Abstract
Cavity disease in white button mushrooms is caused by Burkholderia gladioli pv. agaricicola. We describe the isolation and characterization of six mutants of the strain BG164R that no longer cause this disease on mushrooms. The mutations were mapped to genes of the general secretory pathway (GSP). This is the first report of the association of the type II secretion pathway with a disease in mushrooms. Phenotypes of the six avirulent mutants were the following: an inability to degrade mushroom tissue, a highly reduced capacity to secrete chitinase and protease, and a reduced number of flagella. Using these mutants, we also made the novel observation that the factors causing mushroom tissue degradation, thereby leading to the expression of cavity disease, can be separated from mycelium inhibition because avirulent mutants continued to inhibit the growth of actively growing mushroom mycelia. The GSP locus of B. gladioli was subsequently cloned and mapped and compared to the same locus in closely related species, establishing that the genetic organization of the gsp operon of B. gladioli pv. agaricicola is consistent with that of other species of the genus. We also identify the most common indigenous bacterial population present in the mushroom fruit bodies from a New Zealand farm, one of which, Ewingella americana, was found to be an apparent antagonist of B. gladioli pv. agaricicola. While other investigators have reported enhanced disease symptoms due to interactions between endogenous and disease-causing bacteria in other mushroom diseases, to the best of our knowledge this is the first report of an antagonistic effect.
Burkholderia gladioli, a β-proteobacter, was initially identified as a pathogen of gladiolus (23). It was subsequently associated with diseases in other plants, such as onions, iris, freesia, dendrobium, cymbidium, tulip, green gram, and rice (22). Disease symptoms in the different plant hosts varied from the spotting of foliar parts to scabbing and rotting of storage tissues. In the last decade, different strains of B. gladioli have been demonstrated to have the ability to infect animals, including humans, causing food poisoning and severe pulmonary infections in cystic fibrosis and other immunocompromised human patients (11, 19).
B. gladioli pv. agaricicola is an important pathogen in the mushroom industry. It causes soft rotting symptoms on a number of commercially important mushrooms, such as Lentinula edodes, Pleurotus ostreatus, Flammulina velupies, Pholiota nameko, Hypsizygus marmoreus, and Grifola frondos in Japan and on different cultivated Agaricus species in New Zealand and Europe (14). While soft rot appears to be less prevalent than most of the other mushroom diseases and is only sporadically reported from farms in the United Kingdom and other European countries (9), when it occurs, it can cause devastating effects within very short periods of time. Thus, B. gladioli pv. agaricicola is now considered to be a pathogen that has the potential to cause significant crop losses in the mushroom industry (9, 14).
Cavity disease was first reported in 1992 (13). The causal microorganism was initially identified as Pseudomonas cepacia. The strain, designated CANU-PMS164, was isolated from New Zealand mushrooms with symptoms ranging from mild lesions to deep pitting (13), and it was shown to inhibit the growth of mushroom mycelia in vitro (14). Strain CANU-PMS164 rapidly degraded mushroom sporocarps, which resulted in marked tissue damage within 72 h of infection. Although cavity disease was originally thought to be a novel disease (18), it was later reclassified (as reported by Gill and Tsuneda [14]) as “rapid soft rot disease of edible mushroom.” The causative agent was subsequently renamed Pseudomonas gladioli pv. agaricicola (14) and is now known as B. gladioli pv. agaricicola following the new genus nomenclature of members of the “pseudomallei group” proposed by Yabucchi et al. (35) and accepting the proposal of Lincoln et al. (21) that assigns the mushroom soft rotting bacteria to a third pathovar, “agaricicola” (21, 35).
In this study, we identify four genes necessary for the virulence of B. gladioli pv. agaricicola BG164R, the causative agent of a mushroom soft rot disease (21), also described as cavity disease of the white button mushroom Agaricus bitorquis (13). Avirulent mutants of B. gladioli pv. agaricicola were generated to answer three fundamental questions arising from previous work. First, what are the genes essential for the expression of cavity disease symptoms? Second, are the virulence factors necessary for cavity disease also required to inhibit mycelial growth? Last, why is there such a marked variation in disease severity? In other words, are environmental factors responsible for this variation, or is the differential intensity of disease an attribute of multiple pathovars that exhibit different but overlapping symptoms? We anticipated that identification of the virulence genes and investigating conditions related to disease expression would possibly provide an explanation for the variation in disease severity and explain why the disease is so infrequently reported.
Our findings reconcile the confusing history associated with the observed variation in the intensity of disease expression by the causative agent and advance the understanding of virulence at a molecular level. The ability of B. gladioli pv. agaricicola to inhibit mushroom mycelia has also always been associated with its ability to cause cavity disease (14). From these observations, it has been thought that the pathogen produces both hypha-degrading enzymes and toxins and that cavity disease symptoms are a combined effect of the toxin and the enzymes. The avirulent mutants are affected in the ability to secrete some virulence factors required for the symptoms of cavity disease yet retain inhibitory activity toward fungal mycelia.
(This research was conducted by P. Roy Chowdhury in fulfillment of the requirements for a Ph.D. from the University of Canterbury, Christchurch, New Zealand, 2004.)
MATERIALS AND METHODS
Strains and plasmids.
The main characteristics of the strains, plasmids, and cosmids used are listed in Table 1. All bacteria were grown in Luria-Bertani (LB) medium in either liquid broth or solid agar (24). B. gladioli, Pseudomonas aureofaciens, Burkholderia cepacia, Pseudomonas putida, Pseudomonas fluorescens, Serratia entomophila, and Ewingella americana were cultured at 30°C, while Escherichia coli strains were grown at 37°C. When required, plates were supplemented with the following: ampicillin, 100 μg ml−1; chloramphenicol, 25 μg ml−1; gentamicin, 30 μg ml−1; kanamycin, 50 μg ml−1; nalidixic acid, 15 μg ml−1; rifampin, 50 μg ml−1; tetracycline 15 μg ml−1; and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), 25 μg ml−1.
