Abstract
A temperate bacteriophage (F108) has been isolated through mitomycin C induction of a Pasteurella multocida serogroup A strain. F108 has a typical morphology of the family Myoviridae, presenting a hexagonal head and a long contractile tail. F108 is able to infect all P. multocida serogroup A strains tested but not those belonging to other serotypes. Bacteriophage F108, the first P. multocida phage sequenced so far, presents a 30,505-bp double-stranded DNA genome with cohesive ends (CTTCCTCCCC cos site). The F108 genome shows the highest homology with those of Haemophilus influenzae HP1 and HP2 phages. Furthermore, an F108 prophage attachment site in the P. multocida chromosome has been established to be inside a gene encoding tRNALeu. By using several chromosomal markers that are spread along the P. multocida chromosome, it has been demonstrated that F108 is able to perform generalized transduction. This fact, together with the absence of pathogenic genes in the F108 genome, makes this bacteriophage a valuable tool for P. multocida genetic manipulation.
Pasteurella multocida is a gram-negative bacterium belonging to the class of gammaproteobacteria that is responsible for causing diseases in many species of mammals and birds, resulting in important health problems in the animal production industry (5). Diseases induced by P. multocida include hemorrhagic septicemia in cattle, atrophic rhinitis in swine, and fowl cholera in wild and domestic birds (5). Five different P. multocida serogroups (A, B, D, E, and F) have been described based on the antigenicity of the capsule (3, 5). It is known that serogroup A strains are mainly involved in fowl cholera, whereas serogroups B and E are implicated in hemorrhagic septicemia and serogroup D seems to be responsible for atrophic rhinitis (7, 22, 33, 37). Because of the economic relevance of P. multocida-mediated infections, work in several fields concerning this bacterial species, such as molecular characterization of pathogenic factors and construction of mutants to be used as vaccines, is increasing. Nevertheless, few genetic tools for P. multocida genetic manipulation are available (1, 4, 32).
Temperate bacteriophages are excellent tools for genetic manipulation of bacteria. They may be useful for carrying out generalized transduction and for development of cloning vectors (9, 13, 16, 35). Several P. multocida bacteriophages were described in the past, but they were used only for typing proposals (19). To our knowledge, no transducing bacteriophages have been described for P. multocida so far. Moreover, no P. multocida phage has been completely sequenced.
In this context, we have characterized a temperate bacteriophage (F108) obtained from a P. multocida serogroup A isolate that is able to carry out generalized transduction. This bacteriophage has also been sequenced, and the absence of pathogenic factors in its genome has been demonstrated by both in silico and in vivo experimental methods. These data suggest that bacteriophage F108 may be a suitable genetic tool for P. multocida serogroup A strains.
MATERIALS AND METHODS
Bacteria, plasmids, and media.
Escherichia coli DH5α cells were grown in Luria-Bertani medium (17), and when necessary, ampicillin was added at 50 μg/ml. P. multocida was grown in brain heart infusion (BHI) liquid medium or on BHI or sheep blood agar plates (SBAP). The cultures were always incubated at 37°C. Isolation of spontaneous mutants and transduction experiments were performed by plating overnight cultures on BHI plates using rifampin (100 μg/ml), streptomycin (75 μg/ml), or nalidixic acid (30 μg/ml). DNA extractions, cloning, transformation, and other molecular techniques used in this work were performed as described elsewhere (23).
Isolation and induction of bacteriophage F108.
Several natural isolates of P. multocida from the collection of the Institut de Recerca i Tecnologia Agroalimentàries (IRTA, Spain) were tested. For each P. multocida strain, overnight (ON) liquid cultures were grown and centrifuged, and their supernatant was passed through a 45-μm filter. Each filtered supernatant (lysate) was tested on each P. multocida strain to detect the presence of bacteriophage plaques. Briefly, each P. multocida strain was grown ON on SBAP. Using this fresh plate, a cell suspension (optical density at 600 nm, 0.8) was made in 2 ml of BHI. One hundred microliters of this suspension was added to 3 ml of soft-BHI agar (BHI liquid medium and 0.7% agar; prewarmed at 45°C) and layered on BHI plates. Ten microliters of each lysate was then placed on the plate. After ON incubation at 37°C, growth inhibition was observed only for P. multocida strain PM403 (Table 1) when the PM108 lysate was used. For PFU/ml counts, 100 μl of lysate serial dilutions was added to the warm soft-BHI agar; in this case, and after ON incubation, bacteriophage plaques were observed. Mitomycin C-mediated phage induction was carried out as described elsewhere (19). Briefly, a P. multocida ON culture was diluted 1/100 in fresh BHI medium, and after 2 h of incubation at 37°C, mitomycin C (0.5 μg/ml) was added. Thirty minutes later, the treated culture was centrifuged, and the pellet was resuspended in fresh BHI medium without mitomycin C and incubated 2 h at 37°C. Finally, the culture was centrifuged, the supernatant was filtered, and the lysate was tested for the presence of bacteriophages as described above. Electron microscopic analysis of bacteriophage F108 particles was carried out as previously reported (14).
TABLE 1.
P. multocida strains used in this study
| Strain | Relevant features | Source |
|---|---|---|
| PM108 | Wild type and lysogenic for F108 | I. Badiola (IRTA)a |
| PM403 | Wild type and F108 sensitive | I. Badiola (IRTA) |
| PM1090 | Like PM403, but lysogenic for F108 | This work |
| PM1091 | Like PM403, but Strr | This work |
| PM1092 | Like PM403, but Rifr | This work |
| PM1093 | Like PM403, but Nalr | This work |
IRTA, Institut de Recerca i Tecnologia Agroalimentàries de la Generalitat de Catalunya, Spain.
