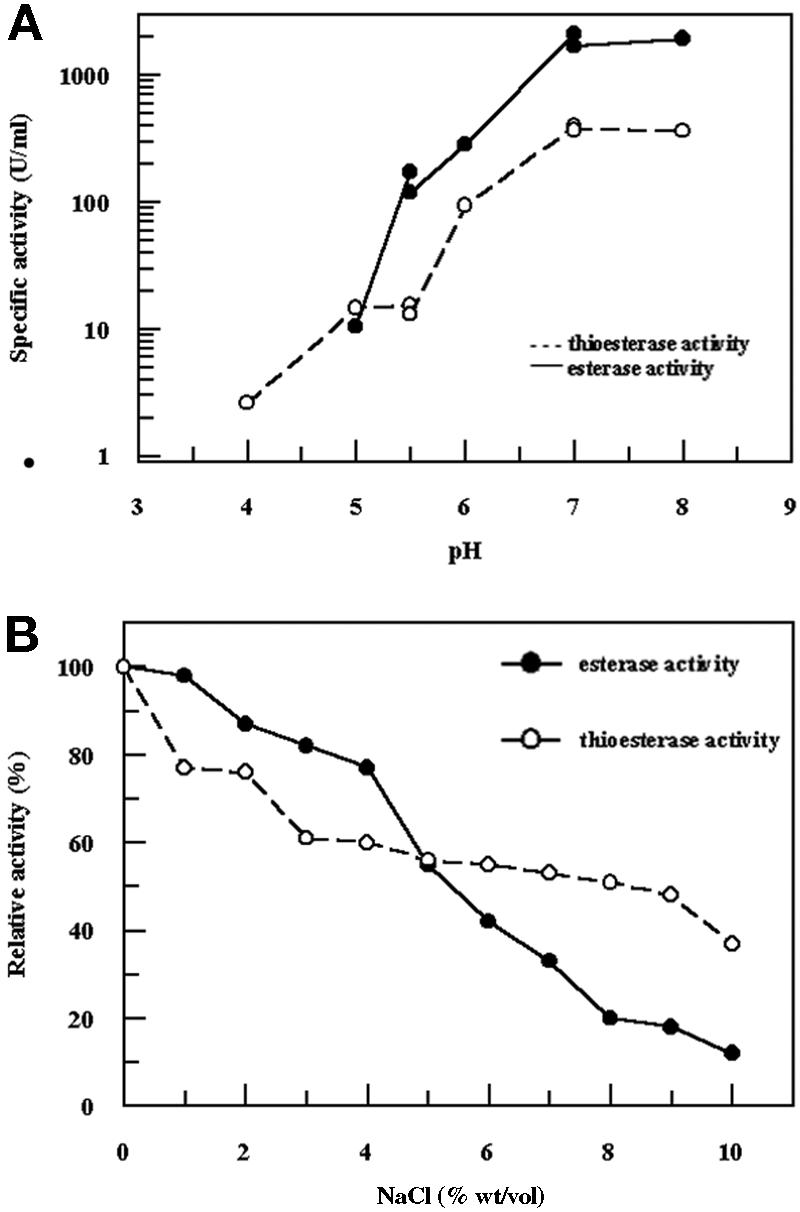
(A) EST2 esterase and thioesterase activities as a function of pH. pNP-hexanoate (100 μM) and S-methyl thiobutanoate (20 mM) were used as the substrates. The buffers used were sodium citrate (pH 4.0 to 5.5), sodium phosphate (pH 5.5 to 7.0), and Tris-HCl (pH 7.0 to 8.0). Assays were performed at 30°C. (B) EST2 esterase and thioesterase activities as a function of NaCl concentration. pNP-hexanoate (100 μM) and S-methyl thiobutanoate (20 mM) were used as the substrates. Assays were performed at 30°C and pH 7.5. The activity is expressed relative to the activity without salt.