TABLE 1.
List of strains, plasmids, cosmids, and vectors used
| Name | Genotype or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| B. gladioli pv. agaricicola | ||
| BG164 | Wild type; Cav+ Af+ Rifs Prot+ | 18 |
| BG164R | Spontaneous Rifr mutant of BG164; Cav+ Af+ Rifr Prot+ | This study |
| BG4-12 | BG164R gspF::mini-Tn5KmlacZ2 Rifr Kmr Cav− Af+ Prot− | This study |
| BG12-88 | BG164R gspK::mini-Tn5KmlacZ2 Rifr Kmr Cav− Af+ Prot− | This study |
| BG12-147 | BG164R gspK::mini-Tn5KmlacZ2 Rifr Kmr Cav− Af+ Prot− | This study |
| BG15-40 | BG164R gspE::mini-Tn5KmlacZ2 Rifr Kmr Cav− Af+ Prot− | This study |
| BG15-87 | BG164R gspD::mini-Tn5KmlacZ2 Rifr Kmr Cav− Af+ Prot− | This study |
| BG16-787 | BG164R gspE::mini-Tn5KmlacZ2 Rifr Kmr Cav− Af+ Prot− | This study |
| BG4-12Cos | BG4-12 complemented with cosmid pCosGSP; Rifr Kanr Tetr Cav+ | This study |
| BG12-88Cos | BG12-88 complemented with cosmid pCosGSP; Rifr Kanr Tetr Cav+ | This study |
| BG15-40Cos | BG15-40 complemented with cosmid pCosGSP; Rifr Kanr Tetr Cav+ | This study |
| BG-LAF3 | BG164R with pLAFR3; Rifr Tetr Cav+ Prot+ | This study |
| BG4-12LAF3 | BG4-12 with pLAFR3; Rifr Kanr Tetr Cav− Prot− | This study |
| E. coli | ||
| S17-1 λpir | thi pro hsdR hsdM+ ΔrecA RP4-2::TcMu− Km::Tn7 | 32 |
| DH5α | supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 thi-1 relA1 recA1 | 16 |
| HB101 | supE44 hsdS20 (rB− mB−) recA13 ara-14 rspL20 proA2 lacY1 galK2 xyl-5 myl-1 | 3 |
| P. aureofaciens | ||
| PA147-2 | Wild type; Af+ Bfm+ Rifr Cmr | 4 |
| PAE639 | PA147-2; yeiJ::mini-Tn5KmlacZ2 Af+ Bfm− Fla− Mot− Rifr Kmr | H. K. Mahanty |
| P. cepacia B111 | Wild type; Chi− | A. L. J. Cole |
| P. putida | Wild type; Prot− | H. K. Mahanty |
| S. entomophila A1MO2 | Derivative of A1 wild type; Ampr Chi+ Path+ | 36 |
| Plasmids and cosmids | ||
| pUTZ2 | pUT containing mini-Tn5KmlacZ2 | 7 |
| pSPRC12 | 8.5-kb SalI fragment containing gspF::mini-Tn5KmlacZ2 from BG4-12 in pBluescript KS+; Apr Kanr | This study |
| pSPRC40 | 5.4-kb SalI fragment containing gspE::mini-Tn5KmlacZ2 from BG15-40 in pBluescript KS+; Apr Kanr | This study |
| pSPRC87 | 6.0-kb SalI fragment containing gspD::mini-Tn5KmlacZ2 from BG15-87 in pBluescript KS+; Apr Kanr | This study |
| pSPRC88 | 5.2-kb SalI fragment containing gspK::mini-Tn5KmlacZ2 from BG15-88 in pBluescript KS+; Apr Kanr | This study |
| pSPRC147 | 5.3-kb SalI fragment containing gspK::mini-Tn5KmlacZ2 from BG12-147 in pBluescript KS+; Apr Kanr | This study |
| pSPRC787 | 5.4-kb SalI fragment containing gspE::mini-Tn5KmlacZ2 from BG12-787 in pBluescript KS+; Apr Kanr | This study |
| pROBE | 1.5-kb NotI fragment containing the kanamycin gene from mini-Tn5KmlacZ2 blunt-end cloned into the EcoRV site of pBluescript KS−; Apr | S. R. Giddens |
| pCosGSP | 23.4-kb Sau3AI partial fragment containing the GSP gene cluster from BG164R into pLAFR3 | This study |
| pGSP ME H-H16.7 | 16.7-kb HindIII subclone from pCosGSP in pME6001; Genr | This study |
| pGSP ME H-H2 | 2-kb HindIII subclone from pCosGSP in pME6001; Genr | This study |
| pGSP ME H-E1.7 | 1.7-kb HindIII-EcoRI subclone from pCosGSP in pME6001; Genr | This study |
| pGSP KS B-B2.2 | 2.2-kb BamHI subclone from pCosGSP in pBluescript KS+; Apr | This study |
| pGSP KS B-B7.2 | 7.2-kb BamHI subclone from pCosGSP in pBluescript KS+; Apr | This study |
| Vectors | ||
| pBluescript | ColE1 ori lacZα/KS polylinker; T3/T7; Apr (KS, M13+) | Stratagene |
| pLAFR3 | pRK290 derivative; RP4(IncP-1) ori λcos; pUC9 multicloning site and lacZα Tcr | 33 |
Cav, cavity disease; Prot, protease; Chi, chitinase; Bfm, biofilm; Af, antifungal.
Mushroom bioassay.
