Abstract
To resolve the fine-scale architecture of anoxic protistan communities, we conducted a cultivation-independent 18S rRNA survey in the superanoxic Framvaren Fjord in Norway. We generated three clone libraries along the steep O2/H2S gradient, using the multiple-primer approach. Of 1,100 clones analyzed, 753 proved to be high-quality protistan target sequences. These sequences were grouped into 92 phylotypes, which displayed high protistan diversity in the fjord (17 major eukaryotic phyla). Only a few were closely related to known taxa. Several sequences were dissimilar to all previously described sequences and occupied a basal position in the inferred phylogenies, suggesting that the sequences recovered were derived from novel, deeply divergent eukaryotes. We detected sequence clades with evolutionary importance (for example, clades in the euglenozoa) and clades that seem to be specifically adapted to anoxic environments, challenging the hypothesis that the global dispersal of protists is uniform. Moreover, with the detection of clones affiliated with jakobid flagellates, we present evidence that primitive descendants of early eukaryotes are present in this anoxic environment. To estimate sample coverage and phylotype richness, we used parametric and nonparametric statistical methods. The results show that although our data set is one of the largest published inventories, our sample missed a substantial proportion of the protistan diversity. Nevertheless, statistical and phylogenetic analyses of the three libraries revealed the fine-scale architecture of anoxic protistan communities, which may exhibit adaptation to different environmental conditions along the O2/H2S gradient.
Anoxic environments have occurred throughout the Earth's history; at the beginning of eukaryote evolution, the oxygen level was about 1% of the present atmospheric level (64). Such environments may harbor eukaryote taxa that (i) have remained isolated from changes in global environmental conditions; (ii) have retained some ancestral characteristics that have been lost in all other extant eukaryotes; and (iii) may be specifically adapted to oxygen-depleted conditions and therefore unlikely to occur in other environments. The relative patchiness and isolation of anoxic systems may provide barriers to gene flow and to dispersal in local genetic radiations. These hypotheses make anaerobic biota a subject of major interest for evolution, ecology, physiology, and diversity research (20). However, gaining insight into the structure of anaerobic communities, which consist almost exclusively of pro- and eukaryotic microbes (20), was and still is hampered by methodological difficulties, since trace amounts of oxygen inhibit or kill many of these microbes. In the past few years, a molecular approach has been successfully used to access microbial communities even in the most extreme environments (3, 5, 40). This approach is based on phylogenetic analysis of eukaryotic 18S rRNA gene sequences (18S rRNA) that have been amplified and cloned from environmental samples (10).
To date, eight environmental 18S rRNA surveys have focused on protistan diversity in aquatic anoxic habitats (16, 17, 41, 43, 71, 72, 73, 75). Most of these surveys detected unexpectedly high species richness, with several highly divergent 18S rRNA gene sequences (16, 17, 41, 72) possibly representing valid novel phylotypes at high taxonomic ranks (6, 61). A meta-analysis of environmental 18S rRNA surveys identified site-specific anoxic sequence groups (61). This seems to contrast with the hypothesis of global dispersal of microbes (22). However, other 18S rRNA surveys of anoxic aquatic environments revealed relatively low phylogenetic richness of protistan communities (43, 75) and no indication of novel high-level eukaryotic lineages (43). This supports the recent hypothesis that there may be few, if any, previously unknown protist phyla or “new kingdoms” (11).
Some reasons for these contrasting statements are evident. First, most of the anoxic clone libraries published to date are heavily undersampled (73), and none of the corresponding studies used statistical analysis to estimate the phylotype richness. Second, most of the previous studies were based on PCR amplicons generated with a single PCR primer set, which captured only a specific fraction of the community (1, 10). Third, in the past, chimeric sequences have been incorporated into phylogenetic analyses, which led to a putatively high level of eukaryote diversity at the mega-evolution level in a few cases (6). Fourth, insufficient taxon sampling in a phylogenetic analysis may produce spurious novel eukaryotic phyla and unwarranted statements about “early branching” eukaryote phyla (11). Finally, in most previous diversity studies of anoxic environments systematic sampling was not performed along an O2/H2S gradient, despite evidence from previous microscopic (19) and molecular (72) analyses that the protistan community structure seems to change significantly along such a gradient.
In the present study we attempted to reduce these biases. We analyzed 1,100 18S rRNA PCR amplicons retrieved from a supersulfidic anoxic fjord in Norway (Framvaren), which makes our study one of the largest environmental molecular diversity surveys available. We used the multiple-primer approach to reduce PCR-related biases (73) and used several state-of-the-art methods to identify potentially chimeric sequences. Furthermore, instead of taking a snapshot of anoxic protistan diversity by sampling a single “spot” within the anoxic system, we analyzed three different samples obtained along the O2/H2S gradient. For the first time, all of these factors were used in concert to obtain better resolution and insight into an anoxic protistan community. To assess phylotype richness in all three communities, we used both parametric abundance models (which recently proved to be useful in a prokaryotic diversity study [31]) and nonparametric estimators (13). Our study significantly increased the number of available environmental 18S rRNA gene sequences, particularly from anoxic sampling sites, and thus led to better phylogenetic resolution for eukaryotic lineages that suffer from low sampling (16).
MATERIALS AND METHODS
Sampling site and procedure.
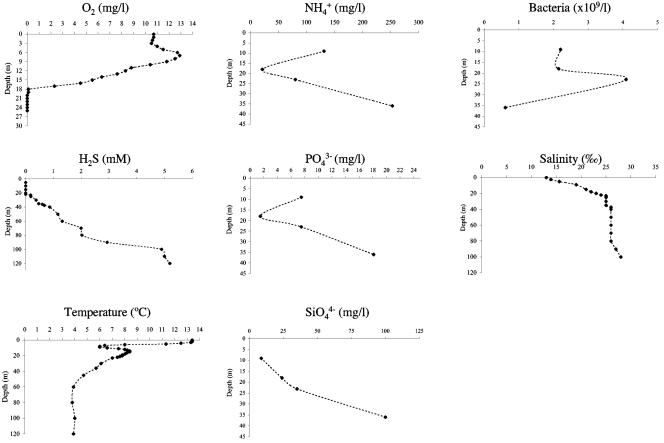
The Framvaren Fjord is located in southwestern Norway. With sulfide levels in the bottom water 25 times greater than those in the Black Sea (68), this fjord represents the extreme of anoxicity and contains the highest levels of H2S (6,000 μM) ever reported for an open anoxic basin (52). Our sampling site was located in the central basin of the Framvaren Fjord at 58°09′N, 06°55′E, with a water depth of 180 m. Physicochemical characteristics at the time of sampling (May 2004) are given in Fig. 1. Samples for protistan diversity studies were collected at three different depths. At 18 m, the oxic-anoxic boundary layer was chemically characterized by an oxidation peak of ammonia (Fig. 1). At 23 m, the pycnocline (Fig. 1) and upper H2S boundary (0.18 mmol liter−1) (Fig. 1) had a peak in bacterial abundance. The water collected from this depth was characterized by a purple-pink color that indicated that phototrophic purple sulfur bacteria were dominant. At 36 m, the H2S concentration (0.6 mM) was twice as high as the concentration in the deep water (>1,000 m) of the Black Sea (78) and 30 times higher than the concentration in the Cariaco bottom water (1,200 m), which is the world's largest truly marine permanently anoxic deep-sea basin (72). This sampling depth was the upper layer of the anoxic intermediate deep water, which was characterized by steep chemical gradients (Fig. 1). The bacterial counts were significantly lower than those in the upper water (Fig. 1).
