Abstract
Overflow metabolism in the form of aerobic acetate excretion by Escherichia coli is an important physiological characteristic of this common industrial microorganism. Although acetate formation occurs under conditions of high glucose consumption, the genetic mechanisms that trigger this phenomenon are not clearly understood. We report on the role of the NADH/NAD ratio (redox ratio) in overflow metabolism. We modulated the redox ratio in E. coli through the expression of Streptococcus pneumoniae (water-forming) NADH oxidase. Using steady-state chemostat cultures, we demonstrated a strong correlation between acetate formation and this redox ratio. We furthermore completed genome-wide transcription analyses of a control E. coli strain and an E. coli strain overexpressing NADH oxidase. The transcription results showed that in the control strain, several genes involved in the tricarboxylic acid (TCA) cycle and respiration were repressed as the glucose consumption rate increased. Moreover, the relative repression of these genes was alleviated by expression of NADH oxidase and the resulting reduced redox ratio. Analysis of a promoter binding site upstream of the genes which correlated with redox ratio revealed a degenerate sequence with strong homology with the binding site for ArcA. Deletion of arcA resulted in acetate reduction and increased the biomass yield due to the increased capacities of the TCA cycle and respiration. Acetate formation was completely eliminated by reducing the redox ratio through expression of NADH oxidase in the arcA mutant, even at a very high glucose consumption rate. The results provide a basis for studying new regulatory mechanisms prevalent at reduced NADH/NAD ratios, as well as for designing more efficient bioprocesses.
Escherichia coli accumulates acetic acid when growing at a high rate of glucose consumption even in the presence of ample oxygen (2, 11, 30). This phenomenon is known as overflow metabolism. Acetate is generated when carbon flux from acetyl-coenzyme A (CoA) is directed to acetate instead of entering the tricarboxylic acid (TCA) cycle (11). This by-product induces a stress response even at extremely low concentrations (21), hinders growth (26), and reduces the production of recombinant proteins (37). Overflow metabolism has been attributed to an enzymatic limitation in the TCA cycle (27). In E. coli the complete oxidation of 1 mol of glucose in glycolysis and the TCA cycle generates 10 mol of NAD(P)H and 2 mol of FADH2 (31):
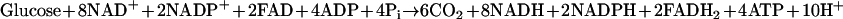 |
If the rate of oxygen utilization is sufficiently high, the reduced cofactors generated by glucose consumption are reoxidized in the electron transport chain, which serves the dual purpose of maintaining an optimal redox environment and generating energy by oxidative phosphorylation. In the absence of oxygen glucose cannot be completely oxidized, and metabolic intermediates accumulate to maintain the redox balance. Even in the presence of oxygen, if the rate of glucose consumption is greater than the capacity to reoxidize the reduced equivalents generated, the response is similar to what is observed under anaerobic conditions (1, 2, 13). Since the flux from acetyl-CoA to acetate does not generate any NADH while the flux from acetyl-CoA through the TCA cycle generates 8NAD(P)H and 2FADH2, carbon flow diversion to acetate could be viewed as a means to reduce or prevent further NAD(P)H accumulation (7, 13). These inferences regarding acetate overflow have been based on physiological observations and in vitro enzyme assays, and the genetic trigger has not been identified. Details of pathways involved in acetate generation and consumption, including specific enzymes and their regulation, have recently been reviewed (39).
Aided by genomic technology, we further investigated the relationship between redox and acetate overflow by transforming wild-type E. coli with water-forming NADH oxidase (encoded by the nox gene from Streptococcus pneumoniae) in order to oxidize residual NADH (3). This enzyme decouples NADH oxidation (oxygen reduction) from respiratory energy generation (10). We report the impact of this perturbation in redox on the physiological and transcriptional activity of cells under steady-state conditions and correlate the results with acetate formation to begin to identify regulatory processes involved in overflow metabolism. Steady-state chemostat cultures (i.e., as opposed to batch cultures) are an important tool for the study of overflow metabolism and transcriptional changes because (i) a chemostat permits the system to be tuned to achieve a desired glucose consumption rate through the selection of dilution rate, in particular glucose consumption rates both above and below the threshold rate which initiates acetate formation, and (ii) steady-state conditions permit collection of transcriptional information which is itself at a steady state. These results provide insights into gene regulation in E. coli as a model facultative anaerobe during the transition from respiratory to respirofermentative metabolism.
MATERIALS AND METHODS
Microorganisms and media.
The E. coli K-12 strains MG1655 and QC2575 (MG1655 ΔarcA::Tet) were used in this study. QC2575 was obtained from D. Touati (l'Institut Jacques Monod, Paris, France). Growth and physiological characteristics were determined using defined media (8) composed of the following (per liter): 5 g glucose, 1.5 g NH4Cl, 0.5 g NaCl, 7.8 g Na2HPO4 · 7H2O, 3.5 g KH2PO4, 0.014 g CaCl2 · 2H2O, 0.246 g MgSO4 · 7H2O, 0.1 ml Antifoam C, 1 mg biotin, 1 mg thiamine, 100 mg ampicillin, and 10 ml trace metal solution. The trace metal solution contained the following (per liter): 16.68 g FeCl3 · 6H2O, 0.36 g ZnSO4 · 7H2O, 0.32 g CuSO4 · 5H2O, 0.2 g MnSO4 · H2O, 0.18 g CoCl2 · 6H2O, 22.4 g EDTA, and 0.1 g NaMoO4 · 2H2O.
Construction of pTrc99A-nox.