The bioassay system was modified from the basic technique of Gandy (12). In routine assays, fresh mushrooms, supplied by Meadow Mushrooms (Christchurch, New Zealand), were surface sterilized, cut into slices of approximately 1.5 to 2 mm thick under sterile conditions, immersed immediately in ice-cold distilled water (to prevent tissue browning), and transferred to plastic lunch boxes lined with wet UV-sterilized paper towels. Either fixed volumes of the test bacterial suspensions or single colonies transferred by toothpicks were used as inocula and incubated for 16 h at 30°C. Within that time frame, wild-type bacteria caused distinct holes of approximately 4 to 5 mm in diameter on the mushroom pieces, while mutants failed to show any similar symptoms.
For screening of mutants, 10,000 transconjugants arising from 66 independent experiments carried out in 16 attempts were screened for the transposition of the mini-Tn5, resulting in “no-cavity” phenotypes.
Transposon mutagenesis.
The transposon mini-Tn5KmlacZ2 (7), borne by pUTZ2 (Table 1) and maintained in E. coli S17-1 λpir (32), was introduced into the recipient, BG164R, by conjugation. The conjugation procedure was as follows. The donor, E. coli S17-1 λpir, and the recipient, BG164R, were grown in liquid broth supplemented with antibiotics for 18 h at the respective temperatures mentioned above. One milliliter of donor and recipient bacteria, harvested by centrifugation (2,000 × g for 3 min in a Bio-Rad mini benchtop centrifuge), was washed with fresh LB broth to remove antibiotics. The recipient was heat shocked at 43°C for 15 min, mixed with the donor in a ratio of 4:1 (1 ml of recipient to 250 μl of donor), and concentrated to 400 μl. Aliquots of 200 μl were applied on a sterilized filter paper placed on top of LB plates as a single droplet without any antibiotics and incubated at 30°C for 4 h. The mating mixture was recovered from filter paper by washing the filter with 1.5 ml of fresh LB broth; the mixture was concentrated to 200 μl, and 100-μl aliquots were spread on plates supplemented with rifampin, kanamycin, and chloramphenicol (15 μg ml−1) and incubated at 30°C for 36 h to select for BG164R transconjugants.
Screening and isolation of avirulent mutants.
Transconjugants arising from independent matings were screened for mutations with an avirulent no-cavity phenotype in the mushroom bioassay. The number of mutations resulting in at least one auxotrophy was determined by transferring transconjugants to minimal agar plates.
Phenotyping.
All phenotypes were determined in comparison to wild-type BG164R.
Morphological changes in mutants were assessed using transmission electron microscopy. Bacterial cells from a 24-h incubation on LB plates were stained with 1% phosphotungstic acid and observed under bright-field conditions with either a Hitachi H-600 electron microscope or a JEOL JEM-1200EX electron microscope.
Motility, or the capacity of mutant bacteria to move away from the point of inoculation on the motility agar plates (0.1% [wt/vol] Bacto tryptone, 0.05% [wt/vol] yeast extract, and 0.5% [wt/vol] NaCl) supplemented with 0.3% agar, was monitored to assess any change in this ability.
Any alteration in the capacity of mutants to interact with the host tissue was monitored with scanning electron microscopy using a Leica s440 SEM, and tissue samples were prepared according to Atkey et al. (2).
The ability of the mutants to produce chitinase was monitored on 1% chitin extract plates (19) incubated at 30°C for 4 days and then scored in terms of the clearing of colloidal chitin around the colonies spotted on the plates. Protease secretion was studied by transferring the test colonies onto 0.1% skim milk agar plates and looking for the capacity of the strains to clear casein around the test colonies.
DNA manipulation and cloning.
The genomic DNA was prepared by the guanidium thiocyanate method (27), and plasmid DNA was prepared by the standard alkaline lysis method (29). For sequencing, plasmid DNA isolated using the alkaline lysis method was further purified by lithium chloride precipitation (29). DNA samples were routinely quantified using a deuterium lamp LKB Ultraspec Plus spectrophotometer. Restriction digestions were set up using the manufacturer's recommendations, analyzed by agarose gel electrophoresis (29), and visualized on a Sigma T2210 UV transilluminator. Gels were photographed using a Kodak Electrophoresis Documentation and Analysis System 120.
For regular cloning, vector DNA was dephosphorylated using calf intestinal phosphatase (Böehringer Mannheim) prior to ligation with the inserts following standard conditions described by Sambrook et al. (29).
Shotgun clones of the mutated genes in BG164R mutants were isolated by transforming DH5α electrocompetent cells (36) with ligation mixtures containing SalI-digested genomic DNA and the vector (pBluescript KS+). Desired clones were isolated by selecting colonies resistant to ampicillin (on vector) and kanamycin (in the transposon). The fact that each mutant had a single transposon insertion was confirmed by a Southern hybridization, in which genomic DNA of the mutants was hybridized to a probe (Table 1) of the kanamycin resistance (nptII) gene originally isolated from Tn5 (data not shown). SalI-generated DNA fragments of variable sizes (Table 1) spanning the transposon insertion sites in the genome of each of the avirulent mutants were “shotgun” cloned into pBluescript KS+.
Construction of a genomic library.
A genomic library of BG164R in cosmid pLAFR3 (33) was constructed and maintained in E. coli DH5α lacking recA activity (16) according to the method outlined by Fleischman et al. (10), except for the extraction of genomic DNA, which was done according to the guanidium thiocyanate method (27). Ligated DNA was packaged in vitro into phage heads using Packagene Lambda DNA Packaging System packaging extract (Promega), following the manufacturer's protocol (Promega Technical Bulletin 005).
Cosmid isolation by colony hybridization.
A total of 3,200 individual clones representing the B. gladioli pv. agaricicola library were screened by colony hybridization (24) with a 1.5-kb EcoRI/SalI probe constructed from pSPRC12. A random primed DNA labeling kit from Böehringer Mannheim was used to label 50 ng of the probe DNA with 32P, following the manufacturer's protocol. Approximately 200,000 cpm of [α-P32]dCTP-labeled probe was used, and membranes were hybridized to the probes for 16 h following the standard techniques of Sambrook et al. (29).
Cosmid mapping by Southern hybridization.