Purification of bacteriophage F108 DNA.
F108 DNA for sequencing was obtained as described elsewhere (23). DNase I (20 μg/ml) and RNase I (25 μg/ml) were added to 10 ml of F108 lysate (1010 to 1011 PFU/ml) and incubated 1 h at 37°C. After 2 h of centrifugation at 132,000 × g (4°C), the supernatant was discarded and the phage pellet was resuspended in 400 ml of buffer A, containing proteinase K (0.2 mg/ml) and sodium dodecyl sulfate (0.5%), and incubated 1 h at 37°C. Afterwards, the sample was treated with buffered phenol and the aqueous layer was saved. Once the white interface was eliminated, the sample was washed with chloroform. The genomic DNA was precipitated in 2 volumes of ethanol with sodium acetate, pH 4.8 (73 nM). The pellet obtained was washed with 70% ethanol, dried, and resuspended in 100 μl Tris-EDTA buffer.
F108 sequencing.
Two strategies were used to sequence F108. First of all, a shotgun subclone library was prepared from purified phage DNA using plasmid pBluescript SK(+). Library plasmids were sequenced using the fmol DNA cycle sequencing system (Roche) by the dideoxy method (24) on an ALF sequencer (Amersham Pharmacia). Sequences obtained from the library clones allowed for the design of oligonucleotides that were used for direct sequencing with the Thermo Sequenase Cy5 dye terminator sequencing kit (Amersham Biosciences) by the dideoxy method, which allows for the closing of gaps between contigs. This direct sequencing showed that the bacteriophage F108 genome is linear with a 7-nucleotide 5′ overhang (data not shown). The full-phage genome was sequenced to sixfold coverage and was assembled using SeqManII (DNAstar). Codon usage was determined by using the www.kazusa.or.jp/codon/countcodon.html facility (18). Open reading frames (ORFs) were identified using Glimmer 2.02 (http://nbc11.biologie.uni-kl.de/glimmer2.02) (8) and FGENESB (http://softberry.com) (34) for automatic annotation and EditSeq (DNAstar) for manual confirmation. Potential ORFs were compared against the NCBI protein databases using the BLASTP nonredundant database (http://www.ncbi.nlm.nih.gov/BLAST/). The cos sequence was identified as described previously (26).
F108 integration.
Analysis of the F108 sequence revealed the presence of a 96-bp region, located upstream of the integrase gene, which shows high homology at the nucleotide level with P. multocida t33 tRNALeu. Oligonucleotides attRF108 (5′-CAAGTTTTCAGCAGACCC-3′), attRPM (5′-ACTTGGTGGTATGTTGGG-3′), attLF108 (5′-AGACAATTGACGCAGACG-3′), and attLPM (5′-ACAACCTTGCCAAGGTTG-3′) were designed and used to identify the F108 integration sites in the chromosomes of 12 independent lysogenic P. multocida strains, as well as to determine the attR and attL regions.
Transduction with F108.
Spontaneous rifampin-, streptomycin-, and nalidixic acid-resistant mutants of strain PM403 were used to obtain donor lysates (Table 1). For transduction experiments, 5 ml of each lysate at 109 PFU/ml (which had previously been treated with DNase I at 10 μg/ml) was added to a 5-ml culture of strain PM403 at an optical density at 600 nm of 0.2, resulting in a multiplicity of infection of about 3 PFU/CFU. After incubation for 30 min at 37°C without agitation and for 45 min at the same temperature with agitation, samples were plated in BHI with the suitable antibiotic. As controls, aliquots of both noninfected PM403 cell suspensions and F108 lysates were plated on the same medium.
Pathogenicity assays.
Female Swiss mice (3 weeks old) obtained from Harlan Iberica Inc. (Barcelona, Spain) and housed under specific-pathogen-free conditions were used for these studies. Bacteria were grown on SBAP prior to infection. The 50% lethal doses (LD50) of PM403 and its F108-lysogenic derivative (strain PM1090) were determined in triplicate as reported previously (12). Basically, groups of five mice were injected intraperitoneally with 0.1 ml of serial 10-fold dilutions of bacteria in buffered peptone water. The concentration of the original bacterial suspensions was determined by the plate count method. The number of animals that survived at 2 weeks postinoculation was recorded, and the LD50 was calculated as described previously (21).
Nucleotide sequence accession number.
The entire nucleotide sequence of bacteriophage F108 has been deposited in GenBank under accession number DQ114220.
RESULTS AND DISCUSSION
General properties of bacteriophage F108.
Thirty-eight different natural isolates of P. multocida (2 from human sources, 11 from sheep, 12 from rabbits, 7 from pigs, 2 from cattle, and 4 from broilers) were tested to detect the putative presence of temperate bacteriophages. After screening of these strains, as described in Materials and Methods, a bacteriophage was obtained only from the PM108 supernatant. This bacteriophage, named F108, formed about 1-mm turbid plaques, and its morphology, determined by electron microscope, is typical of the family Myoviridae. F108 virions present a hexagonal head measuring approximately 50 nm, and the tail is 120 nm long and 20 nm wide (Fig. 1). Sometimes the sheath is found to be contracted, suggesting that an injection mechanism is likely responsible for transferring the phage chromosome into the bacterial host (Fig. 1). Analysis of the F108 host range revealed that this bacteriophage is able to infect several independent strains belonging to P. multocida serogroup A and isolated from rabbits, but not those belonging to serogroups B, D, E, and F. Determination of the adsorption rate of F108 in strain PM403 revealed that about 50% of F108 was adsorbed to the host cells after 5 min, and the adsorption level rose slowly to 98% at 30 min postinfection (data not shown). The F108 lysates are stable when stored at 4°C; no significant loss in infective ability after 3 months was detected by monitoring at intervals of 10 days (data not shown).