FIG. 1.
Characteristics of the sampling site in May 2004.
Samples were taken aboard a small vessel using a tetrafluoroethylene-lined 5-liter Niskin bottle (Hydrobios, Kiel, Germany). To prevent exposure of the anoxic water to atmospheric oxygen, sampling was performed as described previously (72).
Analyses of sampling site characteristics.
Physicochemical parameters were measured onboard using an EOT 196 oxygen probe connected to an Oxi 196 microprocessor (WTW, Germany), an IJ444/Meto temperature probe (GAT, Germany) together with a Portamess 651-2 microprocessor (Knick, Germany), and a Ref211 precision salinity/automatic temperature compensation refractometer (Kuebler, Germany). Hydrogen sulfide, phosphate, ammonium, and silicon levels were determined spectrophotometrically immediately after sampling, based on the methylene blue method (14), the trivalent antimony method (56), the indophenol blue method (36), and the α-silicomolybdic acid method (29), respectively, using a LASA 10/Plus spectrophotometer (Lange, Germany).
For bacterial counts, water samples were preserved with 1% (final concentration) glutaraldehyde. Standard 4′,6′-diamidino-2-phenylindole (DAPI)-stained slides with different volumes of sample water were prepared on black 0.2-μm Poretics polycarbonate membranes for enumeration of prokaryotes by epifluorescence microscopy (58). Protistan abundance was estimated by using fluorescein isothiocyanate/DAPI-stained preparations (0.8-μm-pore-size polycarbonate filter) (65).
DNA extraction and 18S rRNA gene amplification.
Protists were collected on 47-mm Durapore membranes (pore size, 0.65 μm) as described previously (72). Immediately after filtration (5 to 11 liters per filter), membranes were individually frozen (ca. −200°C) in DNA extraction buffer with proteinase K (final concentration, 100 μg ml−1) (71). High-molecular-weight DNA was extracted as described by Stoeck et al. (72). In brief, the samples were heated to 65°C for 2 h in the extraction buffer. Then the lysates were purified by extraction with an equal volume of chloroform-isoamyl alcohol (24:1) and precipitated with 0.7 volume of isopropanol. Potential inhibitors of downstream applications were removed by DNA purification with the resin-based Wizard DNA clean-up system (Promega, Madison, WI). The integrity of the total DNA was checked by agarose gel electrophoresis (0.8%).
Proceeding according to the multiple-primer approach (73), we amplified fragments of the 18S rRNA gene whose sizes ranged from nearly full length to ∼1,100 bp, using four different primer sets (Table 1). PCRs were performed as described previously (72).
TABLE 1.
Primer sequences used in this study for specific amplification of 18S rRNA gene sequences from environmental DNAa
| Primer set | Primer | Sequence (5′-3′) | Reference |
|---|---|---|---|
| 1 | EukA | AAC CTG GTT GAT CCT GCC AGT | 51 |
| EukB | TGA TCC TTC TGC AGG TTC ACC TAC | 51 | |
| 2 | Euk82F | GAA DCT GYG AAY GGC TC | 40 |
| U1391R | GGG CGG TGT GTA CAA RGR | 37 | |
| 3 | Euk360F | CGG AGA RGG MGC MTG AGA | 51 |
| U1517R | ACG GCT ACC TTG TTA CGA CTT | 66 | |
| 4 | 18S-6-Cil-F | AAY CTG GTT GAT CCT GCC AG | 28 |
| 18S-1101-Cil-R | AGG YTR AGG TCT CGT TCG TT | 28 |
Mixed base sites are designated according to the IUB code. F, forward; R, reverse; Cil, ciliate specific; Euk, eukaryote specific; U, universal. The numbers refer to Escherichia coli 16S rRNA gene positions. Primer EukA is a forward primer referring to Saccharomyces cerevisiae 18S rRNA gene positions 2 to 22. Primer EukB is a reverse primer referring to S. cerevisiae 18S rRNA gene positions 1795 to 1772. In the OTU analysis of the pooled data sets only overlapping regions were considered in each case. A figure showing the fragment overlaps between the different primer sets is provided in the supplemental material.
Clone library construction.
The PCR products were used to construct clone libraries from the oxic-anoxic boundary layer (18 m), from just below the redox transition zone (23 m), and from the intermediate anoxic deep water (36 m) with a pGEM-T vector system cloning kit (Promega, Madison, WI). Plasmids were isolated from overnight cultures using a 96-well Directprep kit (QIAGEN, Valencia, CA). Between 350 and 400 clones per depth, nearly evenly distributed among the different primer sets (Table 1), were partially sequenced (M13F sequencing primer) at MWG-Biotech (Ebersheim, Germany), using an Applied Biosystems 3730 DNA Stretch sequence with the XL upgrade and an Applied Biosystems Prism BigDye Terminator version 3.1 cycle sequencing Ready Reaction kit. Initially, partial sequences of each primer set were grouped separately into operational taxonomic units (OTUs) using the program DOTUR (63) based on a 98.0% sequence similarity cutoff value. Selected clones of each OTU were then sequenced bidirectionally. Sequence quality assessments, PHRED and PHRAP analyses, and assembly were performed with the program CodonCode Aligner v. 1.2.4 (CodonCode Corporation, Dedham, MA). Subsequently, complete sequences were finally grouped based on a 98% similarity level using DOTUR (63). Low-quality sequence reads and nontarget metazoan sequences were excluded from the phylogenetic analyses.
Phylogenetic analyses.
Environmental 18S rRNA gene sequences initially were compared to sequences in the GenBank database using gapped BLAST analysis (2) to determine their approximate phylogenetic affiliations. Environmental sequence data together with the closest GenBank matches were compiled in ARB (42) and were aligned using the ARB FastAligner utility. Alignments were manually refined using phylogenetically conserved secondary structures. The conserved and unambiguously aligned positions (the numbers are indicated in Fig. 5 to 8) were used in subsequent phylogenetic analyses. Potentially chimeric sequences were identified using secondary structure predictions, the Chimera_Check command (version 2.7) provided by Ribosomal Database Project II (47), and partial treeing analyses (62). Classification of unique phylotypes was performed by two phylogenetic inference methods: minimum evolutionary distance and maximum likelihood. Trees were constructed using the PAUP* 4.0b10 software package (74). All heuristic searches were performed using random, stepwise addition of taxa with the TBR branch-swapping algorithm. We used the program Modeltest (59) to choose the models of DNA substitution that best fit our data sets from among 56 possible models. Modeltest was run for each individual data set. The DNA substitution models, as well as the parameter settings for each tree constructed, are described in the figure legends. We assessed the relative stability of tree topologies using 1,000 distance bootstrap replicates (500 maximum likelihood bootstrap replicates in the case of the stramenopile tree). For heuristic searches for bootstrap analyses we employed stepwise addition starting trees with simple addition of sequences and TBR branch swapping.