The 1.5-kb SalI-EcoRI fragment from the clone pSPRC12, which served as the gspF probe, had a single BamHI site and hybridized to two bands, 2.3 kb and 7.2 kb (see Fig. 3B), in the BamHI-digested cosmid DNA. Hence, these two bands were placed contiguously on the physical map of the cosmid. The gspK probe, constructed from pSPRC88, hybridized with a 6.5-kb BamHI fragment, as predicted from the restriction map.
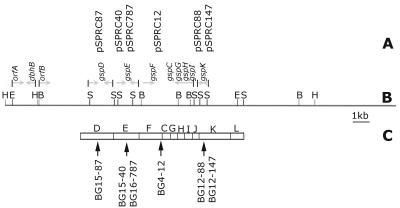
FIG. 3.
(A) Clones aligned to the corresponding fragment of genomic DNA on the cosmid map of pCosBG. (B) Cosmid map. Sites on the map are as follows: B, BamHI; E, EcoRI; H, HindIII; S, SalI. (C) Arrangement of gsp gene cluster in B. cepacia strain KFI, which has 83 to 98% sequence similarity with BG164R gsp genes. The arrows indicate the positions of insertions of the transposon into the GSP operon.
Complementation of mutants.
Avirulent mutants were transformed with the cosmid by triparental matings with an E. coli DH5α donor harboring the cosmid in the presence of a helper plasmid pRK2013 and were maintained in E. coli HB101 (3) following Carruthers et al. (4). The resultant transconjugants were selected from plates supplemented with appropriate antibiotics, purified, and checked for complementation using the assays described above.
DNA sequencing.
The clones were sequenced with IRD41-labeled T3 and T7 primers with a SequiTherm Long-Read Sequencing Kit LC from Epicenter Technologies in a Li-COR automated sequencer following the manufacturer's instructions, exploiting the dideoxy chain termination method of Sangers et al. (30). Sequences were analyzed using DNAMAN, version 4.02 (Lyonnon BioSoft). The BLASTN and BLASTX (www.ncbi.nlm.nih.gov/BLAST) programs were used to search for similar sequences in public databases (1).
Antifungal assay.
Colonies of test bacteria were streaked with a sterile loop approximately 1 cm away from 7-day-old actively growing mushroom mycelia on compost malt agar plates (15) and further incubated for 5 days at 23°C.
Mutant rescue experiments.
Overnight cultures of BG4-12Cos, BG12-88Cos, and BG15-40Cos were diluted 100-fold and grown at 30°C in LB broth in the absence of any antibiotic selection. Aliquots (100 μl each) were aseptically removed from the cultures at 24-h intervals for 3 days, serially diluted 106-fold, spread on LB plates, and incubated for a further 24 h. Individual colonies arising on the plates were screened for kanamycin (on transposon) and tetracycline (on pCosGSP) susceptibilities, indicating loss of both the transposon and the cosmid. All such kanamycin- and tetracycline-susceptible colonies were tested using the mushroom bioassay and scored for strains exhibiting wild-type phenotypes, suggesting the recreation of the wild-type strain by a double-crossover event or allelic exchange between the mutated genes and the complementing fragment of DNA present in pCosGSP. Recreation of wild-type strains was later confirmed by Southern blot analysis.
RESULTS
Selection of avirulent mutants.
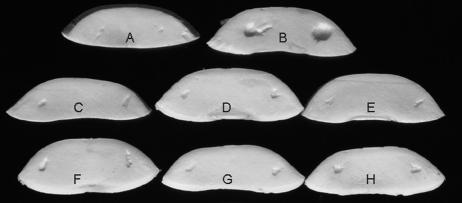
Following mini-Tn5 transposon mutagenesis, 10,000 individual BG164R transconjugants were screened in the modified mushroom bioassay for the cavity-forming phenotype indicative of virulence. On average, 1.5% of transconjugants were auxotrophic mutants. Six prototrophic but no-cavity-forming mutants were isolated (Fig. 1 and Table 1). Generation times of the mutants and parental strain were estimated by measuring the change in optical density (at 600 nm) of the cultures growing in LB medium over a 12-h period. The doubling time of both the wild type and the mutants was approximately 75 min (data not shown).
FIG. 1.
The mushroom bioassay that demonstrates the effect of mutations on cavity disease symptoms: control (A), BG164R (wild type) (B), BG4-12 (C), BG12-88 (D), BG12-147 (E), BG15-40 (F), BG15-87 (G), and BG16-787 (H).
Phenotypes of the BG mutants are consistent with a defect in protein secretion.
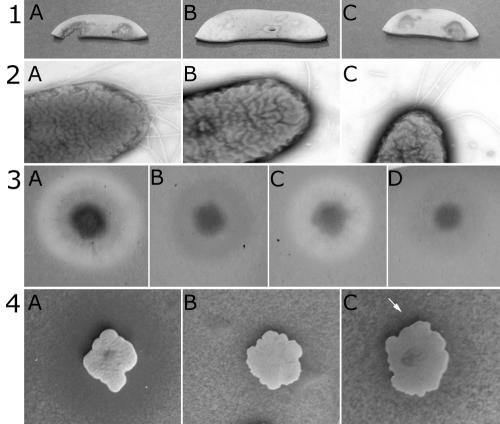
The BG mutants did not degrade mushrooms, had different numbers of flagella, did not secrete protease, and had a highly reduced capacity to secrete chitinase (Fig. 2). In contrast to the five polar flagella in the wild-type strain, four mutants (BG4-12, BG12-147, BG15-87, and BG16-787) out of the six had a single flagellum. Two flagella were visible in transmission electron micrographs of the other two mutants, BG12-88 and BG15-40 (data not shown). Likewise, the mutants and the wild type (BG164R) looked the same on plates with various proportions of agar, ranging from 0.2% to 0.6%, which indicated that there were no differences in motility.
FIG. 2.