FIG. 1.
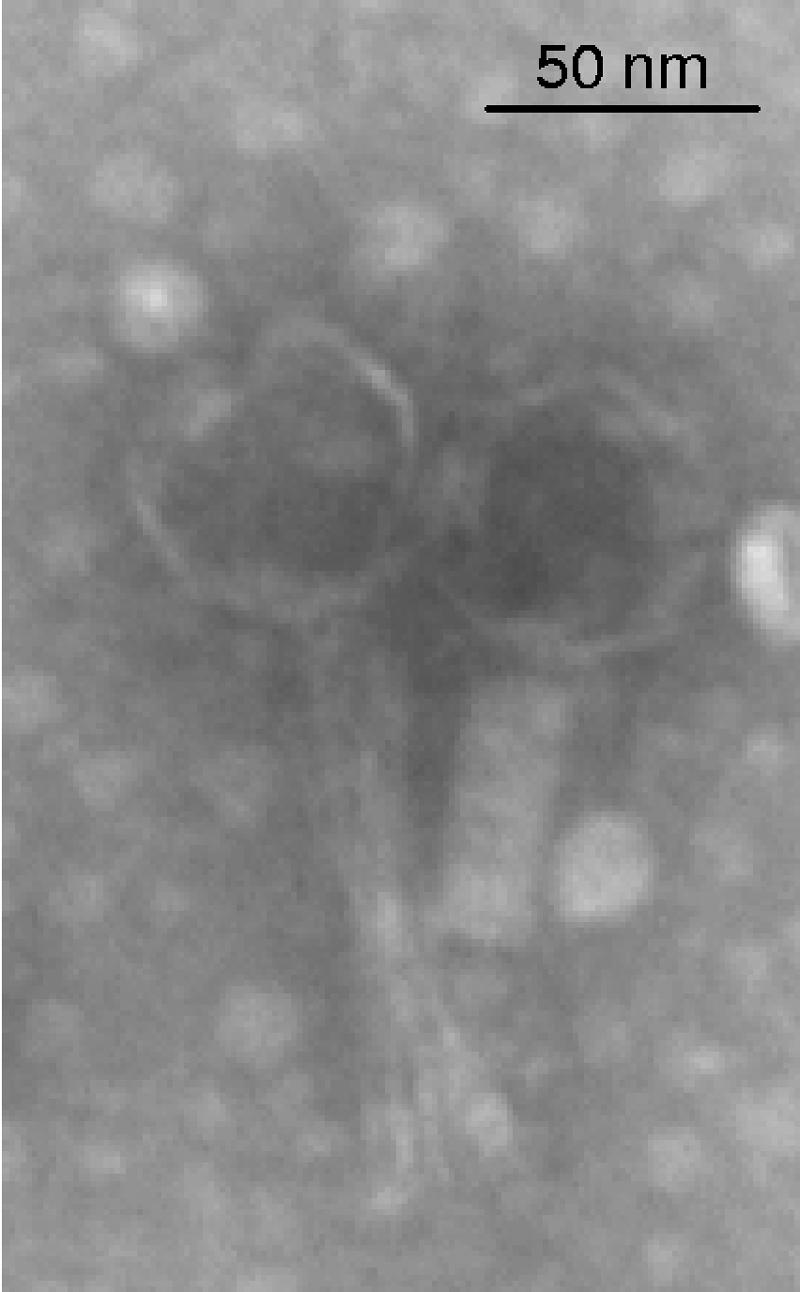
Transmission electron micrograph of bacteriophage F108 particles.
Genome of F108.
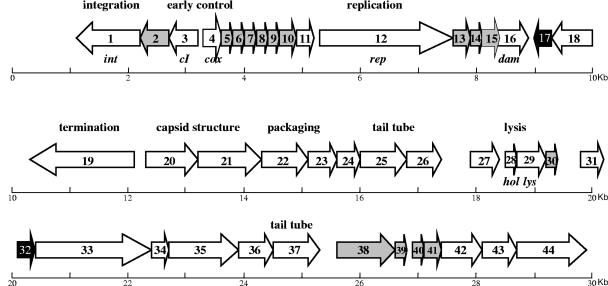
As indicated in Materials and Methods, sequencing of bacteriophage F108 DNA was initiated by a limited-shotgun strategy, followed by extensive primer walking to complete the sequencing process. The bacteriophage genome is a double-stranded DNA linear molecule of 30,505 bp with a G+C content of 42.1%, slightly higher than that of P. multocida (40.3%). No tRNA genes are present. Searches for ORFs revealed 44 ORFs larger than 150 nucleotides (Fig. 2). The codon usage of F108 is rather different from that of its host (Table 2), suggesting that this bacteriophage could have been recently introduced into P. multocida.
FIG. 2.
Schematic representation of the bacteriophage F108 genome. ORFs are numbered consecutively from left to right and are indicated by arrows. Putative functions are also shown. White arrows indicate ORFs that display the highest similarity with H. influenzae bacteriophage HP1 or HP2. Black arrows indicate ORFs that show the highest similarity with other bacteriophages different from HP1 or HP2 (see Table 2). Gray arrows indicate ORFs that share no homology in BLAST-P searches.
TABLE 2.