The Streptococcus pneumoniae nox gene was amplified by PCR using pPANOX7 (M.-C. Trombe, U. Paul Sabatier, Toulouse, France) as template with Pfu DNA polymerase. Primers were designed based on the published S. pneumoniae nox gene sequence (3) and contained a BamHI restriction site and a Shine-Dalgarno sequence at the beginning of the amplified fragment and a PstI restriction site at the end of the amplified fragment (forward primer, 5′-TAC TAT GGA TCC AGG AGG TAA CAG CTA TGA GTA AAA TCG TTG TAG TCG GTG C-3′; reverse primer, 5′-ATA TAG TGA TCG ATA GCA GTC TGC AGT TAT TTT TCA GCC GTA AGG GCA GC-3′ [the underlined sequences are the BamHI, Shine-Dalgarno, ATG start, and PstI sites, respectively]). The resulting 1.4-kb PCR product was gel isolated, digested with BamHI and PstI, and ligated into the pTrc99A expression vector which had been digested with the same two restriction enzymes.
Chemostat cultivation.
Carbon-limited chemostat cultures of 1.5-liter working volume were grown in 2.5-liter vessels (Bioflo II; New Brunswick Scientific, NJ) at 37°C, pH 7.0, and an agitation of 500 rpm. The airflow rate was maintained at 1.5 liter/min using mass flow controllers (Unit Instruments, Orange, CA) to ensure that the dissolved oxygen concentration remained above 40% of saturation at all growth rates studied. Measurements were made after the cells attained a steady state, which required at least 7 volume changes without any perturbation. The biomass formed was quantified by washing the cells with phosphate-buffered saline (pH 7.0) and drying for 12 h at 60°C. Glucose and organic acids in the feed and effluent were measured by high-performance liquid chromatography with a detection limit of about 0.05 g/liter (6). Oxygen uptake rate and CO2 evolution rate were calculated by measuring the effluent concentrations of oxygen and CO2 (Ultramat 23 gas analyzer; Siemens, Germany). Each steady-state growth culture was freshly started from a single colony.
Quantification of NADH/NAD and glycolytic metabolites.
Metabolism was rapidly interrupted by extracting two 10-ml aliquots from a chemostat and plunging them into 40 ml methanol that had been prechilled for 4 h in a dry ice-ethanol bath. The cell pellets were resuspended in 0.2 M HCl (for extracting NAD) or 0.2 M NaOH (for extracting NADH), and the nucleotides were extracted by boiling the cell suspension. A cycling assay (4) which involves the transfer of reducing equivalents from NADH ultimately to 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was used to measure the specific nucleotides (23). The rate of reduction of MTT as measured at 570 nm was proportional to the concentration of NADH or NAD.
The intracellular concentrations of the key metabolites glucose-6-phosphate, fructose-6-phosphate, phosphoenolpyruvate (PEP), pyruvate, and acetyl-CoA were measured enzymatically by using cell extracts prepared by the perchloric acid method (36).
Global transcription profiling.
Changes in the expression of genes at various growth rates were identified using parallel two-color hybridization to whole-genome E. coli MG1655 spotted DNA arrays corresponding to 98.8% of the annotated open reading frames. The design, printing, and probing were previously described in detail (19, 20). After attaining a steady state at a predetermined dilution rate, samples were extracted from the chemostat and placed in RNAprotect buffer (QIAGEN, Valencia, CA), and the cell pellets were frozen at −80°C. Total RNA was extracted by the hot phenol-chloroform method and treated with DNase I in the presence of RNase inhibitor for subsequent labeling by reverse transcription with Cy3-dUTP and Cy5-dUTP fluorescent dyes. Total RNA from the strain containing pTrc99A plasmid was cultured at a dilution rate of 0.1 h−1 and used as the common reference (always labeled with Cy3-dUTP) against which total RNA extracted from cells cultured at five higher equally spaced dilution rates (always labeled with Cy5-dUTP) was hybridized. Similarly, for the strain containing the pTrc99A-nox plasmid, total RNA extracted from cells cultured at a dilution rate of 0.06 h−1 was the reference against which total RNA from cells cultured at five higher equally spaced dilution rates was hybridized. Differential gene expression between the two references (the NOX− strain cultured at a dilution rate of 0.1 h−1 and labeled with Cy3-dUTP, and the NOX+ strain cultured at a dilution rate of 0.06 h−1 and labeled with Cy5-dUTP) was also measured to identify transcriptional changes due to the presence of the S. pneumoniae NADH oxidase. All the hybridizations were performed at least in triplicate by using biologically independent samples as described previously (20) and incubating the labeled mixture on the arrays at 65°C overnight. The slide was subsequently washed and scanned using a GenePix 4000B microarray scanner (Axon Instruments, Union City, CA). The degree of labeling of the two dyes was quantified by measuring the intensity at a wavelength of 532 nm (for Cy3) or 635 nm (for Cy5). The relative expression of a gene was calculated as the base 2 logarithmic ratio of the background subtracted intensity from the Cy5 channel to the background subtracted intensity from the Cy3 channel, and the resulting value was referred to as the expression ratio.
Analysis of gene expression data.
Our goal for the analysis of transcription data was to identify genetic changes that corresponded to physiological observations. Specifically, we were interested in identifying those genes whose expression was sensitive to a perturbation in the redox ratio (i.e., the NADH/NAD ratio). We calculated the Pearson correlation coefficient for the expression ratio of each gene with the redox ratio for NOX+ and NOX− as a function of specific glucose consumption rate. Only those genes whose expression ratios had a high correlation coefficient (R > 0.9 or R < −0.9) with the redox ratio were considered for further analysis. These highly correlated (or anticorrelated) genes were classified into 22 functional categories according to the method of Riley (35). Each functional category was tested for significant overrepresentation (P < 0.05) by using a hypergeometric distribution (15). With a priori information on the distribution of the global gene set among the 22 categories, hypergeometric distribution measures the enrichment of a functional category based on the number of genes of that particular category appearing in the cluster. The P value for each category was calculated according to the following equation:
 |
where M is the total number of genes in the genome, x is the number of common genes, N is the total number of genes in the cluster, and K is the total number of genes in the functional category.