Further biochemical characterization was performed in a buffered system under conditions similar to milk- and cheese-like real conditions. In order to mimic these conditions, esterase and thioesterase activities were tested at pH 5.5 in the presence or absence of 5% NaCl. In this analysis, pure EST2 and EstA enzymes were compared, and the results are shown in Table 2. In the presence of 5% NaCl at 30°C and pH 5.5 the EST2 and EstA esterase activities were dramatically lower (∼5,000- and ∼800-fold, respectively) than the activities at pH 7.5 without NaCl. The thioesterase activities of EST2 and EstA were ∼150- and ∼600-fold lower, respectively. However, it should be noted that the EST2 activities under both conditions were higher than the EstA activities. Moreover, the activities decreased differently for the two enzymes. While the two activities decreased proportionally for EstA, the esterase activity decreased at a higher rate for EST2. In fact, the ratio of esterase activity to thioesterase activity for EST2 at 30°C and pH 7.5 was about 10, while at 30°C and pH 5.5 with 5% NaCl the ratio was about 0.03 (Table 2).
TABLE 2.
Comparison of EST2 and EstA esterase and thioesterase activities determined by using pNP-hexanoate (100 μM) and S-methyl thiobutanoate (20 mM), respectively, as the substrates in milk- and cheese-like conditionsa
| Conditions | Activities with the following substrates (U/mg):
|
|||
|---|---|---|---|---|
|
pNP-hexanoate
|
S-Methyl thiobutanoate
|
|||
| EST2 | EstA | EST2 | EstA | |
| 30°C, pH 7.5 | 2,750.0 ± 150.0 | 90.0 ± 3.5 | 295.0 ± 3.0 | 53.0 ± 1.0 |
| 30°C, pH 7.5, 5% NaCl | 1,510.0 ± 80.0 | 48.0 ± 2.0 | 162.0 ± 1.5 | 20.0 ± 0.3 |
| 30°C, pH 5.5 | 119.0 ± 3.0 | 2.0 ± 0.1 | 13.9 ± 0.3 | 9.6 ± 0.2 |
| 30°C, pH 5.5, 5% NaCl | 0.56 ± 0.04 | 0.110 ± 0.003 | 1.80 ± 0.03 | 0.081 ± 0.007 |
| 20°C, pH 5.5, 5% NaCl | 0.24 ± 0.01 | 0.035 ± 0.004 | 1.10 ± 0.01 | NDb |
Assays were done in duplicate or triplicate, and the data are the means ± standard deviations of two independent experiments.
ND, not detectable.
Ester synthesis in a buffered system.
Cheese ripening is a very complex process, and flavor development is the result of different processes, such as fermentation, proteolytic and lipolytic hydrolysis, and ester and thioester synthesis (1, 5, 12, 13, 37, 43). Part of the work performed in this study was verifying that the thermostable esterase EST2 has the typical synthetic activities found in the esterases/lipases from lactic acid bacteria, as studies have shown that ester and thioester synthesis is catalyzed by esterases/lipases and is important for the development of the characteristic cheese flavors during ripening (5, 11, 26, 28). Nardi et al. (38) demonstrated that EstA is the main enzyme responsible for ester synthesis in L. lactis. Therefore, in this study a new system was developed to measure ester synthesis spectrophotometrically in an aqueous environment under conditions that simulated milk- and cheese-like conditions. First, we examined the reaction mechanism shown in Fig. 2, which is the mechanism generally accepted for esterases and lipases (http://bioweb.ensam.inra.fr/ESTHER/general). The ester degradation reaction is thought to be a typical reversible chemical reaction, and therefore some of the parameters that influence the reaction equilibrium were taken into account, including low pH, low temperature, and reduction in the water activity (aw) (11, 16). In this study the aw was modified by the addition of salt, sugar, and proteins by using concentrations which roughly mimicked milk- and cheese-like conditions, although we did not measure aw variations for the different added compounds and combinations of compounds. All parameters were investigated either separately or in combination.
FIG. 2.
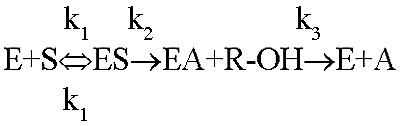
General mechanism of esterases. E, S, ES, EA, R-OH, and A refer to enzyme, substrate, enzyme-substrate complex, acyl enzyme, alcohol, and acid, respectively.