Codon usage in P. multocida and bacteriophage F108
| Amino acid |
P. multocida
|
F108
|
Amino acid |
P. multocida
|
F108
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Codona | Fb | Nc | Codona | Fb | Nc | Codona | Fb | Nc | Codona | Fb | Nc | |||
| Leu | TTA | 53.5 | 35,867 | TTA | 30.8 | 270 | GGC | 16.1 | 10,806 | GGC | 25.1 | 220 | ||
| TTG | 18.1 | 12,096 | TTG | 20.2 | 177 | GGA | 9.8 | 6,597 | GGA | 10.4 | 91 | |||
| CTT | 13.2 | 8,844 | CTT | 15.1 | 132 | GGG | 10.8 | 7,259 | GGG | 12.3 | 108 | |||
| CTC | 9.0 | 6,033 | CTC | 2.6 | 23 | |||||||||
| CTA | 6.8 | 4,547 | CTA | 9.5 | 83 | Ile | ATT | 47.3 | 31,695 | ATT | 39.2 | 343 | ||
| CTG | 8.4 | 5,651 | CTG | 4.8 | 42 | ATC | 16.7 | 11,210 | ATC | 26.0 | 228 | |||
| ATA | 4.4 | 2,930 | ATA | 8.8 | 77 | |||||||||
| Arg | CGT | 24.2 | 16,231 | CGT | 9.8 | 86 | ||||||||
| CGC | 10.0 | 6,669 | CGC | 19.8 | 173 | Phe | TTT | 32.2 | 21,563 | TTT | 32.0 | 280 | ||
| CGA | 3.9 | 2,594 | CGA | 6.9 | 60 | TTC | 12.0 | 8,026 | TTC | 14.3 | 125 | |||
| CGG | 1.9 | 1,277 | CGG | 3.4 | 30 | |||||||||
| AGA | 3.6 | 2,439 | AGA | 11.4 | 100 | Tyr | TAT | 25.0 | 16,778 | TAT | 20.3 | 178 | ||
| AGG | 0.7 | 484 | AGG | 2.1 | 18 | TAC | 7.2 | 4,798 | TAC | 10.7 | 94 | |||
| Ser | TCT | 12.0 | 8,067 | TCT | 6.4 | 56 | His | CAT | 16.7 | 11,158 | CAT | 7.9 | 69 | |
| TCC | 5.2 | 3,499 | TCC | 2.7 | 24 | CAC | 7.0 | 4,695 | CAC | 9.2 | 81 | |||
| TCA | 11.1 | 7,405 | TCA | 13.9 | 122 | |||||||||
| TCG | 5.4 | 3,614 | TCG | 8.8 | 77 | Gln | CAA | 40.5 | 27,146 | CAA | 29.1 | 255 | ||
| AGT | 14.1 | 9,449 | AGT | 9.7 | 85 | CAG | 10.3 | 6,904 | CAG | 13.6 | 119 | |||
| AGC | 8.7 | 5,824 | AGC | 16.2 | 142 | |||||||||
| Asn | AAT | 30.7 | 20,550 | AAT | 35.3 | 309 | ||||||||
| Val | GTT | 16.2 | 10,840 | GTT | 17.0 | 149 | AAC | 12.4 | 8,338 | AAC | 21.5 | 188 | ||
| GTC | 13.1 | 8,758 | GTC | 8.5 | 74 | |||||||||
| GTA | 13.5 | 9,036 | GTA | 10.4 | 91 | Lys | AAA | 50.8 | 34,069 | AAA | 63.6 | 557 | ||
| GTG | 24.9 | 16,653 | GTG | 22.0 | 193 | AAG | 8.2 | 5,500 | AAG | 18.5 | 162 | |||
| Pro | CCT | 11.8 | 7,892 | CCT | 3.2 | 28 | Asp | GAT | 38.6 | 25,855 | GAT | 32.2 | 282 | |
| CCC | 4.3 | 2,879 | CCC | 4.6 | 40 | GAC | 10.3 | 6,900 | GAC | 21.6 | 189 | |||
| CCA | 14.4 | 9,666 | CCA | 6.2 | 54 | |||||||||
| CCG | 8.0 | 5,337 | CCG | 17.2 | 151 | Glu | GAA | 47.3 | 31,682 | GAA | 49.6 | 434 | ||
| GAG | 13.5 | 9,053 | GAG | 18.5 | 162 | |||||||||
| Thr | ACT | 12.0 | 8,033 | ACT | 8.7 | 76 | ||||||||
| ACC | 14.5 | 9,693 | ACC | 3.9 | 34 | Cys | TGT | 8.0 | 5,374 | TGT | 3.8 | 33 | ||
| ACA | 15.3 | 10,239 | ACA | 23.6 | 207 | TGC | 2.9 | 1,911 | TGC | 5.9 | 52 | |||
| ACG | 11.1 | 7,426 | ACG | 20.6 | 180 | |||||||||
| Met | ATG | 23.9 | 16,040 | ATG | 21.5 | 188 | ||||||||
| Ala | GCT | 16.9 | 11,297 | GCT | 8.1 | 71 | ||||||||
| GCC | 17.8 | 11,896 | GCC | 7.3 | 64 | Trp | TGG | 11.7 | 7,823 | TGG | 12.2 | 107 | ||
| GCA | 28.1 | 18,824 | GCA | 26.6 | 233 | |||||||||
| GCG | 22.8 | 15,246 | GCG | 37.3 | 327 | STOP | TAA | 2.3 | 1,516 | TAA | 2.5 | 22 | ||
| TAG | 0.4 | 286 | TAG | 0.8 | 7 | |||||||||
| Gly | GGT | 28.5 | 19,072 | GGT | 12.6 | 110 | TGA | 0.3 | 212 | TGA | 1.7 | 15 | ||
Boldfaced codons are the preferred codons for each amino acid in either the bacterial or the phage genes.
F, frequency per thousand.
N, number of times each codon appears in the genome.