Only those genes from the significantly enriched functional categories were selected to study common regulatory mechanisms governing their expression. Any coregulation among these coexpressed genes was identified by searching for common transcription factor binding sites upstream of their transcription start sites. Sequences 300 bp upstream of the filtered genes were analyzed for common sequence motifs by using the hidden Markov model-based BioProspector software (24).
RESULTS
Physiological response due to NADH oxidase overexpression.
We used two isogenic strains that differ only in the presence of the nox gene, MG1655/pTrc99A (NOX−) and MG1655/pTrc99A-nox (NOX+). In batch cultures, NOX− had a maximum growth rate (μmax) of 0.70 h−1 while NOX+ grew more slowly, with a μmax of 0.51 h−1. Based on these results, seven equally spaced dilution rates were selected for chemostat experiments to assess steady-state physiological and transcriptional responses to the overexpression of the nox gene. Both NOX− and NOX+ exhibited fully respiratory metabolism until a critical dilution rate (or growth rate) was reached, above which respirofermentative metabolism was observed. The value of this critical dilution rate was about 0.4 h−1 for the control, NOX−, and about 0.3 h−1 for NOX+. No glucose was observed in the effluent for a dilution rate of less than 0.4 h−1.
As shown in Fig. 1, acetate overflow is directly related to the rate at which the sole carbon source (glucose) is consumed, with acetate formation occurring only after glucose consumption surpasses some threshold rate. The presence of heterologous NADH oxidase had the effect of increasing the critical glucose consumption rate (qScrit) at which acetate first appeared and thereby delaying the entry of E. coli into respirofermentative overflow metabolism (Fig. 1). This transition between respiratory and respirofermentative metabolism occurred at a qScrit of 0.8 g/g dry cell weight (DCW) h for NOX− and 1.2 g/g DCW h for NOX+. The expression of NADH oxidase therefore increased by 50% the value of qScrit. During respirofermentative metabolism, NOX+ exhibited a lower effluent acetate concentration and a lower specific acetate formation rate (qA) than NOX− at any given qS. Biomass yield (YX/S) from glucose (g dry cell weight/g glucose consumed) was 0.42 to 0.48 g/g for NOX− during respiratory metabolism but decreased during respirofermentative metabolism, consistent with a portion of the glucose carbon being diverted from biomass synthesis to acetate formation. For NOX+, YX/S remained 0.28 g/g at glucose consumption rates above 0.5 g/g DCW h (Fig. 1).
FIG. 1.
Steady-state physiological profiles of E. coli in the presence of heterologous NADH oxidase. YX/S (⋄, ⧫) and qA (○, •) values are compared for NOX− (open symbols and dashed lines) and NOX+ (solid symbols and lines) as functions of the specific glucose consumption rate. The highest dilution rate studied was about 80% of μmax for both strains. The arrows indicate for each strain the critical specific glucose consumption rates at which acetate formation commenced.
The specific oxygen consumption rate (qO2) was twice as great for NOX+ as for NOX− at any given value of qS (Fig. 2), consistent with additional oxygen being required for increased oxidation of NADH to NAD. NOX+ also yielded a specific CO2 evolution rate (qCO2) that was about 50% greater than that of NOX− for any qS (Fig. 2), suggesting greater flux through CO2-forming pathways (e.g., the TCA cycle) for NOX+. The results show that in the presence of NADH oxidase, cells diverted less carbon to biomass and acetate and more carbon to CO2 at any given rate of glucose consumption. A carbon balance for NOX− was within ±8% under all conditions, while for NOX+ the carbon balance was within ±15% (data not shown), assuming identical biomass composition (and thus identical expression of biosynthetic genes). The redox balance closed for NOX− within ±9%, while for NOX+ this balance was only within ±30% (data not shown).
FIG. 2.
Steady-state respiration for NOX− (open symbols and dashed lines) and NOX+ (solid symbols and lines). The steady-state qO2 (▵,▴) and qCO2 (▿,▾) values are shown as functions of qS.
Intracellular response due to NADH oxidase overexpression.
Since the expression of heterologous NADH oxidase in E. coli would be expected to influence the steady-state intracellular NADH and NAD concentrations, the concentrations of each cofactor were determined at each steady state for both strains. For both NOX− and NOX+, the intracellular concentration of NAD changed less than 30%, while the NADH concentration changed more than 10-fold between the lowest and highest glucose consumption rates. Moreover, the NADH concentration increased more quickly for NOX− at lower values of qS than for NOX+. For example, at a qS of about 0.10 g/g DCW h, the NADH concentration was 0.03 μmol/g DCW for both strains, while at a qS of about 1.0 g/g DCW h, the NADH concentration was 0.53 μmol/g DCW for NOX− but only 0.11 μmol/g DCW for NOX+. These changes are reflected in the NADH/NAD ratios (redox ratios) (Fig. 3). At any given value of qS, the redox ratio was always greater for NOX− than for NOX+. The redox ratio remained at 0.01 to 0.02 for both strains during respiratory metabolism but increased just prior to the onset of acetate overflow. Acetate formation for both strains occurred at an identical redox ratio of about 0.06 (Fig. 3). Clearly, this critical redox ratio marked a boundary between respiratory metabolism and respirofermentative metabolism. These results indicate a correlation between the redox ratio and acetate formation. What remains unclear is whether acetate formation is a consequence of the cells achieving the critical redox ratio, whether the increased redox ratio is caused by acetate formation, or whether these two phenomena are independent consequences of some underlying change in metabolism when the glucose consumption rate surpasses qScrit.
FIG. 3.