In order to obtain information about the enzyme behavior in milk- and cheese-like conditions, assays were performed at 20°C, at pH 5.5, and in the presence of 5% NaCl, 4% lactose, or 20 mg/ml BSA. These parameters were used separately and in various combinations, and 50 μM p-nitrophenol and 500 μM hexanoic acid were used as substrates. The synthesis of pNP-hexanoate was monitored by measuring the decrease in p-nitrophenol absorbance at 410 nm, and the results are shown in Table 3. It is worth noting that ester synthesis by EST2 occurred under all conditions tested and that at pH 7.5 the synthetic activity was lower than the activity under the corresponding conditions at pH 5.5. Maximal synthesis occurred at pH 5.5 in the presence of 5% NaCl. The level of synthesis decreased in the presence of BSA. As a control of enzyme activity and stability (Table 3), ester degradation assays were performed under the same conditions used for synthesis by using pNP-hexanoate as the substrate. The results obtained corresponded to the synthesis results because under the same conditions a low level of synthesis corresponded to a high level of degradation and vice versa.
Esterase and thioesterase activities in a cheese matrix and in milk.
After the EST2 synthetic activity in vitro was tested, it was important to demonstrate ester synthesis in a cheese matrix. For this experiment the following procedure was used. Ethanol (20 mg/kg) and hexanoic acid (200 mg/kg) were added as substrates to a cheese paste supplemented with NaCl (1%, wt/wt) and EST2 (0.35 ng/g). The same mixture without enzyme was used as a control. After 24 h of incubation at 17°C, the amount of ethyl-hexanoate synthesized was measured by HS-GC. No synthesis was observed in the absence of EST2, but 2.59 ± 0.50 mg/kg of ethyl-hexanoate was synthesized in the presence of EST2. This indicates that EST2 shows ester synthesis activity in a simulated cheese matrix. For comparison, EstA added at a concentration of 35 μg/10 g to a cheese-like system was reported to produce 32 mg/kg of ethyl hexanoate (11).
The esterase and thioesterase activities of purified EST2 and EstA were also measured following addition to HP milk. HP milk containing ethyl-hexanoate (5 mg/ml) or S-methyl thiobutanoate (150 μg/liter) was incubated in presence of EST2 (3 ng/ml) or EstA (3 ng/ml) at 30°C. At different times after the start of incubation, samples were analyzed by HS-GC to determine residual levels of ester and thioester. The results are shown in Fig. 3. EST2 degraded the ester in milk very efficiently. About 40% of the ester was degraded after 60 min of incubation and 70% was degraded after 16 h, whereas EstA degraded only 40% of the ester after 16 h of incubation. In milk EST2 degraded about 50% of the thioester after incubation, in contrast to the 30% degraded by EstA.
FIG. 3.
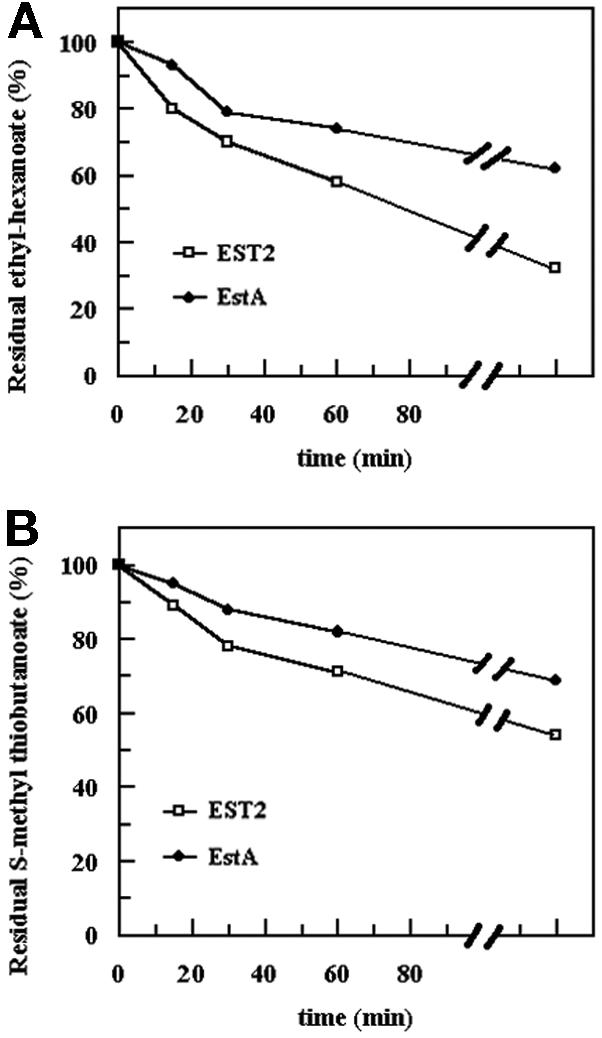
Degradation of esters and thioesters in milk. Ethyl-hexanoate (A; 5 mg/liter) or S-methyl thiobutanoate (B; 150 μg/liter) was added to the HP milk. The residual quantities of nonhydrolyzed esters and thioesters were determined by HS-GC. The data are the means of two or three different determinations.