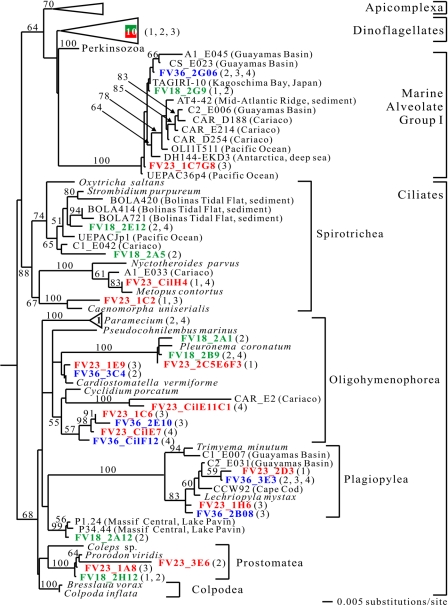
FIG. 5.
Minimum evolution phylogenetic tree of eukaryotic 18S rRNA gene sequences showing the positions of alveolate sequences. The tree was constructed with maximum likelihood criteria by using a GTR+I+G DNA substitution model with the variable-site gamma distribution shape parameter (G) at 0.5647, the proportion of invariable sites (I) at 0.0734, and the base frequencies and rate matrix for the substitution model suggested by Modeltest (59), based on 1,005 unambiguously aligned positions. Distance bootstrap values greater than 50% from an analysis of 1,000 bootstrap replicates are indicated at the nodes. Sequences recovered from the Framvaren Fjord are in bold. The numbers in triangles indicate the numbers of FV library sequences identified in taxonomic units. The numbers in parentheses indicate the primer sets (Table 1), and the colors indicate the three different libraries, as follows: green, FV18 (18 m); red, FV23 (23 m); and blue, FV36 (36 m).
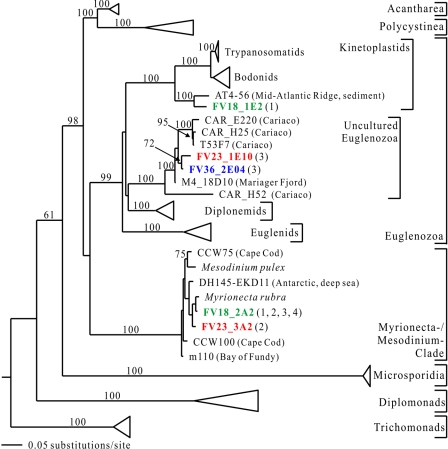
FIG. 8.
Minimum evolution phylogenetic tree of eukaryotic 18S rRNA gene sequences showing the positions of early branching sequences. The tree was constructed with maximum likelihood criteria by using a GTR+I+G DNA substitution model with the variable-site gamma distribution shape parameter (G) at 0.7597, the proportion of invariable sites (I) at 0.0990, and the base frequencies and rate matrix for the substitution model suggested by Modeltest (59), based on 924 unambiguously aligned positions. For additional details see the legend to Fig. 5.
Protistan richness estimates.
To estimate the protistan phylotype richness in each sample, we used two families of statistical procedures (for a summary of the theory see reference 12 and http://www.stat.cornell.edu/∼bunge/). In brief, the first family of procedures consisted of fitting several parametric statistical models to the observed frequency counts by the maximum likelihood method. We considered seven such models: the ordinary Poisson (which assumes equal phylotype abundances), the gamma-mixed Poisson (negative binomial), the inverse Gaussian-, lognormal-, and Pareto-mixed Poisson, and two finite mixture models defined by mixtures (convex combinations) of two or three geometric distributions (exponential-mixed Poissons). At present, there is no convincing theoretical justification for the use of any particular abundance distribution in this application, despite some work in this area (31). Consequently, our approach was empirical; we sought a model that fit the data well, yielded reasonable standard errors, and was mathematically parsimonious (i.e., had a small number of parameters; all the candidates above met this criterion except the mixture of three geometrics, which was not competitive). In some cases no known parametric model fit an entire data set well, so we considered data subsets consisting of the observed frequency counts from 1 up to some maximum value, which we called the right truncation point. We fit all models at all right truncation points and selected the “best of the best” based on goodness of fit (as defined by two chi-square statistics, one a simple measure of discrepancy and the other an asymptotically correct test statistic for model fit), the minimal standard error (among available fitted models), and the maximal data usage (highest right truncation point). Our computer programs are based on a locally adaptive nested EM algorithm and are written in Maple; they are available on our website (http://www.stat.cornell.edu/∼bunge/).
We then applied the second family of procedures, the coverage-based nonparametric estimators ACE and ACE1, to the data sets corresponding to the right truncation point(s) selected as “best” as described above. These estimators are from the studies of Chao (13) and Chao and Bunge (12); ACE1 is a modification of ACE intended for a highly heterogeneous community. To calculate these estimators, their standard errors, and related statistics, we used the software SPADE (http://chao.stat.nthu.edu.tw/). We selected the “best” of these estimators using criteria from the research literature as reported in the SPADE documentation; these procedures did not use parametric models so goodness of fit did not apply. (There is also a recently developed nonparametric maximum likelihood procedure for this problem [8], but we have not applied it yet.) For a discussion of some advantages of parametric models over coverage-based nonparametric estimators in this setting, see reference 31.
Nucleotide sequence accession numbers.
The gene sequences used in this study have been deposited in the GenBank database under accession numbers DQ310187 to DQ310369.
RESULTS AND DISCUSSION
General protistan community composition along an O2/H2S gradient.
We compared three eukaryotic clone libraries along an O2/H2S gradient in the anoxic Framvaren Fjord. These libraries were designated FV18, FV23, and FV36, corresponding to the photic micro-oxic interface (18 m), the lower redox transition zone and upper H2S boundary (23 m), and a highly sulfidic layer with low microbial abundance (36 m) (Fig. 1). Of 1,100 clones analyzed, 239 resulted in failed sequence reads due to insufficient template quality and vectors without an insert. Thus, we obtained a total of 861 partial sequences (ca. 500 to 700 bp), 324 of which were from library FV18, 284 of which were from library FV23, and 253 of which were from library FV36. The libraries contained 18 (18 m), 16 (23 m), and 5 (36 m) nontarget sequences (metazoa, higher plants, bacteria, archaea), which, together with 5 chimeric and 64 low-quality sequences, were excluded from further analyses. The final phylogenetic analyses included a total of 753 protistan sequences. These sequences were grouped into 39 unique phylotypes for the 18-m data set, 60 unique phylotypes for the 23-m data set, 18 unique phylotypes for the 36-m data set, and 92 unique phylotypes for the combined data set.