In vivo molar concentration ratio of NADH/NAD for NOX− (□) and NOX+ (▪) as functions of qS. The critical value of the NADH/NAD ratio at which acetate formation commences is about 0.06 for both NOX− and NOX+ (indicated by vertical lines). qA values are also shown for NOX− (○) and NOX+ (•) as functions of qS.
We measured the steady-state intracellular concentrations of key glycolytic intermediates in order to identify imbalances between glucose consumption and its subsequent metabolism that might occur. Steady-state pools of early glycolytic intermediates (glucose-6-phosphate and fructose-6-phosphate) increased with increasing qS for NOX− (Fig. 4A). Although pyruvate concentration increased, PEP concentration decreased markedly just at the onset of acetate formation, so that the pyruvate:PEP ratio increased from about 0.95 to 25. For NOX+, steady-state concentrations for each metabolite were essentially identical to those for NOX− at the lowest qS. However, the balance between PEP and pyruvate did not vary much with increasing qS (Fig. 4B), with the pyruvate/PEP ratio increasing from about 0.90 to only 1.3.
FIG. 4.
Intracellular concentrations of key glycolysis metabolites glucose-6-phosphate (red), fructose-6-phosphate (green), PEP (yellow), pyruvate (blue), and acetyl-CoA (pink) were measured under steady-state conditions in NOX− (A) and NOX+ (B). qA values are also shown for NOX− (open circles and dashed lines) and NOX+ (filled circles and solid lines) as functions of qS.
Pyruvate and PEP in particular participate in a large number of biochemical reactions, and therefore, these metabolites tightly regulate a large portion of the metabolic network. The observed increase in the steady-state level of pyruvate could indicate increased fluxes in pathways using pyruvate as a substrate or as an enzyme activator, such as those pathways leading to the formation of acetate. Any shift in the pyruvate/PEP ratio suggests a shift in the degree of utilization of pathways that involve pyruvate compared to PEP. The correlation between acetate overflow and pyruvate/PEP ratio is consistent with an elevated intracellular level of pyruvate being a precursor to acetate formation.
Transcriptional response to increasing glucose consumption rate.
Since most physiological events originate at the transcription level, we measured the transcriptional responses to changes in qS for NOX− and NOX+ strains to establish a genetic basis for the observed physiological changes. For each strain, we used a low value for qS (corresponding to dilution rates of 0.1 h−1 for NOX− and 0.06 h−1 for NOX+) as the reference. We first compared the transcription profiles for the two reference cultures (NOX− grown at 0.1 h−1 and NOX+ grown at 0.06 h−1) to identify transcriptional changes only due to the presence of NADH oxidase. There were no significant transcriptional changes between these two reference cultures, suggesting limited influence of NADH oxidase at low qS. This result is consistent with the similar physiological parameters (YX/S, qS, qCO2, and qO2) and redox ratios observed for the two strains at low qS values (Fig. 1 to 3).
Next, we compared the transcriptional changes at higher values for qS relative to the appropriate reference culture for each strain. In general, we did not observe drastic changes in gene expression, but many genes exhibited a reproducible monotonically increasing or decreasing behavior relative to the reference as qS or growth rate increased. For NOX−, the expression of 427 genes varied significantly with qS (P < 0.01), while only 47 genes achieved this level of significance for NOX+ and only 21 genes were common to both subsets. Among the genes whose expression varied significantly for both NOX− and NOX+ were key genes in the biosynthesis of threonine, serine, and nucleotides along with acs, which encodes acetyl-CoA synthetase. Since expression of these genes changed with qS for both strains, their expression is presumably largely glucose consumption rate dependent and relatively insensitive to the redox state of the cell. The average expression ratios of all genes involved in the central metabolic pathways for NOX− and NOX+ relative to their respective reference cultures are shown in Fig. 5 as functions of qS. Transcription profiles of individual genes in these central metabolic pathways for the two strains are shown in Fig. S1 in the supplemental material.
FIG. 5.
Transcriptional profile of central metabolic pathways for NOX− (dashed lines) and NOX+ (solid lines). The mean values of the expression ratios are shown for all genes involved in glycolysis (green), the TCA cycle (red), the pentose phosphate pathway (blue), and respiration (black) as functions of qS. Vertical lines show the demarcation between respiratory and respirofermentative metabolism for NOX− (dotted) and for NOX+ (solid). See Fig. S1 in the supplemental material for detailed expression profiles of individual genes.
The expression of most of the central metabolic genes (genes involved in the glycolysis TCA cycle, the pentose phosphate pathway, and respiration) increased during the respiratory phase of metabolism, but began to decrease just prior to respirofermentative metabolism for both NOX− and NOX+, despite qScrit being 50% higher for NOX+. Regarding some of the key genes of interest to acetate formation, isocitrate dehydrogenase (icd) and citrate synthase (gltA) are inhibited by NADH and have been implicated in the control of flux in the TCA cycle (12, 38), and we also observed repression of these genes for NOX− but induction for NOX+ as the glucose consumption rate increased. Thus, these genes appear to control TCA cycle flux at two levels: through enzyme activity and transcription. Interestingly, some TCA cycle genes (e.g., sucC, sucD) similarly appear to be controlled at both levels, while other TCA cycle genes (e.g., sucB, sdhC) encode enzymes not known to be controlled by the redox ratio but which show similar repression with increasing qS. Induction of the acetate kinase gene (ackA) correlated with the formation of acetate for NOX−, while expression of the phosphotransacetylase gene (pta) was slightly repressed. For NOX+ the expression of ackA and pta increased with qS during respiratory metabolism and remained constant during respirofermentative metabolism (see Fig S1 in the supplemental material). The key acetate consumption gene, acs, was severely (more than fivefold) repressed in both strains with increasing qS. The pyruvate oxidase gene (poxB) was induced for NOX− at low qS values and repressed at high qS values. Genes involved in aerobic respiration, such as the nuo operon (NADH dehydrogenase I chains), were generally repressed for NOX− (Fig. 5; see Fig. S1 in the supplemental material), and this repression was relieved for NOX+. The relative expression of the ndh gene encoding NADH dehydrogenase II, a primary source of NAD turnover under aerobic conditions, increased steadily with qS for NOX− and NOX+ during respiratory growth before saturation under respirofermentative conditions.