Overexpression of EST2 in L. lactis and EST2 hydrolytic activities in a cheese paste model.
In order to test the EST2 esterase and thioesterase activities in a cheese model, an L. lactis strain that overproduced EST2 was constructed. The EST2 gene was subcloned in the vector pNZ8148 and used to transform L. lactis NZ9000 (as described in Material and Methods). This vector had an inducible promoter, PnisA (7), which was tightly regulated by nisin (closed in the absence of nisin and open in a dose-dependent manner in the presence of nisin) in strains containing the nisRK genes. Optimal conditions for EST2 expression were established by induction with different nisin concentrations. An uninduced culture (50 ml) was used as a control, and four cultures were induced with 1, 2, 5, and 10 ng/ml of nisin. After induction the cultures were incubated at 30°C for 90 min. The esterase activity obtained from each culture after harvesting by centrifugation, resuspension, and cell disruption by French cell pressure was normalized for the protein content and expressed in units per milliliter of crude extract (Fig. 4A). The change in expression was very clear because the increase in esterase activity was about 500-fold higher in the induced cultures. No significant difference in the expression level was observed between the cultures induced with different nisin concentrations. The expression was confirmed by Western blot analysis (Fig. 4B). Fifty micrograms of total proteins from each crude extract was loaded onto a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and transferred to a PVDF membrane developed with a specific anti-EST2 antibody (30). A band corresponding to EST2, which was included as a control (lane 1), was observed only in cell extracts induced with nisin, and the level of expression was the same in each extract (Fig. 4B, lanes 2 to 6). EST2 samples containing known concentrations were included as standards, and the levels of expression were about 400 ng/ml of culture.
FIG. 4.
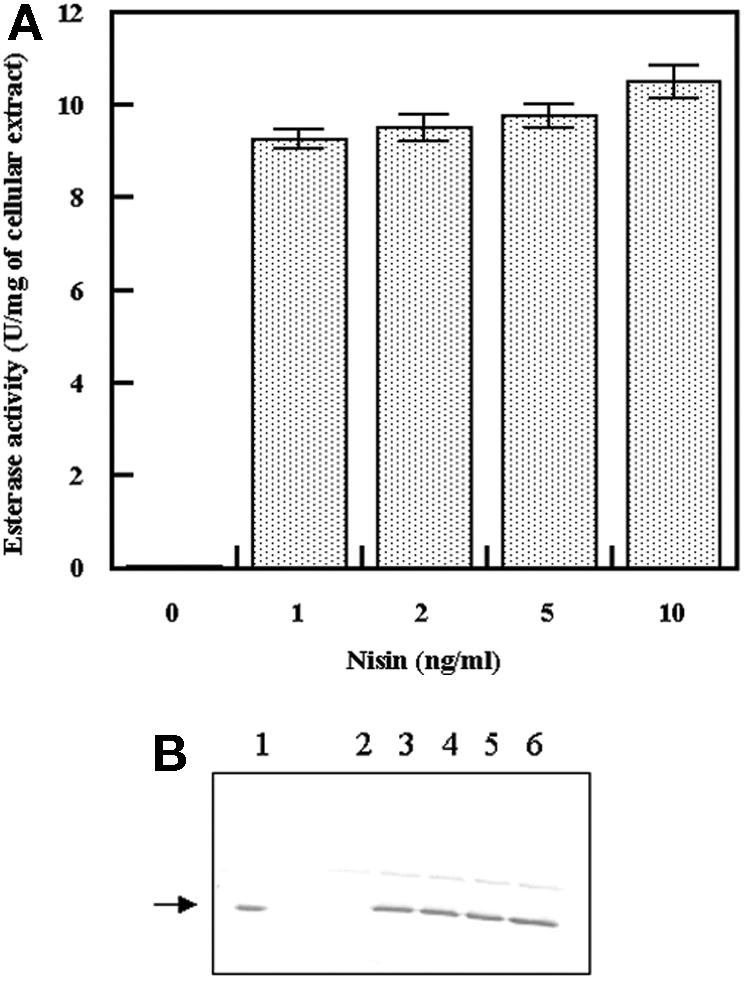