Complementation assays with the wild-type BG164R (A), the representative GSP mutant BG4-12 (B), the complemented mutant BG4-12Cos (C), and BG4-12LAF3 (D). Results shown are for the following: mushroom assay (row 1), transmission electron microscopy observations (row 2), protease assay (row 3), and chitinase assay (row 4). The arrow (row 4) indicates the region of chitin degradation around the complemented BG4-12Cos colony.
The six BG164R mutants formed a continuous sheath on the surface of the mushroom pieces, with occasional breaks under which intact mushroom hyphae could be seen in scanning electron micrographs (data not shown). In comparison, mushroom slices inoculated with the wild-type bacteria remained attached to the skeletal remains of degraded hyphal filaments. Thus, under the standardized assay conditions, mutants failed to degrade the hyphae of mushrooms. Both the secreted casein-degrading protease and chitinase activities were highly reduced in the six BG164R mutants, which strongly suggested that the mutations affected the protein secretion pathway.
Cloning and identification of transposon-tagged genes in cavity disease mutants.
To identify the mutated genes, DNA flanking the transposon was sequenced in all the clones. BLAST analysis of the sequences suggested that the transposon insertions were located in homologs of different general secretory pathway (gsp) genes. The mutations were mapped to the gspF gene (BG4-12), gspK gene (BG12-88 and BG12-147), gspE gene (BG15-40 and BG16-787), and gspD gene (BG15-87) (Fig. 3). The DNA sequences of these four genes were 83 to 98% identical to corresponding gsp genes of two other species belonging to the genus Burkholderia, namely, B. cepacia strain KF1 (GenBank accession number AB050004.1) and Burkholderia pseudomallei strain 1026b (GenBank accession number AF110185).
Each mutation could be complemented using a single genomic clone carried by the cosmid pCosGSP. The genomic fragment of 23.4 kb was isolated from a pLAFR3-based genomic library of B. gladioli BG164R. pCosGSP was introduced by triparental mating, and the presence of the cosmid in the mutants was confirmed by subsequent reisolation and restriction analysis. The cosmid restored the virulent cavity-causing phenotype in each mutant. Since all the mutations mapped to the gsp operon (Fig. 3), only three of four mutants—BG4-12 (gspF), BG12-88 (gspK), and BG15-40 (gspE)—were used for further experiments. The extent of degradation of milk proteins and colloidal chitin by the mutants and the numbers of flagella (Fig. 2) were restored back to the levels of the wild type when mutants were complemented in trans by the cosmid pCosBG. All three representative gsp mutants could be rescued by recombination of the cosmid with the chromosome, which was confirmed by Southern hybridization (data not shown), and when they were compared in a bioassay to the original mutants BG4-12, BG12-88, and BG15-40, no differences in the intensity of cavity formation were observed.
Mapping of the gsp genes in the cosmid pCosGSP.
The relative order of the genes coding for the different components of the type II GSP machinery appears to be highly conserved, as does the size of the gsp operons in the different species of Burkholderia, which varies from 9 to 14 kb (8) (GenBank accession number AB050004). The gspF, gspK, and gspE genes also define the ends and middle of the GSP cluster in closely related members of Burkholderia (28). The cosmid pCosGSP probably carried the full GSP locus of BG164R, because it complemented these three genes in the different BG164R mutants. The GSP locus in B. gladioli was also physically mapped by Southern hybridization, and the relative order of the different SalI fragments of DNA cloned from the mutants could be arranged along the 23.4-kb cosmid pCosGSP.
The genomic insert in the cosmid was later subcloned (pGSP ME H-H16.7, pGSP ME H-H2, pGSP ME1.7, pGSP KS B-B 2.2, and pGSP KS B-B 7.2) as smaller fragments, and the ends of each fragment were sequenced. The above data, together with the end-sequencing data of the cosmid subclones, were used to map (Fig. 3B) the entire 23.4-kb cosmid, pCosGSP, which revealed the presence of the entire gsp gene cluster of BG164R on the cosmid.
Factors involved in the inhibition of mushroom mycelia are different from those involved in the expression of cavity disease.
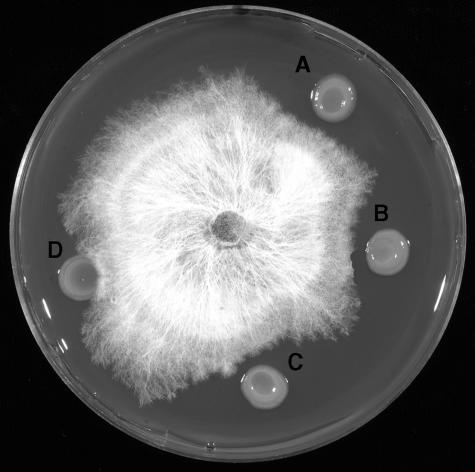
The no-cavity-forming gsp mutants, BG4-12 and BG15-40, still retained the capacity to inhibit mushroom mycelia (Fig. 4). The extent of inhibition was similar to that expressed by the wild-type cavity-forming strain, BG164R, thereby indicating that the two processes are independent of each other.
FIG. 4.
Inhibition of mushroom mycelia by BG164R and its no-cavity-forming mutants: BG164R (A), BG4-12 (B), BG15-40 (C), and P. putida (D).
Qualitative observations on disease progression.
As indicated above, cavity disease symptoms can vary from infection to infection. This could be due to any number of reasons. For example, differences in the severity of symptoms could be due to different pathogen concentrations. These concentrations could be determined by environmental conditions and/or the effect of other microbes. We manipulated these variables using the mushroom bioassay.
It is normally expected that a small number of pathogenic bacteria would initially colonize a mushroom and multiply to reach the threshold concentration to cause disease. To test this hypothesis, mushrooms were inoculated with various concentrations of BG164R, and disease progression was observed at regular intervals over 5 days (Table 2). Surprisingly, no disease symptoms were observed when as few as 20 cells of BG164R were used. This was in contrast to the results presented by Atkey et al. (2), who initiated soft rot disease within 72 h using only 3 to 5 cells of the strain RR3.