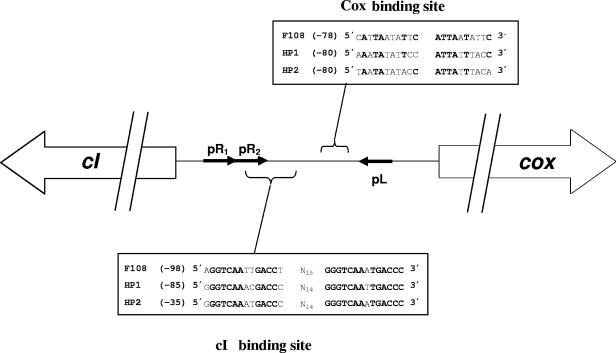
The characteristics of F108 ORFs and their corresponding predicted proteins are described in Table 3. Bacteriophage F108 shows the highest similarity and organization resemblance with Haemophilus influenzae bacteriophages HP1 and HP2 (10, 36), which, in turn, are similar to each other (36). Similar sites recognized by the products of the HP1 and HP2 cI and cox genes (encoding the lytic cycle repressor and the cI repressor with excisionase activity, respectively) (10, 11, 36) are also present in the promoters of these two genes in F108 (Fig. 3). Furthermore, as expected from the high similarity of the F108 terminase to that of HP2, F108 presents a cos region (CTTCCTCCCC) identical to that described for this bacteriophage (27).
TABLE 3.
Description of bacteriophage F108 ORFs, gene products, and functional assignments
| ORF | Product | Start position | End position | Predicted function | Protein E valuea | Best BLASTP hit |
|---|---|---|---|---|---|---|
| ORF1 | Int | 2230 | 1208 | Integrase | 3 × 10−139 | Integrase (HP2 phage) |
| ORF2 | 2685 | 2233 | ||||
| ORF3 | cI | 3260 | 2685 | Repressor protein | 3 × 10−77 | cI repressor (HP2 phage) |
| ORF4 | Cox | 3390 | 3590 | cI regulator | 1.9 × 10−3 | Hypothetical protein (Edwardsiella ictaluri) |
| ORF5 | 3643 | 3810 | ||||
| ORF6 | 3834 | 4019 | ||||
| ORF7 | 3985 | 4167 | ||||
| ORF8 | 4220 | 4441 | ||||
| ORF9 | 4474 | 4662 | ||||
| ORF10 | 4696 | 4947 | ||||
| ORF11 | 4953 | 5228 | 1 × 10−09 | Orf8 (S2 phage) | ||
| ORF12 | Rep | 5328 | 7637 | DNA polymerase | 0.0 | Hypothetical protein Haso02002083 (Haemophilus somnus 2336) |
| ORF13 | 7651 | 7902 | ||||
| ORF14 | 7905 | 8177 | ||||
| ORF15 | 8191 | 8445 | ||||
| ORF16 | Dam | 8418 | 8933 | Methylase | 2 × 10−62 | Dam (HP2 phage) |
| ORF17 | 9271 | 8996 | 9 × 10−13 | Hypothetical protein Haso02001707 (H. somnus 2336) | ||
| ORF18 | 10006 | 9347 | Portal protein | 3 × 10−90 | Probable portal protein (HP1 phage) | |
| ORF19 | 12195 | 10384 | Terminase | 0.0 | Orf16 (HP2 phage) | |
| ORF20 | 12400 | 13284 | Scaffold | 5 × 10−45 | Orf17 (HP2 phage) | |
| ORF21 | 13288 | 14319 | Capsid | 2 × 10−70 | Major capsid protein precursor (HP1 phage) | |
| ORF22 | 14326 | 15111 | Packaging protein | 8 × 10−83 | Hypothetical protein Hflu203001355 (H. influenzae R2866) | |
| ORF23 | 15175 | 15627 | Packaging protein | 5 × 10−51 | Orf20 (HP2 phage) | |
| ORF24 | 15624 | 16136 | 3 × 10−58 | Orf21 (HP2 phage) | ||
| ORF25 | 16153 | 16845 | 3 × 10−44 | Orf22 (HP1 phage) | ||
| ORF26 | 16864 | 17445 | Tail sheath | 6 × 10−74 | Orf23 (HP2 phage) | |
| ORF27 | 17998 | 18450 | Tail tube | 9 × 10−50 | Orf24 (HP1 phage) | |
| ORF28 | Hol | 18530 | 18742 | Holin | 2 × 10−06 | Hypothetical protein Haso02000478 (H. somnus 2336) |
| ORF29 | Lys | 18729 | 19280 | Lysis | 3 × 10−36 | Lysozyme precursor (HP1 phage) |
| ORF30 | 19277 | 19429 | ||||
| ORF31 | 19800 | 20108 | 3 × 10−25 | Orf26 (HP2 phage) | ||
| ORF32 | 20117 | 20287 | 1 × 10−15 | Hypothetical protein Hflu203001344 (H. influenzae R2866) | ||
| ORF33 | 20301 | 22424 | 0.0 | Orf27 (HP1 phage) | ||
| ORF34 | 22428 | 22766 | 2 × 10−34 | Orf28 (HP1 phage) | ||
| ORF35 | 22759 | 23925 | 0.0 | COG3299 protein (H. influenzae R2866) | ||
| ORF36 | 23922 | 24518 | 1 × 10−68 | Orf30 (HP2 phage) | ||
| ORF37 | 24556 | 25422 | Tail fibers | 1 × 10−61 | Orf31 (HP2 phage) | |
| ORF38 | 25656 | 26672 | ||||
| ORF39 | 26656 | 26871 | ||||
| ORF40 | 26904 | 27137 | ||||
| ORF41 | 27091 | 27411 | ||||
| ORF42 | 27422 | 28177 | 9 × 10−80 | Orf33 (HP1 phage) | ||
| ORF43 | 28174 | 28746 | 5 × 10−48 | Orf34 (HP1 phage) | ||
| ORF44 | 28743 | 29924 | 5 × 10−169 | Orf35 (HP2 phage) |
Most significant alignment obtained with BLAST-P.