(A) Esterase activities with cellular extracts of L. lactis NZ9000 containing plasmid pNZ7408 after induction with different nisin concentrations. The assays were performed at 30°C and pH 7.5, using pNP-hexanoate as the substrate. (B) Western blot analysis of the cellular extracts used for the experiment whose results are shown in panel A. Lane 1, 200 ng of purified EST2; lane 2, 50 μg of uninduced L. lactis NZ9000/pNZ7408 crude extract; lanes 3 to 6, 50 μg of L. lactis NZ9000/pNZ7408 crude extract induced with 1, 2, 5, and 10 ng/ml nisin, respectively.
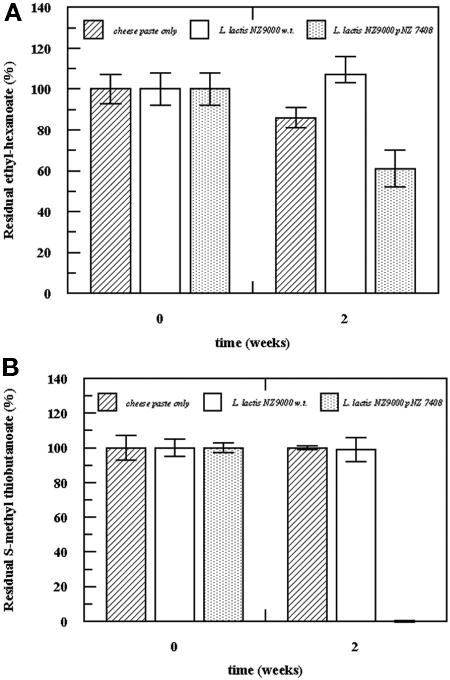
Cells of L. lactis NZ9000 and L. lactis NZ9000 harboring the pNZ7408 plasmid were added at a final OD600 of 0.5 to a cheese paste supplemented with additional ethyl-hexanoate (5 mg/kg) or S-methyl thiobutanoate (150 μg/kg). Cells containing pNZ7408 were induced with nisin at a final concentration of 2 ng/g. The control contained only substrates and L. lactis NZ9000 cells. The cheese paste samples were incubated for 2 weeks at 17°C, and the residual amounts of ester and thioester were determined. Figures 5A and B show that the ester and thioester were degraded only in the presence of EST2. L. lactis overproducing EST2 degraded 40% of the ester and all of the thioester, while in the controls no degradation of the ester or thioester was evident. This indicates that EST2 is also very active in the degradation of esters and thioesters under simulated cheese-like conditions.
FIG. 5.
Degradation of ester (A) and thioester (B) in cheese paste model system. Cells of wild-type L. lactis NZ9000 (w.t.) and L. lactis NZ9000 containing pNZ7408 (induced with 2 ng/ml nisin) at a final OD600 of 0.5 were added to cheese paste supplemented with 5 mg/kg of ethyl-hexanoate or 150 μg/kg of S-methyl thiobutanoate. Only sodium phosphate buffer was used in the control cheese paste. The cheese paste samples were incubated at 17°C, and samples were taken at zero time and 2 weeks. The residual amounts of ester and thioester were determined by HS-GC. The error bars indicate the standard deviations of triplicate experiments.