TABLE 2.
Cavity formation as a function of inoculum size
| Dilution factor | Estimated inoculum size (no. of cells) | Symptoms observed after incubation (h) fora:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 16 | 32 | 48 | 64 | 80 | 96 | 112 | 120 | ||
| 100 | 24 × 106 | * | ** | **+ | x | x | x | x | x |
| 102 | 24 × 104 | — | * | ** | **+ | x | x | x | x |
| 104 | 24 × 102 | — | — | — | — | H | H | H | A |
| 106 | 24 | — | — | — | — | — | — | — | A |
| 108 | — | — | — | — | — | — | — | A | |
*, cavity formation; **, massive tissue degradation; **+, complete tissue degradation of mushrooms; —, no reaction observed; H, hypersensitive reaction in 30% of the mushroom slices; A, assay terminated after mushroom slice started degenerating; x, tissue disappearance due to excess degradation by the pathogen.
Disease symptoms were consistently seen in bioassays using larger numbers of bacteria, so we tested whether the indigenous flora was inhibiting establishment of the pathogen. The indigenous flora was isolated from sporocarp samples. Three distinct types of bacteria were identified by different colony morphologies, and Biolog tests and 16S rRNA sequence analysis using the universal primers U16A and U16B followed. Two (PRC121 and PRC122) were identified as different strains of P. fluorescens (GenBank accession numbers DQ383803 and DQ383804), while the third (PRC120) was E. americana (GenBank accession number DQ383802). Different combinations of the pathogen, the three isolated mushroom bacteria, a mutant, and a nonpathogenic Pseudomonas strain (PA147-2) were diluted to the same optical density from saturated cultures, mixed in equal proportions, and spotted on mushroom slices, and qualitative changes of the mushroom slices were observed over a period of 3 days (Table 3). All combinations in which strain PRC120 was present appeared to have a significantly reduced cavity-forming capacity on mushroom slices, which indicated a possible inhibition of BG164R by PRC120. Following this qualitative test, a quantitative assay was done with only strains BG164R and PRC120, which showed that disease establishment was inhibited for at least 72 h when mushrooms were inoculated with 1:1 mixtures of BG164R and PRC120 totaling approximately 107 cells of each strain. The observation was in contrast to observations recorded in Table 2, where 2.4 × 107 BG164R cells could cause cavity disease within 16 h. This indicated that BG164R was being inhibited by strain PRC120. Antagonism between BG164R and PRC120 was further demonstrated in an in vitro plate assay. In this assay, PRC120 was spotted onto a lawn of BG164R. The lawn of BG164R around the PRC120 colony was inhibited, producing a clear zone without growth. The observation confirmed the ability of this isolate of E. americana to inhibit the cavity disease pathogen, BG164R.
TABLE 3.
Interactions between bacteria on mushrooms influence severity of cavity disease
| Strain or combination | Observations noted after interaction for:
|
||
|---|---|---|---|
| 24 h | 48 h | 72 ha | |
| BG164R | Distinct cavity | Slice degraded | — |
| PA147-2 | No cavity | No cavity | No cavity |
| BG15-40 | No cavity | No cavity | No cavity |
| BG164R+PRC120 | Hypersensitive reaction | Indent | Prominent indent |
| BG164R+PRC121 | Distinct cavity | Slice degraded | — |
| BG164R+PRC122 | Distinct cavity | Slice degraded | — |
| PA147-2+PRC120 | No cavity | No cavity | No cavity |
| PA147-2+PRC121 | No cavity | No cavity | No cavity |
| PA147-2+PRC122 | No cavity | No cavity | No cavity |
| BG15-40+PRC120 | No cavity | No cavity | No cavity |
| BG15-40+PRC121 | No cavity | No cavity | No cavity |
| BG15-40+PRC122 | No cavity | No cavity | No cavity |
| BG164R+PRC120+PRC121 | No cavity | Hypersensitive reaction | Indent |
| BG164R+PRC120+PRC122 | No cavity | Hypersensitive reaction | Indent |
| BG164R+PRC121+PRC122 | Distinct cavity | Slice degraded | — |
| PA147-2+PRC120+PRC121 | No cavity | No cavity | No cavity |
| PA147-2+PRC120+PRC122 | No cavity | No cavity | No cavity |
| PA147-2+PRC121+PRC122 | No cavity | No cavity | No cavity |
| BG15-40+PRC120+PRC121 | No cavity | No cavity | No cavity |
| BG15-40+PRC120+PRC122 | No cavity | No cavity | No cavity |
| BG15-40+PRC121+PRC122 | No cavity | No cavity | No cavity |
| BG164R+PRC120+PRC121+PRC122 | Indent | Prominent indent | Cavity |
| PRC120 | No cavity | No cavity | No cavity |
| PRC121 | No cavity | No cavity | No cavity |
| PRC122 | No cavity | No cavity | No cavity |
| Controlb | No cavity | No cavity | No cavity |
—, mushroom slice fully degraded.
Mushroom slice without bacteria.
DISCUSSION
We have found that the type II protein secretion system, or GSP, of B. gladioli pv. agaricicola BG164R is necessary for mushroom cavity disease. The GSP is apparently responsible for secreting the protease and chitinase activities that are necessary for disease symptoms. It could not be known in advance that secretion of these proteins was associated with cavity disease expression and that the GSP itself was necessary for the disease.