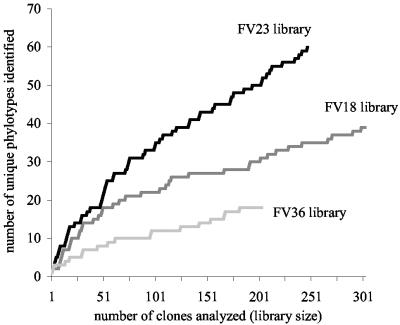
The Framvaren clone libraries significantly enlarge the inventory of protistan diversity in anoxic marine environments. Only a few sequences are closely related to sequences found in previous studies of anoxic systems. Although our data set is among the largest 18S rRNA inventories published to date, the number of sequences that we analyzed is insufficient to reveal the protistan richness in our samples (Fig. 2). We therefore used statistical methods to estimate the protistan phylotype richness. In general, all of the methods (parametric and nonparametric) yielded comparable estimates (taking standard errors into account) for each data set studied here (Table 2). It is interesting that in every case the preferred parametric model was the inverse Gaussian-mixed Poisson model, although this (unfortunately) does not imply that this model is universally applicable to microbial (or other richness) data. The Pareto model tended to underfit the number of singletons (phylotypes observed once), and the lognormal and two-mixed geometric/exponential models tended to have slightly larger standard errors. (The Poisson, negative binomial, and three-mixed geometric/exponential models were noncompetitive in this analysis, and the results are not reported here.) Finally, we noted that the preferred coverage-based nonparametric estimator was ACE1 in every case.
FIG. 2.
Sampling saturation profile (phylotype accumulation curve). The number of phylotypes is plotted as a function of the number of clones sampled. Clone samples were randomly resampled to completion without replacement to quantify coverage of phylotype diversity. Phylotypes were defined to encompass clones that exhibited at least 98.0% sequence similarity based on a pairwise comparison of the 18S rRNA gene sequences.
TABLE 2.
Protistan diversity estimates along a vertical O2/H2S gradient in the anoxic Framvaren Fjorda
| Library | Parameter | Parametric model
|
Nonparametric estimator
|
||||
|---|---|---|---|---|---|---|---|
| Inverse Gaussian | Lognormal | Pareto | Two-mixed exponential | ACE | ACE1 | ||
| FV18 | Phylotype estimate | 63.8 | 65.4 | 63.0 | 68.4 | 65.5 | 85.4 |
| SE | 14.6 | 20.8 | 11.8 | 14.9 | 13.1 | 28.6 | |
| Naïve goodness of fit | 0.2595 | 0.1877 | 0.1889 | 0.3615 | NPb | NP | |
| Asymptotically correct goodness of fit | 0.3613 | 0.3798 | 0.3739 | NAc | NP | NP | |
| FV23 | Phylotype estimate | 146.5 | 138.9 | 129.7 | 143.2 | 118.5 | 167.6 |
| SE | 46.2 | 56.6 | 29.8 | 47.1 | 22.7 | 53.6 | |
| Naïve goodness of fit | 0.4884 | 0.3744 | 0.3006 | 0.5071 | NP | NP | |
| Asymptotically correct goodness of fit | 0.4616 | 0.4103 | 0.3607 | 0.4094 | NP | NP | |
| FV36 | Phylotype estimate | 27.4 | 27.7 | 27.6 | 31.9 | 26.3 | 30.7 |
| SE | 8.1 | 10.7 | 6.4 | 11.7 | 6.5 | 11.5 | |
| Naïve goodness of fit | 0.1362 | 0.1017 | 0.0866 | 0.1458 | NP | NP | |
| Asymptotically correct goodness of fit | NA | NA | NA | NA | NP | NP | |
Phylotype groups are based on 98.0% sequence similarity. Four of seven parametric models tested gave statistically well-supported results for all data sets.
NP, estimation not possible.
NA, not available.
The estimates are quite different for the three libraries that were constructed. The highest phylotype richness was calculated for the FV23 library from the upper H2S boundary (147 phylotypes [standard error, 46 phylotypes]), corroborating observations obtained by light and fluorescence microscopy that revealed remarkably diverse morphologies (not shown). This was not unexpected, as it was hypothesized that chemoautotrophy, the dominant microbial process in such environments, supports a secondary microbial food web and stimulates the growth of bacterivorous protists (77). In contrast, 64 phylotypes (standard error, 15 phylotypes) were estimated for the H2S-free oxic-anoxic boundary layer (FV18), and only 27 phylotypes (standard error, 8 phylotypes) were estimated for the highly sulfidic waters (FV36). The hypothesis that there was equal richness in all three communities was rejected by a conservative, asymptotically valid, Bonferroni-corrected test at a level of α = 0.05. As the numbers were restricted to the time of sampling and the volumes of sample water used for DNA extraction, it is likely that they underestimated the true richness in this environment. Still, these snapshots strongly indicate that the protistan communities in this vertical physicochemical gradient in the water column differ from each other.
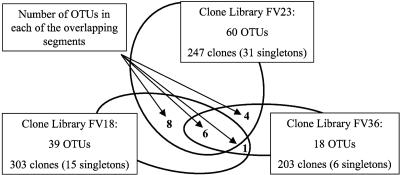
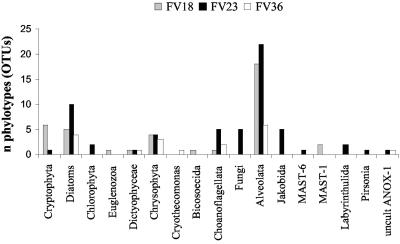
This communal division is supported by two further results: (i) the overlap of phylotypes between the three libraries is only marginal (1 to 8 OTUs) (Fig. 3) and (ii) the distributions of the 17 major eukaryotic phyla detected in the three libraries are very unequal (Fig. 4). Two of the libraries (FV18 and FV23) were dominated by alveolates (mainly ciliates), followed by stramenopiles (mainly diatoms and chrysophytes). In the FV36 library, alveolates were second only to stramenopiles. Cryptophytes were abundant only in the deep suboxic library (FV18). Other noteworthy taxonomic groups are fungi and jakobid flagellates, each represented by five phylotypes exclusively in the FV23 library. Also, the number of choanoflagellates peaked in this community. As our libraries were not sampled to saturation (Fig. 2), we did not wish to base comparisons of libraries on rare phylotypes that appeared only once or twice in libraries (e.g., MAST-6 and Euglenozoa) (Fig. 4).
FIG. 3.
Phylotypes (OTUs) shared by the three 18S rRNA clone libraries along the vertical O2/H2S gradient in Framvaren Fjord. The area of an oval is proportional to the size of the corresponding clone library. The numbers in the overlapping areas are the numbers of OTUs shared by the relevant libraries, and the overlap area is proportional to the amount.
FIG. 4.
Taxonomic distribution of 18S rRNA phylotypes retrieved from three protistan communities along the vertical O2/H2S gradient in Framvaren Fjord. Phylotypes were defined to encompass clones that exhibited at least 98.0% sequence similarity based on a pairwise comparison of the 18S rRNA gene sequences.
The sequences belonging to each phylum detected were generally highly diverse. Only a few of the clones were closely related to named taxa in established clades (e.g., FV18_1C7 was 99.64% similar to Gyrodinium fusiforme).
Phylogeny.
Most of the sequences represented novel phylogenetic lineages at different taxonomic levels. Below we discuss the detailed phylogeny of the sequences from Framvaren waters. For this reason we divided the eukaryotic tree of life into four different partial trees.
(i) Alveolates.