In conclusion, it appears that recombinant EST2 is more active than EstA in terms of esterase and thioesterase activities, as well as synthesis. As EST2 also has high thermostability and exhibits resistance to organic solvents, it could be successfully used in biotechnological applications in the dairy industry to develop characteristic flavors and/or to accelerate cheese ripening. Overexpression of EST2 in E. coli does not pose any particular problems in terms of yield and offers the advantage of rapid purification following thermal denaturation of host proteins (30). Therefore, recombinant EST2 could be tested in the future as an additive in cheese manufacturing by following the rules prescribed for food additives. On the other hand, overexpression of EST2 in L. lactis cells used as a starter or adjunct culture could have the advantage of better distribution in the cheese curd and more balanced enzymatic action. However, public concern about the use of genetically modified organisms and unclear European legislation restrict rather than prohibit use (40) of this manipulated strain.
Acknowledgments
This work was financially supported in part by the Ministry of University and Research of Italy (MIUR). L.M. was supported by a Ph.D. grant from University of Naples Federico II.
REFERENCES
- 1.Ayad, E. H. E., A. Verheul, W. J. M. Engels, J. T. M. Wouters, and G. Smit. 2001. Enhanced flavour formation by combination of selected lactococci from industrial and artisanal origin with focus on completion of a metabolic pathway J. Appl. Microbiol. 90:59-67. [DOI] [PubMed] [Google Scholar]
- 2.Barbieri, G., M. Bolzoni, M. Careri, A. Mangia, G. Parolari, S. Spagnoli, and R. Virgili. 1994. Study of the volatile fraction of Parmesan cheese. J. Agric. Food Chem. 42:1170-1176. [Google Scholar]
- 3.Bolotin, A., P. Wincker, S. Manger, O. Taillon, K. Malarme, J. Weissenbach, S. D. Ehlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacteria Lactococcus lactis ssp. lactis IL1403 genome. Genome Res. 11:731-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chich, J.-F., K. Marchesseau, and J.-C. Gripon. 1997. Intracellular esterase from Lactococcus lactis subsp. lactis NCDO 763: purification and characterization. Int. Dairy J. 7:169-174. [Google Scholar]
- 5.Cristiani, G., and V. Monnet. 2001. Food and micro-organisms and aromatic ester synthesis. Sci. Aliments 21:211-230. [Google Scholar]
- 6.Darland, G., and T. D. Brock. 1971. Bacillus acidocaldarius sp. nov., an acidophilic thermophilic spore-forming bacterium. J. Gen. Microbiol. 67:9-15. [Google Scholar]
- 7.De Ruyter, P. G. G. A., O. P. Kuipers, and W. M. De Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Simone, G., S. Galdiero, G. Manco, D. Lang, M. Rossi, and C. Pedone. 2000. A snapshot of a transition state analogue of a novel thermophilic esterase belonging to the subfamily of mammalian hormone-sensitive lipase. J. Mol. Biol. 303:761-771. [DOI] [PubMed] [Google Scholar]
- 9.Dixon, M., and E. C. Webb. 1979. Effect of pH on V, p. 148-154. In Enzymes, 3rd ed. Longman, London, United Kingdom.
- 10.El Soda, M., S. A. Madkor, and P. S. Tong. 2000. Adjunct cultures: recent developments and potential significance to the cheese industry. J. Dairy Sci. 83:609-619. [DOI] [PubMed] [Google Scholar]
- 11.Fenster, K. M., S. A. Rankin, and J. L. Steele. 2003. Accumulation of short n-chain ethyl esters by esterases of lactic acid bacteria under conditions simulating ripening Parmesan cheese. J. Dairy Sci. 86:2818-2825. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez, L., M. M. Beethuyzen, J. Brown, R. J. Siezen, T. Coolbear, R. Holland, and O. P. Kuipers. 2000. Cloning, characterization, controlled overexpression, and inactivation of the major tributyrin esterase gene of Lactococcus lactis. Appl. Environ. Microbiol. 66:1360-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, P. F., and T. Cogan. 2000. Cheese: scientific highlights of the 20th century, p. 83-121. In T. M. Cogan, P. L. H. McSweeney, and T. P. Guinee (ed.), 6th Cheese Symposium. Teagasc, Dublin, Ireland.