The relative expression of intermediate metabolic genes involved in the biosynthesis of amino acids and nucleotides increased with qS, before either stabilizing or slightly decreasing at high qS values for both NOX− and NOX+ (see Fig. S1 in the supplemental material). Moreover, we did not observe significant differences in the expression profiles of these genes between the strains at any given value of qS, except for those involved in methionine and glycine biosynthesis. The relative expression of metABCEHJL genes either remained steady or decreased with qS for NOX−, while these genes were significantly upregulated for NOX+. The glycine biosynthesis genes, particularly glyA, were repressed for NOX−, but the repression appeared to be reduced for NOX+. Purine and pyrimidine nucleotide biosynthesis genes monotonically increased with qS for both strains. We also observed a repression in most of the transport genes at high qS values for both NOX− and NOX+. Among the genes encoding for symport or antiport proteins we found a few genes of the multifacilitator family that were upregulated (such as yhfC, yhaU, codB, uraA, and proP) while all others (including yjcG, lacY, gltS, gntT, dctA, tatC, melB, and nupG) were repressed (see Fig. S1 in the supplemental material). Genes belonging to the ATP binding cassette transporters and the PEP-dependent phosphotransferase systems for the uptake of several sugars (including glucose) were also strongly repressed for both NOX− and NOX+ as qS increased (see Fig. S1 in the supplemental material). There was no particular trend observed in most unclassified genes except the gene encoding b4249 (a putative oxidoreductase), which was induced for NOX− but was repressed for NOX+ as qS increased.
Identifying coregulation among coexpressed genes.
The approach governing our data analysis methodology was to first group coexpressed genes and then evaluate these gene groups for common regulatory mechanisms (see Materials and Methods). Genes involved in the biosynthesis of amino acids, cofactors, macromolecules, and nucleotides along with central and intermediary metabolic genes were positively correlated with qS (R > 0.9) for both NOX− and NOX+. Among the genes that were negatively correlated with qS (R < −0.9) were those responsible for the degradation of small molecules, transport proteins, and unclassified genes. Interestingly, we observed that only the genes involved in the biosynthesis of amino acids, cofactors, and nucleotides along with the central and intermediary metabolic genes were correlated (R > 0.9) with the redox ratio for NOX− (Fig. 6). There were also several partially classified genes in this subset, suggesting that the expression of a majority of these genes depends on the rate of glucose consumption and/or redox. While we identified strongly overrepresented sequences upstream of genes correlated with qS, we could not relate these sequences with any of the known promoter binding sites. However, a significantly overrepresented (P < 10−170) sequence (Fig. 6) upstream of the genes correlated with the redox ratio for NOX− was identified (by BioProspector) as the binding site for ArcA (25, 29, 33). The identification of an ArcA binding site upstream of genes that were correlated with the redox ratio (for NOX−) is consistent with a recent discovery that cellular redox state is the signal for the activation of ArcB signal transduction (9, 28). Table S2 in the supplemental material provides a complete list of genes in NOX− that showed a reduction in expression by high NADH/NAD (negatively correlated with redox ratio) and which were determined (by BioProspector) to have a binding site for ArcA. In light of these results, we speculated that the strong repression observed for several TCA cycle and respiratory genes in NOX− at high qS values might be relieved by deleting arcA.
FIG. 6.
(Left) Hierarchical clustering of genes (rows) that are correlated (R > 0.9 or R < −0.9) with the redox ratio (NADH/NAD) in NOX− as a function of increasing qS (columns). (Right) Significantly overrepresented functional categories are shown in the table, along with the number of genes in each category and the P value of its significance as calculated using a hypergeometric distribution. Several key genes involved in the TCA cycle, respiration, and biosynthesis exhibited a strong negative correlation with the redox ratio. A large portion of the genes negatively correlated to the redox ratio were partially classified, revealing redox-dependent regulation of many of these genes.
Characterization of arcA mutant.
The identification of ArcA binding sites upstream of genes correlated with the redox ratio prompted us to characterize the phenotypes of QC2575/pTrc99A (ARCA− NOX−) and QC2575/pTrc99A-nox (ARCA− NOX+). In batch culture, the μmax for ARCA− NOX− (0.73 h−1) was similar to that for NOX−, but the μmax for ARCA− NOX+ (0.63 h−1) was 20% greater than the value for NOX+. We performed accelerostat experiments (18) for ARCA− NOX− and ARCA− NOX+ to provide a pseudo-steady-state representation of physiological changes over a range of dilution rates (0.20 h−1 to 0.54 h−1). An accelerostat approximates the environment inside a chemostat, and these experiments began at steady state (after seven volume changes at a dilution rate of 0.2 h−1). Once a steady state had been established, the dilution rate was slowly increased at a constant acceleration rate of 0.01 h−2. For these experiments, the yield YX/S was about 30% lower for ARCA− NOX+ than for ARCA− NOX− (Fig. 7). The most striking result of deleting arcA was the absence of acetate for ARCA− NOX+ even at the highest dilution rate studied. The value of qScrit for ARCA− NOX− was 0.9 g/g DCW h, while we did not observe acetate even when qS was equal to 1.5 g/g DCW h for ARCA− NOX+ (Fig. 7). The values of qO2 and qCO2 were 40% greater for ARCA− NOX− than for NOX−, while they remained constant at about 27 mmol/g DCW h and 22 mmol/g DCW h, respectively, for ARCA− NOX+ (Fig. 8). Although parameters obtained with a steady-state chemostat may differ from those obtained with a pseudo-steady-state accelerostat, we have similarly observed no acetate formation for ARCA− NOX+ in batch fermentations (data not shown).