It is no surprise that the most abundant and diverse taxonomic group identified in the alveolates (33 unique phylotypes) was the ciliates (22 unique phylotypes) (Fig. 4 and 5). These organisms are major consumers of bacteria, and many of them have independently adapted to an anoxic lifestyle (20). Known groups of anaerobic and micro-oxic ciliates include the families Plagiopylidae, Strombiidae, Nyctotheridae, Cycliidae, and Prorodontidae, all of which have been retrieved from anoxic Framvaren waters.
We detected ciliate phylotypes belonging to 4 of 10 major classes defined by Lynn and Small (45). Some of these sequences were closely related to known species or genera (e.g., FV23_2C5E6F3 exhibited 98.7% sequence similarity to Pleuronema coronatum), while others were very distant from sequenced species and formed novel clades in previously described classes (e.g., the novel oligohymenophorean clade FV23_1C6, FV36_2E10, FV23CilE7, and FV36_CilF12). In the case of the FV18_2A12 sequence, it was not even possible to place it in a particular class. Together with two other environmental sequences (38), this sequence formed a well-supported, deeply branching clade, which appears to be a sister group of the established ciliate class Plagiopylea and may deserve designation at the same taxonomic level. This is remarkable because the ciliates are among the best-described protistan groups, and after two centuries of research on their systematics (18, 23, 24, 25, 26, 34) some investigators contend that most, if not all, ciliate species have already been described (21, 22). Whatever the correct taxonomic status of the clade that was discovered, its very existence suggests that it is too early to establish limits for protistan diversity even for the best-studied taxa.
In contrast to previous studies of anoxic systems (71, 72, 73), we could not detect apicomplexan or perkinsozoan sequences. However, we recovered 10 unique dinoflagellate phylotypes, all of which were relatively closely (92.8 to 99.6%) related to named species with a global distribution in a wide range of different habitats.
In all three libraries we detected sequences (FV18_2G9, FV23_1C7G8, and FV36_2G06) branching in uncultured marine alveolate group I (40). This sequence clade is an enigma, as we have no idea about its cellular identity and ecological role. However, it is a frequent constituent of 18S rRNA clone libraries of anoxic marine systems (71, 72, 73).
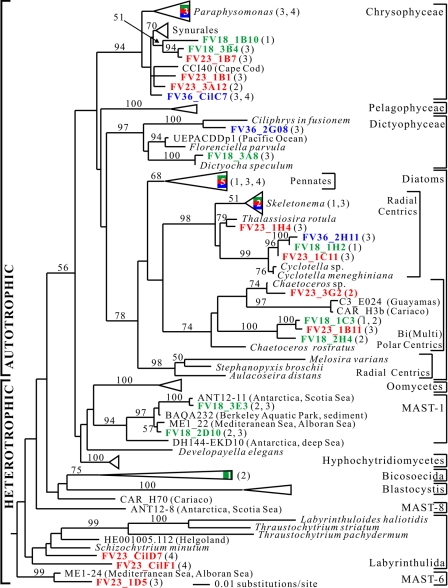
(ii) Stramenopiles.
Our phylogenetic analysis supported the monophyly of phototrophic stramenopiles (Fig. 6). Enlarging the stramenopile sequence database should help resolve the controversy regarding the evolution of this group (summarized in references 7 and 50). We recovered numerous phylotypes (n = 23) related to the phototroph stramenopiles from all three sampling depths (Fig. 6). At first, this finding may seem unusual, as algal photosynthesis at depths below 18 m is restrained by light availability. However, some of these organisms, especially the chrysophytes and diatoms, in fact have heterotrophic capability and are able to prosper in oxygen-depleted environments without light (15, 30). For example, this is the case for the diatom Cyclotella meneghiniana (44), to which some of our clones (FV18_1H2, FV23_1C11B9, and FV36_2H11) are closely related (97.7 to 98.4%).
FIG. 6.
Maximum likelihood tree of eukaryotic 18S rRNA gene sequences showing the positions of stramenopile sequences. The tree was constructed by using a GTR+I+G DNA substitution model with the variable-site gamma distribution shape parameter (G) at 0.5436, the proportion of invariable sites (I) at 0.2751, and the base frequencies and rate matrix for the substitution model suggested by Modeltest (59), based on 1,361 unambiguously aligned positions. Maximum likelihood bootstrap values greater than 50% from an analysis of 500 bootstrap replicates are indicated at the nodes. For additional details see the legend to Fig. 5.
These results illustrate an evident bias of environmental 18S rRNA clone libraries: as a rule, DNA extracted from environmental material does not allow identification of active members of the extant microbial community being studied. A solution to this problem might be to target RNA molecules directly instead of their genes. As opposed to rRNA genes, significant quantities of rRNA are produced only in active microbes (46).
The heterotrophic stramenopiles detected in our study were restricted to the chemocline (18 m) and lower redox transition zone (23 m) (Fig. 6). Two sequences found exclusively below the chemocline (FV23_CilD7 and FV23_CilF1) are related to the globally distributed osmotroph labyrinthulids, which typically inhabit deep-sea environments (60). Some of them are known to be anaerobes (57). Clone FV23_1D5 belongs to the uncultured stramenopile cluster MAST-6, which is characterized by novel stramenopiles that are found sporadically and are probably minor components of marine picoeukaryote assemblages (50). Five other phylotypes detected exclusively at the suboxic chemocline branched within the MAST-1 uncultured stramenopile cluster. Originally, MAST-1 was assumed to be a sister of the oomycetes (49, 54). This assumption was not supported by the results of our study (bootstrap value, <50%) or a previous analysis (50). This cluster was recently characterized as a group of truly planktonic marine aerobic organisms (50), which may explain the restriction of MAST-1 clones to the suboxic chemocline. The placement of the MAST clusters among diverse groups of organisms, including osmotrophs, phagotrophs, free-living species, and parasites, makes it nearly impossible to speculate about the types of organisms that they represent. Their phylogenetic affiliation is unresolved, and they may represent novel clades at a high evolutionary level. This makes the MAST clusters a primary target for culturing efforts.
(iii) Opisthokonts and diverse bikonts.
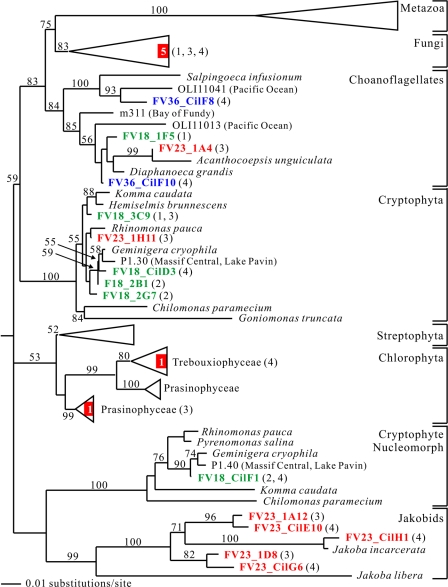
We discovered five fungal phylotypes (one ascomycete, two chytridiomycetes, and two basidiomycetes) in our FV libraries (Fig. 4 and 7). The relatively low fungal diversity is in agreement with the results of environmental 18S rRNA surveys for other anoxic marine sampling sites (41, 72, 75). However, this is in sharp contrast to clone libraries retrieved from oxygen-depleted freshwater habitats, one of which is even dominated by fungal clones (43, 69). Furthermore, we identified seven choanoflagellate phylotypes, one of which was quite divergent (FV36_ CilF8) (Fig. 4 and 7). Choanoflagellates are phylogenetically among the closest relatives of animals (35). Even though these organisms are cosmopolitan in aquatic habitats, it was not until recently that they have been detected in anoxic environments (69), which suggests that their diversity in such habitats is grossly undersampled.