- 14.Fuji, T., N. Nagasawa, A. Iwamatsu, T. Bogaki, Y. Tamai, and M. Hamaci. 1994. Molecular cloning, sequence analysis, and expression of the yeast alcohol acetyltransferase gene. Appl. Environ. Microbiol. 60:2786-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gobbetti, M., P. F. Fox, and L. Stepaniac. 1996. Esterolytic and lipolytic activities of mesophilic and thermophilic lactobacilli. Ital. J. Food Sci. 2:127-135. [Google Scholar]
- 16.Ha, J. K., and R. C. Lindsay. 1992. Influence of aw on volatile free fatty acids during storage of cheeses bases lipolyzed by kid goat pregastric lipase. Int. Dairy J. 2:179-195. [Google Scholar]
- 17.Ha, J. K., and R. C. Lindsay. 1993. Release of volatile branched-chain and other fatty acids from ruminant milk fats by various lipases. J. Dairy Sci. 76:677-690. [DOI] [PubMed] [Google Scholar]
- 18.Hemilä, H., T. T. Koivula, and I. Palva. 1994. Hormone-sensitive lipase is closely related to several bacterial proteins, and distantly related to acetylcholinesterase and lipoprotein lipase: identification of a superfamily of esterases and lipases. Biochim. Biophys. Acta 1210:249-253. [DOI] [PubMed] [Google Scholar]
- 19.Holm, C., T. G. Kirchgessner, K. L. Svenson, G. Fredrikson, S. Nilsson, C. G. Miller, J. E. Shively, C. Heinzmann, R. S. Sparker, T. Mohandas, A. J. Lusis, P. Belfrage, and M. C. Schotz. 1988. Hormone-sensitive lipase: sequence, expression, and chromosomal localization to 19 cent-q13.3. Science 241:1503-1506. [DOI] [PubMed] [Google Scholar]
- 20.Hotelier, T., L. Renault, X. Cousin, V. Negre, P. Marchot, and A. Chatonnet. 2004. ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins. Nucleic Acids Res. 32(database issue):D145-D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kehadr, E. E., J. C. Vuillemard, and S. A. El Deeb. 2000. Accelerated cheddar cheese ripening with encapsulated proteases. Int. J. Food Sci. Technol. 35:483-495. [Google Scholar]
- 22.Kehadr, E. E., J. C. Vuillemard, and S. A. El Deeb. 2002. Acceleration of cheddar cheese lipolysis by using liposome-entrapped lipases. J. Food Sci. 67:485-492. [Google Scholar]
- 23.Kehadr, E. E., J. C. Vuillemard, and S. A. El Deeb. 2003. Impact of liposome-encapsulated enzyme cocktails on cheddar cheese ripening. Food Res. Int. 36:241-252. [Google Scholar]
- 24.Krejci, E., N. Duval, Clatonnet, A. P. Vincens, and J. Maussoliè. 1991. Cholinesterase-like domains in enzymes and structural proteins: functional and evolutionary relationships and identification of a catalytically essential aspartic acid. Proc. Natl. Acad. Sci. USA 88:6647-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lamberet, G., B. Auberger, and J. L. Bergere. 1997. Aptitude of cheese bacteria for volatile S-methyl thioester synthesis. II. Comparison of coryneform bacteria, Micrococcaceae and some lactic acid bacteria starters. Appl. Microbiol. Biotechnol. 47:393-397. [Google Scholar]
- 27.Langin, D., H. Laurell, L. Stenson-Holst, P. Belfrage, and C. Holm. 1993. Gene organization and primary structure of human hormone-sensitive lipase: possible significance of a sequence homology with a lipase of Moraxella TA144, an antarctic bacterium. Proc. Natl. Acad. Sci. USA 90:4897-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, S.-Q., R. Holland, and V. L. Crow. 1998. Ethyl butanoate formation by dairy lactic acid bacteria. Int. Dairy J. 8:651-657. [Google Scholar]
- 29.Manco, G., E. Adinolfi, F. M. Pisani, V. Carratore, and M. Rossi. 1997. Identification of an esterase from Bacillus acidocaldarius with sequence similarity to a hormone sensitive lipase subfamily. Protein Pept. Lett. 4:375-382. [Google Scholar]