FIG. 7.
Physiological characterization of ARCA− NOX− (open symbols and dashed lines) and ARCA− NOX+ (solid symbols and lines) in accelerostat cultures. YX/S (⋄,⧫) and qA (○, •) values are compared as functions of specific glucose consumption rate. The steady-state values of these parameters, obtained for NOX− (dashed lines without symbols) and NOX+ (solid lines without symbols) by using chemostats, are also shown.
FIG. 8.
Respiration of ARCA− NOX− (open symbols and dashed lines) and ARCA− NOX+ (solid symbols and lines) in accelerostat cultures. qO2 (▵,▴) and qCO2 (▿,▾) values are compared as functions of specific glucose consumption rate. The steady-state values of these parameters, obtained for NOX− (dashed lines without symbols) and NOX+ (solid lines without symbols) by using chemostats, are also shown.
Encouraged by these results, we measured pseudo-steady-state gene expression in ARCA− NOX− relative to that in NOX− when both strains were grown at a specific growth rate of 0.4 h−1 (the critical growth rate for NOX−). The most important transcriptional changes in response to deletion of arcA occurred in genes involved in the TCA cycle and respiration. The expression of these genes increased over sevenfold for ARCA− NOX− (see Table S3 in the supplemental material), presumably leading to the observed qCO2 and qO2 values being greater than those for NOX−. The fivefold increase in expression of the ptsG gene did not translate into a higher qS value for ARCA− NOX− than for NOX−, providing further evidence that glycolysis is not transcriptionally limited. Interestingly, the ptsG gene has also been demonstrated to be under ArcA control (17). Among the 110 genes showing significant difference between the strains (P < 0.01), 30 of them are not classified while 21 are partly classified, including some regulatory genes (such as ispH [P = 0.008]). Furthermore, we analyzed the 300 bp upstream of each gene whose expression of the ArcA binding box was statistically significant (P < 0.01) in ARCA− NOX− and compared it to NOX−. BioProspector identified a binding site for ArcA upstream of about 60% of these genes. A comprehensive list of all genes with P values of <0.01 is given in Table S3 in the supplemental material.
DISCUSSION
The primary physiological consequences of providing additional means to oxidize excess NADH were reduction of acetate formation and biomass yield and a 50% increase in qScrit. An increase in qScrit at the expense of biomass formation indicates faster NADH turnover (i.e., both generation and consumption). Higher qO2 and qCO2 values for NOX+ also indicate higher glycolytic and TCA cycle flux. In the current study, increased NADH turnover due to overproduced NADH oxidase led to a 70% increase in glucose uptake at any given dilution rate (see Fig. S1 in the supplemental material), revealing a strong link between the rate of glycolysis and NAD availability. This result is in accordance with the view that control of glycolysis principally resides outside the pathway (32). A previous study (16) with ATP synthase mutants similarly increased the rate of glycolysis. More recently, increasing ATP hydrolysis by overexpressing F1-ATPase in E. coli was shown to increase the ADP pool and qS values by 70% with a concomitant reduction in YX/S (22), leading to the conclusion that demand for ATP could control glycolytic flux. Our experiments with overproducing NADH oxidase similarly increased qS values and also reduced the intracellular redox ratio, and E. coli responded by upregulating genes involved in the TCA cycle and PDH complex, pathways that synthesize NADH and generate CO2. These results provide experimental evidence to support the theory that glycolytic flux is controlled by the cellular demand for global cofactors such as NADH and ATP.
Although the rapid generation and subsequent oxidation of NADH in the NOX+ strain essentially introduces a futile NAD turnover, it reveals two very important metabolic events correlated with overflow metabolism in E. coli: both the redox ratio and the pyruvate/PEP ratio are correlated with the appearance of acetate. First, the redox ratio at the onset of acetate overflow was, surprisingly, identical for NOX− and NOX+ (Fig. 3), indicating a relationship between the redox state of the cell and overflow metabolism. Since numerous reactions utilize or generate NADH, the redox ratio and overflow metabolism are likely to be the consequences of a complex network of metabolic events. The importance of NADH/NAD in by-product formation in E. coli has been previously demonstrated through increased reduction of NAD, which resulted not only in increased acetate but also in the appearance under aerobic conditions of typical fermentation products (5). In our study, TCA cycle genes which were generally repressed for NOX− with increasing qS values commonly showed less repression upon introduction of NADH oxidase. Considering that acetate overflow has thus far been assumed to be due to rate-limiting enzymes of the TCA cycle or the electron transport chain attaining maximum reaction velocity (1, 7, 13, 27), our results provide evidence that acetate overflow occurs as a consequence of transcriptional repression of the TCA cycle and respiratory genes (see Fig. S1 in the supplemental material). The introduction of NADH oxidase appears to delay the attainment of the critical redox ratio and limit acetate formation.
Also, the pyruvate/PEP ratio appears to be related to acetate formation (Fig. 4). As pyruvate is the branch point between respiration and fermentation and a precursor to several macromolecules, its level is highly regulated. In E. coli, PEP is a cosubstrate for glucose uptake and for the principal anaplerotic pathway during growth on glucose. Since acetate is produced from pyruvate directly (via pyruvate oxidase) or indirectly (via the PDH complex and acetate pathway), a 25-fold increase in the pyruvate/PEP ratio would shift the thermodynamic equilibrium towards pyruvate utilization by these pathways. Although an increase in the pyruvate/PEP ratio may not directly cause acetate overflow, the observed shift in the control strain NOX− does signal the onset of a bottleneck at the entrance to the TCA cycle, which we have shown can be modulated by redox. The introduction of NADH oxidase served to decrease the pyruvate/PEP ratio and by mass action would make acetate formation less favorable. These results provide circumstantial evidence for considering pyruvate to be one of the candidate signaling metabolites for inducing the phosphorylation of ArcB (14). NADH was previously proposed as a possible signal (14), and more recent evidence (9) indicates that the cellular redox state is the signal for the activation of Arc regulation while pyruvate is an allosteric activator (9).