The GSP of gram-negative bacteria is the main terminal branch of a two-step secretion process. It generally consists of 12 invariant proteins spanning the cell envelope to form the core secreton, plus some associated proteins present in specific cases for the efficient functioning of the secretory machinery (26). The GSP apparatus in the different bacteria studied shows a high species specificity. The gsp operon of B. pseudomallei (1026b) is a well-characterized and mapped operon in the genus Burkholderia. The gspC gene in B. pseudomallei 1026b is transcribed in a direction opposite to the other genes in the gsp gene cluster (8). The two boundaries of this gsp operon were identified by constructing mutants specifically in the orfC and the orfD genes present in the left and right ends of the gene cluster, respectively, which did not have any effect on secretion. We present a physical map of the gsp operon of B. gladioli pv. agaricicola which demonstrates that the organization of the gsp genes in B. gladioli is similar to that of B. pseudomallei 1026b.
In all members of the genus Burkholderia studied so far, the virulence factors that are secreted through the GSP include proteases, lipases, and phospholipase C. The most common virulence factors in all of these cases have been the different types of proteases (5, 8, 20, 31). Although the above-mentioned secreted factors have been reported to be dependent on the GSP for secretion, there are two different schools of thought about their roles in pathogenesis. In some cases, the secreted products could be directly linked to pathogenesis (31, 8), while in others they were not (12). Reports on the secretion of virulence factors by the GSPs in the different Burkholderia strains have always been correlated to the pathogenicity in animal models. This is the first report linking the type II secretion system to a disease in mushrooms. We have shown through the characterization of avirulent mutants that the GSP in BG164R is necessary for secretion of the cavity disease virulence factors, which may or may not be the protease and chitinase activities monitored in this study.
The complementation experiments confirm that each tested mutant carried a mutation in only the 23.4-kb region corresponding to the gsp locus. Therefore, we expect that the virulence factors themselves are probably still produced by BG164R avirulent mutants. We also agree with Gill and Tsuneda (14), who hypothesized that cavity disease is a manifestation of the combined action of more than one factor. The secreted proteins are capable of causing disease symptoms only when present together on the mushroom. Their hypothesis is strengthened by the finding that gsp mutants were avirulent because the inability to secrete certain proteins would affect several virulence factors simultaneously.
Gill and Tsuneda (14) also proposed that the expression of cavity disease is a combined effect of mushroom tissue-degrading enzymes and toxic compounds that inhibit the growth of the mycelia. We provide evidence that the mechanism of inhibition of mushroom mycelia is different from that of degradation of sporocarps, since the avirulent mutants still inhibited mushroom mycelia.
The avirulent mutants isolated in the course of this study had mutations in gspD, gspE, gspF, and gspK. The role and localization of these proteins are known by comparison to their description in other species. The GspD protein (BG15-40) is the only outer membrane-associated protein forming the secreton. The GspF and GspK proteins (BG4-12, BG12-88, and BG12-147) are associated with the inner membrane (26). The GspE protein (BG15-40 and BG15-87) is cytoplasmic. It possesses a conserved ATP-binding motif providing an autokinase activity; evidently it energizes the secretion process or assembly of the secretory apparatus (34).
This is the first observation associating a reduction in flagella number to mutations in the gsp genes. The only gene that is known to be linked to flagella number is the fleN gene in Pseudomonas aeruginosa. The FleN protein regulates flagella number by acting as a negative regulator of fleQ, the transcriptional activator of the flagellar synthesizing genes (6). The fleN mutants of P. aeruginosa strains PAK and PAO1 had an increase in flagella number. Interestingly, Dasgupta et al. (6) also suggested that motility and flagella number were not necessarily linked. All our mutants were motile (data not presented) despite a reduction in the number of flagella.
The occurrence and severity of cavity disease are highly variable, and the reason for the variability has gone unexplained until now. We provide evidence that antagonistic interactions between B. gladioli and endogenous flora account for this variability. Healthy New Zealand mushrooms appear to be colonized consistently by E. americana. This bacterium significantly impaired B. gladioli from creating the conditions necessary for cavity disease symptoms. Possibly E. americana prevents B. gladioli from reaching the necessary population density to cause symptoms, either passively through resource competition or actively by expression of a toxin. Other mushroom diseases, such as brown blotch caused by Pseudomonas tolaasii (25) and internal stipe necrosis caused by E. americana (18), require a very high number of cells (∼108) for their initiation under test conditions. Thus, either quantitative or qualitative differences in the native flora from mushroom to mushroom may account for all or some of the variability in the disease.
E. americana PRC120 is the first report of a New Zealand isolate of E. americana from mushrooms, and it has been deposited in the New Zealand Culture Collection (accession number of NZRM 4225). Interestingly, PRC120 isolated from button mushrooms in New Zealand did not cause any necrotic symptoms (17, 18) in our assay.
While we present evidence that E. americana inhibits cavity disease on New Zealand mushrooms, it remains formally possible that other bacteria, not as consistently distributed, augment the ability of B. gladioli to establish the conditions necessary for the disease. Moquet et al. (25) in their study of blotch disease observed a species-specific difference in symptom intensity to disease caused by P. tolaasii. Inglis et al. (18) mentioned a variation in the intensity of disease symptoms by E. americana and reported an interaction between P. fluorescens and E. americana in the formation of internal stipe necrosis of mushrooms. They also suggested a contributory role played by P. fluorescens in the expression of disease symptoms. To the best of our knowledge, this is the first report of an interaction on mushrooms that is antagonistic.
Acknowledgments
P.R.C. was supported by NZAID and University of Canterbury doctoral scholarships.