FIG. 3.
Sequence comparison of the cI and Cox binding sites of bacteriophages F108, HP1, and HP2. White arrows indicate the cI and cox genes. Black arrows represent either the lysogeny promoter (pL) or the two lysis promoters (pR1 and pR2) from which the cI and cox genes are transcribed, respectively. Boxes contain the sequence of either the Cox or the cI binding site for each bacteriophage. The distance relative to either the cI or the cox translational start codon is given in parentheses.
Despite the similarity of F108 to previously described H. influenzae bacteriophages, 15 ORFs of phage F108 (14.2% of its genome) display no amino acid similarity to other proteins in the GenBank nonredundant database after BLAST-P searches. These 15 ORFs are not dispersed in the F108 genome but rather are placed together, forming groups of 3 to 6 genes (Fig. 2). In comparing the F108 gene distribution to that of other, similar bacteriophages, it can be noted that usually these nonhomologous genes are replacing others, of practically equal size, that were placed in the same modular region. Moreover, in accordance with the bacteriophage modular gene acquisition theory (2, 25), the orf38-to-orf41 set seems to be inserted into the tail module of bacteriophage F108, which is rather similar to that of phages HP1 and HP2. This insertion clearly caused both the deletion of the 3′ end of orf37 in F108, which is shorter than its HP1/HP2 homolog (orf31), and the complete elimination of HP1/HP2 orf32.
Detailed analysis of the F108 phage genome reveals the absence of genes encoding known pathogenic factors. In agreement, the pathogenicity of P. multocida cells is not increased when they are lysogenic for F108. Thus, when analyzed as described in Materials and Methods, both PM403 and its lysogenic derivative (PM1090) display the same LD50 (1.37 × 107 CFU/animal).
Identification of bacteriophage F108 integration site.
As cited above, temperate bacteriophages may be used to construct insertional plasmids which are widely used to obtain strains with new biotechnological characteristics (9, 13, 35). With the aim of further enabling the construction of this kind of vector for P. multocida, the F108 phage integration site was determined.
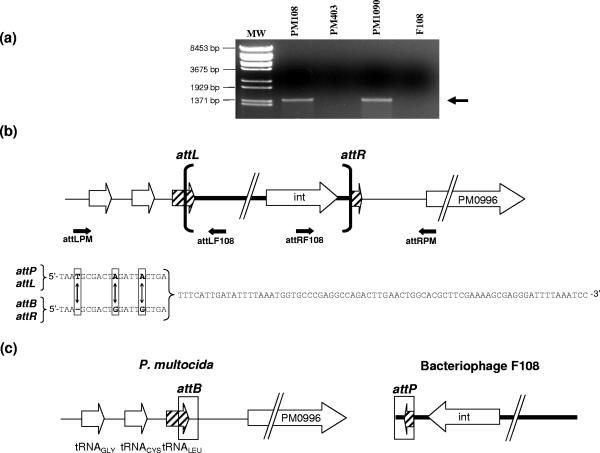
When the whole-genome sequence of F108 was used as query in a BLAST-N analysis, a 95-bp region common to the P. multocida chromosome sequence (15) was detected. In the bacteriophage DNA, this region is upstream of the integrase-encoding gene. Moreover, this region is between bp 1169740 and 1169646 of the P. multocida PM70 genome sequence, which carries the 3′ end of the t33 tRNALeu and the promoter region of the PM0996 locus, encoding a hypothetical protein. In order to determine whether this region was the F108 integration site, PCR experiments were carried out with two sets of primers designed to read out from the prophage sequence to the genome of the lysogenic cell (Fig. 4a). The results obtained indicate that a PCR band is detected with each one of two pairs of primers when DNA from lysogenic cells is analyzed but not when DNA from either isolated phage or nonlysogenic cells is used (Fig. 4a). The same results were obtained when 10 additional lysogenic strains isolated in independent experiments were tested (data not shown).
FIG. 4.
(a) Determination of the bacteriophage F108 integration site. PCR analyses were performed using attRF108 and attRPM oligonucleotides and chromosomal DNAs from PM108 (original source of bacteriophage F108), PM403 (a non-F108-lysogenic strain), PM1090 (F108 lysogenic strain, obtained in this work), and bacteriophage F108. The black arrow indicates the presence of the 1,446-bp amplification product. Lambda BstXI-digested DNA was used as a size marker (MW). (b) Diagram showing the F108 integration site in a P. multocida-F108 lysogenic strain. attR and attL positions are shown. Small arrows indicate the locations of oligonucleotides used to determine either attR or attL sequences. The phage F108 genome is enclosed by brackets. Comparison of attL, attR, attB, and attP sequences is also shown. Differences in those regions are boxed and indicated by arrows. Each sequence was determined, at least twice, for both coding and noncoding strands. (c) Schematic representation of the bacterial (attB) and bacteriophage (attP) integration regions in the P. multocida and bacteriophage F108 genomes, respectively. Both attB and attP are boxed. PM0996 encodes a hypothetical protein.
Examination of the phage genome sequence revealed that the phage attachment sequence (attP) was identical to that of attL (Fig. 4b and c). Furthermore, sequencing of the region of PM1090 chromosomal DNA in which the phage is integrated revealed that attB was the same as attR (Fig. 4b and c). It is worth noting that the t33 tRNALeu sequence is completely restored after bacteriophage insertion (Fig. 4b).
F108-mediated transduction of chromosomal markers.