The strong correlation (R > 0.9) between the redox ratio and the expression of genes involved in central and intermediary metabolism and the biosynthesis of amino acids, cofactors, and nucleotides demonstrates the important regulatory control exerted by the redox state. The identification of binding sites for the ArcA protein upstream of many of these genes suggests redox-dependent regulation of the ArcAB system and is consistent with recent studies which propose that the redox state triggers the Arc system (9). Our analysis does not rule out the possibility of secondary regulation, and therefore the relationships between redox state, ArcA, and acetate overflow could be indirect. Although ArcA-mediated repression has been reported for many individual genes (operons), their integrated effect on induction of overflow metabolism has been largely overlooked. The reduced redox ratio for NOX+ may delay significant activation of the Arc system. For the ARCA− strains, several of the TCA cycle and respiratory genes were induced and qCO2 was elevated, demonstrating greater TCA cycle flux. This view is in accordance with recent 13C-labeling studies using Arc mutants of E. coli and showing increased TCA flux under both aerobic and anaerobic conditions (34). The resulting higher rate of NADH formation appears to be accommodated at least partly by the elevated qO2 which results from derepression of the respiratory chain in these strains. Importantly, although the qScrit was about 10% greater for ARCA− NOX− compared to NOX−, acetate formation was even more pronounced for ARCA− NOX− at higher levels of qS. One possible explanation for this observation is that the heightened TCA cycle flux resulting from the absence of ArcA-mediated repression elevated NADH accumulation to a level beyond the capacity of the (derepressed) respiratory chain. Without a transcriptional mechanism to prevent further NADH formation in the TCA cycle, acetate formation may have occurred through some other mechanism (such as inhibition of citrate synthase). The overexpression of NADH oxidase in the arcA strain seems sufficient to provide another outlet for NADH oxidation and prevent acetate formation even at high glucose consumption rates.
In summary, our results using steady-state chemostats support a model in which an increase in the redox ratio contributes to a repression of the TCA cycle and to acetate formation, and they suggest that this overflow is due to transcriptional limitation. Providing another outlet for NADH turnover relieves TCA cycle gene repression and delays acetate formation. An arcA mutation delays the onset of acetate formation (only so far) through the maintenance of both TCA cycle flux and respiration. Moreover, a strain with an arcA mutation and heightened NADH oxidase activity appears able to both maintain an elevated TCA cycle flux and alleviate NADH buildup, thereby preventing acetate formation altogether. Considering the deleterious impact of acetate on growth (26) and recombinant protein production (37) and the wide variety of genetic and process approaches proposed to reduce acetate formation, our findings provide evidence at the level of transcription for the cause of acetate overflow as well as offer a means to overcome it.
Supplementary Material
Acknowledgments
We thank Sidney Kushner and Jens Nielsen for technical advice on the manuscript. We also acknowledge Sarah Lee and Kristopher DeWitt for technical support.
This project was supported by the U.S. DOE (grant number DE-FG36-01ID14007), which provided a fellowship to G.N.V., and the Georgia Experiment Stations. D.P.S. was supported in part by the NSF (grant number QSB 0222636 to Friedrich Srienc and A.B.K.).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Andersen, K. B., and K. von Meyenburg. 1977. Charges of nicotinamide adenine nucleotides and adenylate energy charge as regulatory parameters of the metabolism in Escherichia coli. J. Biol. Chem. 252:4151-4156. [PubMed] [Google Scholar]
- 2.Andersen, K. B., and K. von Meyenburg. 1980. Are growth rates of Escherichia coli in batch cultures limited by respiration? J. Bacteriol. 144:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auzat, I., S. Chapuy-Regaud, G. Le Bras, D. Dos Santos, A. D. Ogunniyi, I. Le Thomas, J. R. Garel, J. C. Paton, and M. C. Trombe. 1999. The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol. Microbiol. 34:1018-1028. [DOI] [PubMed] [Google Scholar]
- 4.Bernofsky, C., and M. Swan. 1973. An improved cycling assay for nicotinamide adenine dinucleotide. Anal. Biochem. 53:452-458. [DOI] [PubMed] [Google Scholar]
- 5.Berrios-Rivera, S. J., G. N. Bennett, and K. Y. San. 2002. Metabolic engineering of Escherichia coli: increase of NADH availability by overexpressing an NAD+-dependent formate dehydrogenase. Metabol. Eng. 4:217-229. [DOI] [PubMed] [Google Scholar]
- 6.Eiteman, M. A., and M. J. Chastain. 1997. Optimization of the ion-exchange analysis of organic acids from fermentation. Anal. Chim. Acta 338:69-70. [Google Scholar]
- 7.El-Mansi, E. M. T., and W. H. Holms. 1989. Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J. Gen. Microbiol. 135:2875-2883. [DOI] [PubMed] [Google Scholar]
- 8.Emmerling, M., M. Dauner, A. Ponti, J. Fiaux, M. Hochuli, T. Szyperski, K. Wuthrich, J. E. Bailey, and U. Sauer. 2002. Metabolic flux responses to pyruvate kinase knockout in Escherichia coli. J. Bacteriol. 184:152-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgellis, D., O. Kwon, and E. C. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 10.Higuchi, M., M. Shimada, Y. Yamamoto, T. Hayashi, T. Koga, and Y. Kamio. 1993. Identification of two distinct NADH oxidases corresponding to H2O2-forming oxidase and H2O-forming oxidase induced in Streptococcus mutans. J. Gen. Microbiol. 139:2343-2351. [DOI] [PubMed] [Google Scholar]