We are indebted to H. K. Mahanty for his mentorship in this project. We thank A. L. J. Cole for providing the strain BG164, Mark Silby for critical review of the manuscript, and Scott Godfrey and Mark Braithwaite (MAF-New Zealand) for the identification of the mushroom isolates PRC120, PRC121, and PRC122.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhand, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 24:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkey, P. T., T. R. Fermor, and S. P. Lincoln. 1992. Electron microscopy of the infection process of rapid soft rot disease of the edible mushroom Agaricus bitorquis. Mycol. Res. 96:717-722. [Google Scholar]
- 3.Boyer, H. W., and D. Rolland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459. [DOI] [PubMed] [Google Scholar]
- 4.Carruthers, F. L., A. J. Conner, and H. K. Mahanty. 1994. Identification of a genetic locus in Pseudomonas aureofaciens involved in fungal inhibition. Appl. Environ. Microbiol. 60:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbett, C. R., M. N. Burtnick, C. Kool, D. E. Woods, and P. A. Sokol. 2003. An extracellular zinc metalloprotease gene of Burkholderia cepacia. Microbiology 149:2263-2271. [DOI] [PubMed] [Google Scholar]
- 6.Dasgupta, N., S. K. Arora, and R. Ramphal. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lorenzo, V., M. Herrero, U. Jakunzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertional mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeShazer, D., P. J. Brett, M. N. Burtnick, and D. E. Woods. 1999. Molecular characterization of genetic loci required for secretion of exoproducts in Burkholderia pseudomallei. J. Bacteriol. 181:4661-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fermor, T., and S. Lincoln. August 2001. Rotten mushrooms. International Society for Mushroom Science. [Online.] http://www.isms.biz/article13.htm.
- 10.Fleischmann, R., M. McCormick, and B. H. Howard. 1987. Preparation of a genomic library. Methods Enzymol. 151:405-416. [DOI] [PubMed] [Google Scholar]
- 11.Foley, P. L., J. J. Lipuma, and S. H. Feldman. 2004. Outbreak of otitis media caused by Burkholderia gladioli infection in immunocompromised mice. Comp. Med. 54:93-99. [PubMed] [Google Scholar]
- 12.Gandy, D. G. 1968. A technique for screening bacteria causing brown blotch of cultivated mushrooms. Rep. Glasshouse Crops Res. Inst. 1968:150-154. [Google Scholar]
- 13.Gill, W. M., and A. L. J. Cole. 1992. Cavity disease of Agaricus bitorquis caused by Pseudomonas cepacia. Can. J. Microbiol. 38:394-397. [Google Scholar]
- 14.Gill, W. M., and A. Tsuneda. 1997. The interaction of soft rot bacterium Pseudomonas gladioli pv. agaricicola with Japanese cultivated mushrooms. Can. J. Microbiol. 43:639-648. [Google Scholar]
- 15.Godfrey, S. A. C. 2003. Molecular investigation of pseudomonads causative of Agaricus bisporus blotch disease in New Zealand mushroom farms. Ph.D. thesis. University of Canterbury, Christchurch, New Zealand.
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Inglis, P. W., and J. F. Peberdy. 1997. Production and purification of a chitinase from Ewingella americana, a recently described pathogen of the mushroom, Agaricus bisporus. FEMS Microbiol. Lett. 157:189-194. [Google Scholar]
- 18.Inglis, P. W., J. L. Burden, and J. F. Peberdy. 1996. Evidence for the association of the enteric bacterium Ewingella americana with internal stipe necrosis of Agaricus bisporus. Microbiology 142:3253-3260. [Google Scholar]
- 19.Jiao, Z., Y. Kawamura, N. Mishima, R. Yang, N. Li, X. Liu, and T. Ezaki. 2003. Need to differentiate lethal toxin-producing strains of Burkholderia gladioli, which cause severe food poisoning: description of B. gladioli pathovar cocovenenans and an emended description of B. gladioli. Microbiol. Immunol. 47:915-925. [DOI] [PubMed] [Google Scholar]
- 20.Lee, M.-A., and Y. Liu. 2000. Sequencing and characterisation of a novel serine metalloprotease from Burkholderia pseudomallei. FEMS Microbiol. Lett. 192:67-72. [DOI] [PubMed] [Google Scholar]
- 21.Lincoln, S. P., T. R. Fermor, D. E. Stead, and J. E. Sellwood. 1991. Bacterial soft rot of Agaricus bitorquis. Plant Pathol. 40:136-144. [Google Scholar]
- 22.Matsuyama, N. 1998. Presumptive identification of several phytopathogenic bacteria by novel diagnostic tests. J. Agric. Kyushu Univ. 43:337-343. [Google Scholar]
- 23.McCulloch, L. 1921. Bacterial disease of gladiolus. Science 54:115-116. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Moquet, F., M. Mamoun, and J. M. Oliver. 1996. Pseudomonas tolaasii and tolaasin: comparison of symptom induction on a wide range of Agaricus bisporus strains. FEMS Microbiol. Lett. 142:99-103. [Google Scholar]
- 26.Peabody, C. R., Y. J. Chung, M. Yen, D. Vidal-Ingigliardi, A. P. Pugsley, and M. H. Saier, Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archeal flagella. Microbiology 149:3051-3072. [DOI] [PubMed] [Google Scholar]
- 27.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 28.Pugsley, A. P., O. Francetic, O. M. Possot, N. Sauvonnet, and K. R. Hardie. 1997. Recent progress and future directions in studies of the main terminal branch of the general secretory pathway in gram-negative bacteria—a review. Gene 192:13-19. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sexton, M. M., A. L. Jones, W. Chaowagul, and D. E. Woods. 1994. Purification and characterization of a protease from Pseudomonas pseudomallei. Can. J. Microbiol. 40:903-910. [DOI] [PubMed] [Google Scholar]
- 32.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 33.Staskawicz, B., D. Dahlbeck, N. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thanassi, D. G., and S. J. Hultgren. 2000. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 12:420-430. [DOI] [PubMed] [Google Scholar]
- 35.Yabuchi, E., Y. Kosako, H. Oyaizu, I. Yano, H. Hotta, Y. Hashimoto, T. Ezaki, and M. Arakawa. 1992. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36:1251-1275. [DOI] [PubMed] [Google Scholar]
- 36.Zabarovsky, E. R., and G. Winberg. 1990. High efficiency electroporation of ligated DNA into bacteria. Nucleic Acids Res. 18:5129. [DOI] [PMC free article] [PubMed] [Google Scholar]