There are practically no data about the ability of phages presenting cohesive ends to carry out generalized transduction. In fact, this question has been extensively analyzed only for the E. coli bacteriophage λ (28). Thus, it has been reported that production of generalized transduction particles by λ requires inactivation of its redB gene, encoding an exonuclease (24). Moreover, it has been largely demonstrated that the host chromosome is packaged in generalized transducing particles when the headful cutting system of phage DNA concatemers recognizes pseudo-pac sequences in the bacterial DNA (29, 30, 31). Furthermore, the cos-based packaging strategy implies a high-level specificity of DNA recognition which makes it difficult to produce generalized transducing particles.
Although, as described above, phage F108 presents cos ends, two specific characteristics make it different from bacteriophage λ. The first is the absence of a redB-homologous gene. The second is the fact that the F108 cos sequence is shorter than that of λ, increasing the putative presence of cos-like sequences in the whole P. multocida genome. In fact, mathematical evaluation of the a priori probability of finding a particular 10-bp sequence (like the F108 cos region) puts it at 1/410, whereas the probability goes down to 1/412 for a 12-bp sequence (such as the λ cos region).
As a consequence of these two factors, analysis of the generalized transduction ability of phage F108 was carried out using three different chromosomal markers, gyrA, rpoB, and rpsL, which are spread along the P. multocida genome (Table 4). The results indicate that F108 is able to transduce all of these markers, showing frequencies of 10−7 to 10−8 transductants per CFU. Reversion of these markers was also determined in the absence of F108 infection as a control. In all cases, transduction frequencies are about 100-fold higher than spontaneous rates (Table 4). It is known that in H. influenzae, a member of the Pasteurellaceae family, rifampin-resistant mutants present an amino acid substitution in the β subunit of the RNA polymerase (encoded by the rpoB gene), located at codon 513, 516, 518, 526, or 533 (6). The chromosomal DNA sequences of the rpoB genes from 10 P. multocida rifampin-resistant transductants revealed that all of them showed the same substitution (Asp-516→Val-516) as the rifampin-resistant strain PM1092, used as a donor in the F108-mediated transduction experiments.
TABLE 4.
F108 bacteriophage transduction frequencies of several chromosomal markers encoding antibiotic resistance
| Donor strain | Recipient strain | Gene marker | Gene locationa | Antibiotic resistance phenotype | Spontaneous frequency of mutants resistant to selected markerb | Multiplicity of infection | Transduction frequencyb,c |
|---|---|---|---|---|---|---|---|
| PM1091 | PM403 | rpsL | 1543395 to 1543769 | Strr | 9.1 × 10−10 | 1.4 | 1.6 × 10−8 |
| PM1092 | PM403 | rpoB | 1960620 to 1956592 | Rifr | 1.7 × 10−9 | 3.6 | 1 × 10−7 |
| PM1093 | PM403 | gyrA | 991251 to 993932 | Nalr | <3.4 × 10−10 | 1.4 | 5.3 × 10−8 |
P. multocida PM70 chromosomal coordinates for each gene.
Spontaneous resistance and transduction frequencies are means of three independent experiments in each case. All values were reproducible to within an error of ±10%.
Calculated as the number of transductants per survivor cells obtained from the P. multocida cell-F108 mixture after adsorption of phage particles was allowed for 30 min.
In conclusion, the results presented here clearly show that bacteriophage F108, the first P. multocida phage sequenced, not only can serve as a valuable tool for genetic manipulation of P. multocida and closely related bacteria but also is the first phage described that, presenting a cos-mediated package system, is able to carry out generalized transduction without the introduction of any specific mutation.
Acknowledgments
This work was funded by grants RTA 03-065 of the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA) (Spain), AGL2005-03574 of the Ministerio de Educación y Ciencia (Spain) and 2005SGR-533 from the Departament d’Universitats, Recerca i Societat de la Informació (DURSI) de la Generalitat de Catalunya. J. Aranda and S. Campoy were recipients of a predoctoral fellowship from the Universitat Autònoma de Barcelona and a postdoctoral contract from INIA-IRTA, respectively.
We are deeply grateful to Ignacio Badiola and Montserrat Saco for generously providing us with P. multocida strains and to our English-teaching university colleague, Chuck Simmons, for help in the language revision and correction of this article. We acknowledge Joan Ruiz and Pilar Cortés for excellent technical assistance.