- 11.Hollywood, N., and H. W. Doelle. 1976. Effect of specific growth rate and glucose concentration on growth and glucose metabolism of Escherichia coli K-12. Microbios 17:23-33. [PubMed] [Google Scholar]
- 12.Holms, H. 1996. Flux analysis and control of the central metabolic pathways in Escherichia coli. FEMS Microbiol. Rev. 19:85-116. [DOI] [PubMed] [Google Scholar]
- 13.Holms, H. 2001. Flux analysis: a basic tool of microbial physiology. Adv. Microb. Physiol. 45:271-340. [DOI] [PubMed] [Google Scholar]
- 14.Iuchi, S., A. Aistarkhov, J. M. Dong, J. S. Taylor, and E. C. C. Lin. 1994. Effects of nitrate respiration on expression of the Arc-controlled operons encoding succinate dehydrogenase and flavin-linked l-lactate dehydrogenase. J. Bacteriol. 176:1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakt, L. M., L. Cao, K. S. Cheah, and D. K. Smith. 2001. Assessing clusters and motifs from gene expression data. Genome Res. 11:112-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen, P. R., and O. Michelsen. 1992. Carbon and energy metabolism of atp mutants of Escherichia coli. J. Bacteriol. 174:7635-7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong, J.-Y., Y.-J. Kim, N. Cho, D. Shin, T.-W. Nam, S. Ryu, and Y.-J. Seok. 2004. Expression of ptsG encoding the major glucose transporter is regulated by ArcA in Escherichia coli. J. Biol. Chem. 279:38513-38518. [DOI] [PubMed] [Google Scholar]
- 18.Kasemets, K., M. Drews, I. Nisamedtinov, K. Adamberg, and T. Paalme. 2003. Modification of A-stat for the characterization of microorganisms. J. Microbiol. Methods 55:187-200. [DOI] [PubMed] [Google Scholar]
- 19.Khodursky, A. B., B. J. Peter, N. R. Cozzarelli, D. Botstein, P. O. Brown, and C. Yanofsky. 2000. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:12170-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khodursky, A. B., J. A. Bernstein, B. J. Peter, V. Rhodius, V. F. Wendisch, and D. P. Zimmer. 2003. Escherichia coli spotted double-strand DNA microarrays: RNA extraction, labeling, hybridization, quality control, and data management. Methods Mol. Biol. 224:61-78. [DOI] [PubMed] [Google Scholar]
- 21.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koebmann, B. J., H. V. Westerhoff, J. L. Snoep, D. Nilsson, and P. R. Jensen. 2002. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol. 184:3909-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonardo, M. R., P. R. Cunningham, and D. P. Clark. 1993. Anaerobic regulation of the adhE gene, encoding the fermentative alcohol dehydrogenase of Escherichia coli. J. Bacteriol. 175:870-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, X., D. L. Brutlag, and J. S. Liu. 2001. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001:127-138. [PubMed] [Google Scholar]
- 25.Liu, X., and P. De Wulf. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 279:12588-12597. [DOI] [PubMed] [Google Scholar]
- 26.Luli, G. W., and W. R. Strohl. 1990. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl. Environ. Microbiol. 56:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majewski, R. A., and M. M. Domach. 1990. Simple constrained-optimization view of acetate overflow in E. coli. Biotechnol. Bioeng. 35:732-738. [DOI] [PubMed] [Google Scholar]
- 28.Malpica, R., B. Franco, C. Rodríguez, O. Kwon, and D. Georgellis. 2004. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. USA 101:13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGuire, A. M., P. DeWulf, G. M. Church, and E. C. Lin. 1999. A weight matrix for binding recognition by the redox-response regulator ArcA-P of Escherichia coli. Mol. Microbiol. 32:219-221. [DOI] [PubMed] [Google Scholar]
- 30.Meyer, H.-P., C. Leist, and A. Fiechter. 1984. Acetate formation in continuous culture of Escherichia coli K12 D1 on defined and complex media. J. Biotechnol. 1:355-358. [Google Scholar]
- 31.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach. Sinauer Associates, Sunderland, Mass.
- 32.Oliver, S. 2002. Metabolism: demand management in cells. Nature 418:33-34. [DOI] [PubMed] [Google Scholar]
- 33.Pellicer, M. T., A. S. Lynch, P. DeWulf, D. Boyd, J. Aguilar, and E. C. Lin. 1999. A mutational study of the ArcA-P binding sequences in the aldA promoter of Escherichia coli. Mol. Gen. Genet. 261:170-176. [DOI] [PubMed] [Google Scholar]
- 34.Perrenoud, A., and U. Sauer. 2005. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J. Bacteriol. 187:3171-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riley, M. 1998. Genes and proteins of Escherichia coli K-12 (GenProtEC) Nucleic Acids Res. 26:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaefer, U., W. Boos, R. Takors, and D. Weuster-Botz. 1999. Automated sampling device for monitoring intracellular metabolite dynamics. Anal. Biochem. 270:88-96. [DOI] [PubMed] [Google Scholar]
- 37.Swartz, J. R. 2001. Advances in Escherichia coli production of therapeutic proteins. Curr. Opin. Biotechnol. 12:195-201. [DOI] [PubMed] [Google Scholar]
- 38.Underwood, S. A., M. L. Buszko, K. T. Shanmugam, and L. O. Ingram. 2002. Flux through citrate synthase limits the growth of ethanologenic Escherichia coli KO11 during xylose fermentation. Appl. Environ. Microbiol. 68:1071-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.