FIG. 7.
Minimum evolution phylogenetic tree of eukaryotic 18S rRNA gene sequences showing the positions of opisthokont clones and several bikont clones of the crown radiation. The tree was constructed with maximum likelihood criteria by using a GTR+I+G DNA substitution model with the variable-site gamma distribution shape parameter (G) at 0.5877, the proportion of invariable sites (I) at 0.1415, and the base frequencies and rate matrix for the substitution model suggested by Modeltest (59), based on 1,316 unambiguously aligned positions. For additional details see the legend to Fig. 5.
Two other groups of bikont organisms represented in our FV libraries are cryptophytes and jakobids (Fig. 4 and 7). We found five phylotypes belonging to the latter group, all of which were restricted to the anoxic water layer right below the chemocline. Four of these clones were highly divergent. Jakobids are free-living heterotroph flagellates. They are evolutionarily very interesting because of their bacterium-like mitochondrial genome, suggesting that they might represent primitive descendants of early eukaryotes. Morphologically, they are excavate organisms and thus are expected to branch at the base of the eukaryotic tree of life (4). However, our phylogenetic analyses identified the jakobids as a sister group of the cryptophytes (nucleomorph). Most likely, this was due to the phylogenetic inference method. At this point, the phylogenetic position of jakobids in the eukaryotic tree of life remains uncertain (70).
We also recovered seven cryptophyte phylotypes from the 18-m oxic-anoxic interface and one cryptophyte phylotype from the anoxic 23-m layer (Fig. 4 and 7). This distribution may have been due to the sensitivity of cryptophytes to oxygen depletion (69).
(iv) Early branching lineages.
Four phylotypes were phylogenetically affiliated with early branching eukaryote groups (Fig. 8). One of them, occurring in the FV18 and FV23 libraries (FV18_2A2 and FV23_3A2), was closely related to Myrionecta rubra. M. rubra and Mesodinium pulex, the only two named species in this sequence clade, were classified as ciliates previously (27, 32). However, they have several distinct morphological characteristics (39, 76). In phylogenetic analyses, due to their divergent sequences (33), long-branch attraction often places this sequence clade at the base of the eukaryotic tree.
Three other interesting sequences branched within the Euglenozoa (Fig. 8). One of them, FV18_1E2, was related to AT4-56 from anoxic hydrothermal sediment (41). This sequence is especially important for kinetoplastid phylogeny. Until recently, based on 18S rRNA gene analyses, trypanosomatids were assumed to have emerged from the bodonids (9, 67). At the same time, these analyses showed that there is a huge evolutionary distance between kinetoplastids, diplonemids, and euglenids, making correct inference of the phylogenetic relationships between these groups very difficult. However, AT4-56, together with FV18_1E2, emerged robustly at the base of the kinetoplastids, breaking the long branch which leads to diplonemids and euglenids. Using this sequence as a close outgroup in phylogenetic analyses resulted in a much more stable and resolved kinetoplastid phylogeny (55). Thus far, AT4-56 is the only representative of this basal kinetoplastid “group.” Now the Framvaren FV18_1E2 sequence confirms that this lineage is a real biological entity.
Two other sequences (FV23_1E10 and FV36_2E04) branched in an uncultured euglenozoan sequence cluster (Fig. 8). Thus far, this cluster consists exclusively of sequences from anoxic marine environments (72, 73; A. Zündorf, A. Behnke, J. Bunge, K. Barger, and T. Stoeck, unpublished data; this study). This suggests that we are dealing with a group of strictly anaerobic organisms. This environmental sequence cluster exhibits well-supported affiliation with diplonemids, whose phylogenetic position in the euglenozoa remains unsettled (48, 53, 79). Taxonomic sampling was shown to be a major factor affecting the topology of the 18S rRNA euglenozoan tree (53). Concerted efforts are necessary to enlarge euglenozoan sampling, and retrieving novel sequences is a high priority. The novel sequence clade discovered in anoxic marine systems not only may be a group of hitherto unknown organisms but also may be very valuable for resolving euglenozoan phylogeny better.
What did we learn about the nature of protistan communities in anoxic aquatic environments from this study?
An improved methodology for chimeric sequence identification indeed seems to decrease the number of putatively novel kingdom-level sequences, suggesting in the past there was overestimation of the level of eukaryote diversity at the mega-evolution level. Nevertheless, anoxic environments like the Framvaren Fjord in Norway are a source of highly divergent, potentially novel, and distinct clades of organisms (e.g., the novel class-level ciliate clade). Thus, it is presumptuous to argue that there are no unknown protist phyla left, as diversity research has barely touched the anoxic reservoirs on our planet. Furthermore, anoxic systems contain sequences that are very important for reconstruction of eukaryote phylogeny (e.g., the kinetoplastid sequences AT4-56 and FV18_1E2). They also harbor phylogenetic clades of specifically adapted organisms, which are unlikely to occur in other environments (e.g., the novel anoxic euglenozoan clade). Moreover, there is evidence that primitive descendants of early eukaryotes are prospering in anoxic environments (e.g., the Framvaren jakobid clones).
Our study revealed the fine-scale architecture of protistan communities along an O2/H2S gradient. This should be taken into account when 18S rRNA clone libraries from different sampling sites are compared, as the vast majority of such libraries target only an unspecified spot in the anoxic system being studied. Undersampling by virtually all eukaryotic 18S rRNA clone libraries, together with only marginal global coverage of anoxic sampling sites, restricts conclusions to a defined sampling site at a defined sampling time. Thus, we encourage workers performing future diversity surveys in such environments to address basic questions about the diversity, biogeography, and nature of anoxic protistan communities.
Supplementary Material
Acknowledgments
We thank the crew of Nordstranda (Helvik, Norway) for their hospitality during field work in the Framvaren Fjord. We thank C. Beardsley for support during sampling. We thank three anonymous reviewers, whose careful reading and detailed comments enabled us to significantly improve the manuscript.
This study was funded by grant STO414/2-2 from the Deutsche Forschungsgemeinschaft to T.S. This research was conducted using the resources of the Cornell Theory Center, which receives funding from Cornell University, New York State, federal agencies, foundations, and corporate partners.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaral-Zettler, L. A., F. Gómez, E. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Eukaryotic diversity in Spain's River of Fire. Nature 417:137. [DOI] [PubMed] [Google Scholar]
- 4.Baldauf, S. L., D. Bhattacharya, J. Cockrill, P. Hugenholtz, J. Pawlowski, and G. B. Simpson. 2004. The tree of life: an overview, p. 43-75. In J. Cracraft and M. J. Donoghue (ed.), Assembling the tree of life. Oxford University Press, Oxford, United Kingdom.