- 30.Manco, G., E. Adinolfi, F. M. Pisani, G. Ottolina, G. Carrea, and M. Rossi. 1998. Overexpression and properties of a new thermophilic and thermostable esterase from Bacillus acidocaldarius with sequence similarity to hormone-sensitive lipase subfamily. Biochem. J. 332:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manco, G., E. Giosuè, D. S. Auria, P. Herman, G. Carrea, and M. Rossi. 2000. Cloning, overexpression, and properties of a new thermophilic and thermostable esterase with sequence similarity to hormone-sensitive lipase subfamily from the archaeon Archaeoglobus fulgidus. Arch. Biochem. Biophys. 373:182-192. [DOI] [PubMed] [Google Scholar]
- 32.Manco, G., L. Mandrich, and M. Rossi. 2001. Residues at the active site of the esterase 2 from Alicyclobacillus acidocaldarius involved in substrate specificity and catalytic activity at high temperature. J. Biol. Chem. 276:37482-37490. [DOI] [PubMed] [Google Scholar]
- 33.Mandrich, L., L. Merone, M. Pezzullo, L. Cipolla, F. Nicotra, M. Rossi, and G. Manco. 2005. Role of the N terminus in enzyme activity, stability and specificity in thermophilic esterases belonging to the HSL family. J. Mol. Biol. 345:501-512. [DOI] [PubMed] [Google Scholar]
- 34.McSweeney, P. L. H., P. F. Fox, J. A. Lucey, K. N. Jordan, and T. M. Cogan. 1993. Contribution of the indigenous microflora to the maturation of cheddar cheese. Int. Dairy J. 3:613-634. [Google Scholar]
- 35.Mendìa, C., F. C. Ibanez, P. Torre, and Y. Barcina. 1999. Effect of pasteurization on the sensory characteristics of a ewe's cheese. J. Sens. Stud. 14:415-424. [Google Scholar]
- 36.Moio, L., and F. Addeo. 1998. Grana Padano cheese aroma. Int. Dairy Res. 65:317-333. [Google Scholar]
- 37.Molimard, P., and H. E. Spinnler. 1996. Compounds involved in the flavor of surface mold-ripened cheeses: origin and properties. J. Dairy Sci. 79:169-184. [Google Scholar]
- 38.Nardi, M., C. Fiez-Vandal, P. Tailliez, and V. Monnet. 2002. The EstA esterase is responsible for the main capacity of Lactococcus lactis to synthesize short chain fatty acid esters in vitro. J. Appl. Microbiol. 93:994-1002. [DOI] [PubMed] [Google Scholar]
- 39.Ortigosa, M., P. Torre, and J. M. Izco. 2001. Effect of pasteurization of ewe's milk and use of a native starter culture on the volatile components and sensory characteristics of Roncal cheese. J. Dairy Sci. 84:1320-1330. [DOI] [PubMed] [Google Scholar]
- 40.Pedersen, M. B., S. L. Iversen, K. I. Soerensen, and E. Johansen. 2005. The long and winding road from the research laboratory to industrial applications of lactic acid bacteria. FEMS Microbiol. Rev. 29:611-624. [DOI] [PubMed] [Google Scholar]
- 41.Quian, M., and G. Reineccius. 2002. Identification of aroma compounds in Parmigiano-Reggiano cheese by gas chromatography/olfactometry. J. Dairy Sci. 85:1362-1369. [DOI] [PubMed] [Google Scholar]
- 42.Smit, G., A. Braber, W. van Spronsen, G. van der Berg, and F. A. Exterkate. 1995. Ch-easy model: a cheese-based model to study cheese ripening, p. 185-190. In Bioflavour 95. Dijon, Paris, France.
- 43.Smit, G., B. A. Smit, and W. J. Engels. 2005. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29:591-610. [DOI] [PubMed] [Google Scholar]
- 44.Uren, J. R. 1987. Cystathionine β-lyase from Escherichia coli. Methods Enzymol. 143:483-486. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson, M., T. Guinee, D. O'Callaghan, and P. Fox. 1992. Effects of commercial enzymes on proteolysis and ripening in cheddar cheese. Lait 72:449-458. [Google Scholar]