REFERENCES
- 1.Adler, B., R. Chancellor, P. Homchampa, M. Hunt, C. Ruffolo, R. Strugnell, and D. Wapling. 1996. Immunity and vaccine development in Pasteurella multocida infections. J. Biotechnol. 44:139-144. [DOI] [PubMed] [Google Scholar]
- 2.Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-490. [DOI] [PubMed] [Google Scholar]
- 3.Boyce, J. D., J. Y. Chung, and B. Adler. 2000. Pasteurella multocida capsule: composition, function and genetics. J. Biotechnol. 83:153-160. [DOI] [PubMed] [Google Scholar]
- 4.Cárdenas, M., A. R. Fernández de Henestrosa, S. Campoy, A. M. Pérez de Rozas, J. Barbé, I. Badiola, and M. Llagostera. 2001. Virulence of Pasteurella multocida recA mutants. Vet. Microbiol. 80:53-61. [DOI] [PubMed] [Google Scholar]
- 5.Carter, G. R. 1967. Pasteurellosis: Pasteurella multocida and Pasteurella hemolytica. Adv. Vet. Sci. 11:321-379. [PubMed] [Google Scholar]
- 6.Cruchaga, S., M. Pérez-Vazques, F. Román, and J. Campos. 2003. Molecular basis of rifampicin resistance in Haemophilus influenzae. J. Antimicrob. Chemother. 52:1011-1014. [DOI] [PubMed] [Google Scholar]
- 7.Davies, R. L., R. MacCorquodale, S. Baillie, and B. Caffrey. 2003. Characterization and comparison of Pasteurella multocida strains associated with porcine pneumonia and atrophic rhinitis. J. Med. Microbiol. 52:59-67. [DOI] [PubMed] [Google Scholar]
- 8.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupont, L., B. Boizet-Bonhoure, M. Coddeville, F. Auvray, and P. Ritzenthaler. 1995. Characterization of genetic elements required for site-specific integration of Lactobacillus delbrueckii subsp. bulgaricus bacteriophage mv4 and construction of an integration-proficient vector for Lactobacillus plantarum. J. Bacteriol. 177:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esposito, D., W. P. Fitzmaurice, R. C. Benjamin, S. D. Goodman, A. S. Waldman, and J. J. Scocca. 1996. The complete nucleotide sequence of bacteriophage HP1 DNA. Nucleic Acids Res. 24:2360-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esposito, D., J. C. Wolson, and J. J. Scocca. 1997. Reciprocal regulation of the early promoter region of bacteriophage HP1 by the Cox and CI proteins. Virology 234:267-276. [DOI] [PubMed] [Google Scholar]
- 12.Fernández de Henestrosa, A. R., I. Badiola, M. Saco, A. M. Pérez de Rozas, S. Campoy, and J. Barbé. 1997. Importance of the galE gene on the virulence of Pasteurella multocida. FEMS Microbiol. Lett. 154:311-316. [DOI] [PubMed] [Google Scholar]
- 13.Ginsburg, D. S., and M. P. Calos. 2005. Site-specific integration with φC31 integrase for prolonged expression of therapeutic genes. Adv. Genet. 54:179-187. [DOI] [PubMed] [Google Scholar]
- 14.Llagostera, M., J. Barbé, and R. Guerrero. 1986. Characterization of SE1, a new general transducing phage of Salmonella typhimurium. J. Gen. Microbiol. 132:1035-1041. [DOI] [PubMed] [Google Scholar]
- 15.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapu. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mederle, I., I. Bourguin, D. Ensergueix, E. Badell, J. Moni-Peireira, B. Gicquel, and N. Winter. 2002. Plasmidic versus insertional cloning of heterologous genes in Mycobacterium bovis BCG: impact on in vivo antigen persistence and immune responses. Infect. Immun. 70:303-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1991. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen, J. P., and V. T. Rosdahl. 1990. Development and epidemiological applications of bacteriophage typing system for typing Pasteurella multocida. J. Clin. Microbiol. 28:103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pulliger, G. D., T. Bevir, and A. J. Lax. 2004. The Pasteurella multocida toxin is encoded within a lysogenic bacteriophage. Mol. Microbiol. 51:255-269. [DOI] [PubMed] [Google Scholar]
- 21.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 22.Rhoades, K. R., and R. B. Rimler. 1987. Capsular groups of Pasteurella multocida isolated from avian hosts. Avian Dis. 31:895-898. [PubMed] [Google Scholar]
- 23.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Sanger, F., S. Nicklen, and S. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shackelton, L. A., and E. C. Holmes. 2004. The evolution of large DNA viruses: combining genomic information of viruses and their hosts. Trends Microbiol. 12:458-465. [DOI] [PubMed] [Google Scholar]
- 26.Siboo, I. R., A. B. A. Bensing, and P. M. Sullam. 2003. Genomic organization and molecular characterization of SM1, a temperate bacteriophage of Streptococcus mitis. J. Bacteriol. 185:6968-6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skowronek, K., and A. Piekarowicz. 1996. Determination of the cos sequence of the mature genome S2/HP1 type B bacteriophage of Haemophilus influenzae. Gene 172:71-73. [DOI] [PubMed] [Google Scholar]
- 28.Sternberg, N. 1986. The production of generalized transducing phage by bacteriophage lambda. Gene 50:69-85. [DOI] [PubMed] [Google Scholar]
- 29.Sternberg, N., and R. Hoess. 1983. The molecular genetics of bacteriophage P1. Annu. Rev. Genet. 17:123-154. [DOI] [PubMed] [Google Scholar]
- 30.Reference deleted.
- 31.Susskind, M. M., and D. Botstein. 1978. Molecular genetics of bacteriophage P22. Microbiol. Rev. 42:385-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabatabaei, M., Z. Liu, A. Finucane, R. Parton, and J. Coote. 2002. Protective immunity conferred by attenuated aroA derivatives of Pasteurella multocida B:2 strains in a mouse model of hemorrhagic septicemia. Infect. Immun. 70:3355-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thurston, J. R., R. B. Rimler, M. R. Ackermann, and N. F. Cheville. 1992. Use of rats to compare atrophic rhinitis vaccines for protection against effects of heat-labile protein toxin produced by Pasteurella multocida serogroup D. Vet. Immunol. Immunopathol. 33:155-162. [DOI] [PubMed] [Google Scholar]
- 34.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 35.Van Mellaert, L., L. Mei, E. Lammertyn, S. Schacht, and J. Anné. 1998. Site-specific integration of bacteriophage VWB genome into Streptomyces venezuelae and construction of VWB-based integrative vector. Microbiology 144:3351-3358. [DOI] [PubMed] [Google Scholar]
- 36.Williams, B. J., M. Golomb, T. Phillips, J. Brownlee, M. V. Olson, and A. L. Smith. 2002. Bacteriophage HP2 of Haemophilus influenzae. J. Bacteriol. 184:6893-6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson, M. A., R. B. Rimler, and L. J. Hoffman. 1992. Comparison of DNA fingerprints and somatic serotypes of serogroup B and E Pasteurella multocida isolates. J. Clin. Microbiol. 30:1518-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]