- 5.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bérney, C., J. Fahrni, and J. Pawlowski. 2004. How many novel eukaryotic ‘kingdoms’? Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackwell, W. H., and M. J. Powell. 2000. A review of group affiliation of Stramenopiles, additional approaches to the question. Evol. Theory 12:49-88. [Google Scholar]
- 8.Bohning, D., and D. Schon. 2005. Nonparametric maximum likelihood estimation of population size based on the counting distribution. J. R. Stat. Soc. C Appl. 54:721-738. [Google Scholar]
- 9.Callahan, H. A., R. W. Litaker, and E. J. Noga. 2002. Molecular taxonomy of the suborder Bodonina (Order Kinetoplastida), including the important fish parasite, Ichthyobodo necator. J. Eukaryot. Microbiol. 49:119-128. [DOI] [PubMed] [Google Scholar]
- 10.Caron, D. A., P. Countway, and M. V. Brown. 2004. The growing contributions of molecular biology and immunology to protistan ecology: molecular signatures as ecological tools. J. Eukaryot. Microbiol. 51:38-48. [DOI] [PubMed] [Google Scholar]
- 11.Cavalier-Smith, T. 2004. Only six kingdoms of life. Proc. R. Soc. Lond. B 271:1251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao, A., and J. Bunge. 2002. Estimating the number of species in a stochastic abundance model. Biometrics 58:531-539. [DOI] [PubMed] [Google Scholar]
- 13.Chao, A. 2005. Species richness estimation, p. 7907-7916. In C. Balakrishnan, B. Read, and B. Vidakovic (ed.), Encyclopedia of statistical sciences, 2nd ed. Wiley, New York, N.Y.
- 14.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 15.Davidson, K., and E. H. John. 2001. The grazing response of the heterotrophic microflagellate Paraphysomonas vestita when ingesting phytoplankton prey. Protistology 2:22-23. [Google Scholar]
- 16.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgcomb, V. P., D. T. Kysela, A. Teske, A. de Vera Gomez, and M. L. Sogin. 2002. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. USA 99:7658-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehrenberg, C. C. 1838. Die Infusionsthierchen als Vollkommene Organismen. Leopold Voss, Leipzig, Germany.
- 19.Fenchel, T., C. Bernard, G. Esteban, B. J. Finlay, P. J. Hansen, and N. Iversen. 1995. Microbial diversity and activity in a Danish fjord with anoxic deep water. Ophelia 43:45-100. [Google Scholar]
- 20.Fenchel, T., and B. J. Finlay. 1995. Ecology and evolution in anoxic worlds. Oxford University Press, Oxford, United Kingdom.
- 21.Finlay, B. J., J. O. Corliss, G. F. Esteban, and T. Fenchel. 1996. Biodiversity at the microbial level: the number of free-living ciliates in the biosphere. Q. Rev. Biol. 71:221-237. [Google Scholar]
- 22.Finlay, B. J. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061-1063. [DOI] [PubMed] [Google Scholar]
- 23.Foissner, W., H. Blatterer, H. Berger, and F. Kohmann. 1991. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems—Band I: Cyrtophorida, Oligotrichida, Hypotrichia, Colpodea. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft, 1/91. Bartels und Wernitz Druck, Munich, Germany.
- 24.Foissner, W., H. Berger, and F. Kohmann. 1992. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems—Band II: Peritrichia, Heterotrichida, Odontomastida. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft, 5/92. Bartels und Wernitz Druck, Munich, Germany.
- 25.Foissner, W., H. Berger, and F. Kohmann. 1994. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems—Band III: Hymenostomata, Protomatida, Nassulida. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft, 1/94. Bartels und Wernitz Druck, Munich, Germany.
- 26.Foissner, W., H. Berger, H. Blatterer, and F. Kohmann. 1995. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems—Band IV: Gymnostomatea, Loxodes, Suctoria. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft, 1/95. Bartels und Wernitz Druck, Munich, Germany.
- 27.Foissner, W., H. Berger, and J. Schaumburg. 1999. Identification of limnetic planktonic ciliates. Informationsberichte des Bayer. Landesamtes für Wasserwirtschaft, 3/99. Bartels und Wernitz Druck, Munich, Germany.
- 28.Fried, J., W. Ludwig, R. Psenner, and K. H. Schleifer. 2002. Improvement of ciliate identification and quantification: a new protocol for fluorescence in situ hybridization (FISH) in combination with silver stain techniques. Syst. Appl. Microbiol. 25:555-571. [DOI] [PubMed] [Google Scholar]
- 29.Grasshoff, K. 1964. On the determination of silica in sea water. Deep Sea Res. 11:597-604. [Google Scholar]
- 30.Hellebust, J. A., and J. Lewin. 1977. Heterotrophic nutrition, p. 169-197. In D. Werner (ed.), The biology of diatoms. University of California Press, Berkeley.
- 31.Hong, S.-H., J. Bunge, S.-O. Jeon, and S. Epstein. 2005. Predicting microbial species richness. Proc. Natl. Acad. Sci. USA 103:117-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jankowski, A. W. 1976. Revision of the classification of the cyrtophorids, p. 167-168. In A. P. Markevich and I. Yu (ed.), Materials of the II All-Union Conference of Protozoology, part I. General protozoology. Naukova Dumka, Kiev, Ukraine.
- 33.Johnson, M. D., T. Tengs, D. W. Oldach, C. F. Delwiche, and D. K. Stoecker. 2004. Highly divergent SSU rRNA genes found in the marine ciliates Myrionecta rubra and Mesodinium pulex. Protist 155:347-359. [DOI] [PubMed] [Google Scholar]
- 34.Kahl, D. M. 1935 1930. Urtiere oder Protozoa. I. Wimpertiere oder Ciliata (Infusoria), eine Bearbeitung der freilebenden und ectocomensalen Infusorien der Erde, unter Ausschluss der marinen Tintinnidae. Gustav Fischer, Jena, Germany.
- 35.King, N., and S. B. Carroll. 2001. A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution. Proc. Natl. Acad. Sci. USA 98:15032-15037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koroleff, F. 1969. Direct determination of ammonia in natural waters as indophenol blue, p. 19-22. In Information on techniques and methods for seawater analysis. Interlaboratory report 3. International Council for Exploration of the Sea, Charlottenlund, Denmark.
- 37.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-148. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, N.Y.
- 38.Lefranc, M., A. Thénot, C. Lepère, and D. Debroas. 2005. Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl. Environ. Microbiol. 71:5935-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindholm, T., P. Lindroos, and A. C. Mörk. 1988. Ultrastructure of the photosynthetic ciliate Mesodinium rubrum. Biosystems 21:141-149. [DOI] [PubMed] [Google Scholar]
- 40.López-García, P., F. Rodríguez-Valera, C. Pedros-Alió, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 41.López-García, P., H. Philippe, F. Gail, and D. Moreira. 2003. Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc. Natl. Acad. Sci. USA 100:697-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludwig, W., O. Strunk, R. Westram, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo, Q., L. R. Krumholz, F. Z. Najar, A. D. Peacock, B. A. Roe, D. C. White, and M. S. Elshahed. 2005. Diversity of the microeukaryotic community in sulfide-rich Zodletone Spring (Oklahoma). Appl. Environ. Microbiol. 71:6175-6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lylis, J. C., and F. R. Trainor. 1973. The heterotrophic capabilities of Cyclotella meneghiniana. J. Phycol. 9:365-369. [Google Scholar]
- 45.Lynn, D. H., and E. B. Small. 2000. Phylum Ciliophora, p. 371-656. In J. J. Lee, G. F. Leedale, and P. Bradbury (ed.), An illustrated guide to the protozoa, vol. 1. Allen Press, Lawrence, KS. [Google Scholar]
- 46.MacGregor, B. J., D. P. Moser, B. J. Baker, E. W. Alm, M. Maurer, K. H. Nealson, and D. A. Stahl. 2001. Seasonal and spatial variability in Lake Michigan sediment small-subunit rRNA concentrations. Appl. Environ. Microbiol. 67:3908-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maslov, D. A., S. Yasuhira, and L. Simpson. 1999. Phylogenetic affinities of Diplonema within the Euglenozoa as inferred from the SSU rRNA gene and partial COI protein sequences. Protist 150:33-42. [DOI] [PubMed] [Google Scholar]
- 49.Massana, R., L. Guillou, B. Díez, and C. Pedrós-Alió. 2002. Unveiling the organism behind the novel eukaryotic ribosomal DNA sequences from the ocean. Appl. Environ. Microbiol. 68:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Massana, R., J. Castresana, V. Balagué, L. Guillou, K. Romari, A. Groisillier, K. Valentin, and C. Pedros-Alió. 2004. Phylogenetic and ecological analysis of novel marine stramenopiles. Appl. Environ. Microbiol. 70:3528-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Medlin, L., H. J. Elwood, S. Stickel, and M. L. Sogin. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA coding regions. Gene 71:491-499. [DOI] [PubMed] [Google Scholar]
- 52.Millero, F. J. 1991. The oxidation of H2S in Framvaren Fjord. Limnol. Oceanogr. 36:1007-1014. [Google Scholar]
- 53.Moreira, D., P. López-García, and F. Rodriguez-Valera. 2001. New insights into the phylogenetic position of diplonemids: G+C content bias, differences of evolutionary rate and a new environmental sequence. Int. J. Syst. Evol. Microbiol. 51:2211-2219. [DOI] [PubMed] [Google Scholar]
- 54.Moreira, D., and P. López-García. 2002. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 10:31-38. [DOI] [PubMed] [Google Scholar]
- 55.Moreira, D., P. López-Gracía, and K. Vickerman. 2004. An updated view of kinetoplastid phylogeny using environmental sequences and a closer outgroup: proposal for a new classification of the class Kinetoplastea. Int. J. Syst. Evol. Microbiol. 54:1861-1875. [DOI] [PubMed] [Google Scholar]
- 56.Murphy, J. G., and J. Riley. 1962. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 27:31-36. [Google Scholar]
- 57.Naqvi, S. W. A. 1994. Denitrification processes in the Arabian Sea. Proc. Indian Acad. Sci. Earth Planet. Sci. 103:279-300. [Google Scholar]
- 58.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 59.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 60.Raghukumar, S. 2002. Ecology of the marine protists, the Labyrinthulomycetes (Thraustochytrids and Labyrinthuloids). Eur. J. Protistol. 38:127-145. [Google Scholar]
- 61.Richards, T. A., and D. Bass. 2005. Molecular screening of free-living microbial eukaryotes: diversity and distribution using a meta-analysis. Curr. Opin. Microbiol. 8:240-252. [DOI] [PubMed] [Google Scholar]
- 62.Robison-Cox, J. F., M. M. Bateson, and D. M. Ward. 1995. Evaluation of nearest-neighbor methods for detection of chimeric small-subunit rRNA sequences. Appl. Environ. Microbiol. 61:1240-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schopf, J. W., and C. Klein. 1992. The proterozoic biosphere: a multidisciplinary study. Cambridge University Press, New York, N.Y.
- 65.Sherr, E. B., D. A. Caron, and B. F. Sherr. 1993. Staining of heterotrophic protists for visualization via epifluorescence microscopy, p. 213-228. In P. F. Kemp (ed.), Handbook of methods in aquatic microbial ecology. Lewis, Boca Raton, Fla.
- 66.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strain. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simpson, A. G., J. Lukeŝ, and A. J. Roger. 2002. The evolutionary history of kinetoplastids and their kinetoplasts. Mol. Biol. Evol. 19:2071-2083. [DOI] [PubMed] [Google Scholar]
- 68.Skei, J. 1988. Framvaren—environmental setting. Mar. Chem. 23:209-218. [Google Scholar]
- 69.Slapeta, J., D. Moreira, and P. López-García. 2005. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc. Biol. Sci. 272:2073-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stechmann, A., and T. Cavalier-Smith. 2002. Rooting the eukaryote tree using a derived gene fusion. Science 297:89-91. [DOI] [PubMed] [Google Scholar]
- 71.Stoeck, T., and S. S. Epstein. 2003. Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl. Environ. Microbiol. 69:2657-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stoeck, T., G. Taylor, and S. S. Epstein. 2003. Novel eukaryotes from a permanently anoxic Cariaco Basin (Caribbean Sea). Appl. Environ. Microbiol. 69:5656-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stoeck, T., B. Hayward, G. T. Taylor, R. Varela, and S. S. Epstein. 2006. A multiple PCR-primer approach to access the microeukaryotic diversity in environmental samples. Protist 157:31-43. [DOI] [PubMed] [Google Scholar]
- 74.Swofford, D. L. 2001. PAUP*. Phylogenetic analysis using parsimony (and other methods), 4.0b6. ed. Sinauer Associates, Sunderland, Mass.
- 75.Takishita, K., H. Miyake, M. Kawato, and T. Maruyama. 2005. Genetic diversity of microbial eukaryotes in anoxic sediment around fumaroles on a submarine caldera floor based on the small-subunit rDNA phylogeny. Extremophiles 9:185-196. [DOI] [PubMed] [Google Scholar]
- 76.Taylor, F. J. R., D. J. Blackbourn, and J. Blackbourn. 1971. The red-water ciliate Mesodinium rubrum and its “incomplete symbionts”: a review including new ultrastructural observations. J. Fish. Res. Bd. Can. 28:391-407. [Google Scholar]
- 77.Taylor, G. T., M. I. Scranton, M. Iabichella, T.-Y. Ho, R. C. Thunell, F. Muller-Karger, and R. Varela. 2001. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol. Oceanogr. 46:148-163. [Google Scholar]
- 78.Volkov, I. I., A. G. Rozanov, and T. P. Demidova. 1992. Inorganic reduced sulfur and manganese in the water column of the Black Sea, p. 38-50. In M. E. Vinogradov (ed.), Winter ecosystem of the central Black Sea. Institute of Oceanology, Russian Academy of Sciences, Moscow, Russia. (In Russian.)
- 79.Von der Heyden, S., E. E. Chao, K. Vickerman, and T. Cavalier-Smith. 2004. Ribosomal RNA phylogeny of bodonid and diplonemid flagellates and the evolution of euglenozoa. J. Eukaryot. Microbiol. 51:402-416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.