Abstract
A closely related group of alphaproteobacteria were found to be present in seven genera of marine sponges from several locations and were shown to be transferred between sponge generations through the larvae in one of these sponges. Isolates of the alphaproteobacterium were cultured from the sponges Axinella corrugata, Mycale laxissima, Monanchora unguifera, and Niphates digitalis from Key Largo, Florida; Didiscus oxeata and Monanchora unguifera from Discovery Bay, Jamaica; an Acanthostronglyophora sp. from Manado, Indonesia; and Microciona prolifera from the Cheasapeake Bay in Maryland. Isolates were very similar to each other on the basis of 16S rRNA gene sequence (>99% identity) and are closely related to Pseudovibrio denitrificans. The bacterium was never isolated from surrounding water samples and was cultured from larvae of M. laxissima, indicating that it is a vertically transmitted symbiont in this sponge. Denaturing gradient gel electrophoresis, 16S rRNA gene clone library analysis, and fluorescent in situ hybridization with probes specific to the alphaproteobacterium confirmed the presence of this bacterium in the M. laxissima larvae. The alphaproteobacterium was densely associated with the larvae rather than being evenly distributed throughout the mesohyl. This is the first report of the successful culture of a bacterial symbiont of a sponge that is transferred through the gametes.
Sponges are simple multicellular invertebrates that filter large quantities of seawater and acquire nutrients by phagocytosis of captured bacteria. Bacteria embedded in sponge mesohyl tissue were first described in electron microscope studies by Levi and Levi (30). Sponges have been shown to harbor a large number of bacteria, in some cases up to 60% of the tissue volume (46). Microorganisms detected so far in sponges include archaea, heterotrophic bacteria, cyanobacteria, red and green algae, dinoflagellates, and diatoms (19, 20, 51). Although bacteria are a food source for sponges, not all bacteria in the mesohyl are broken down for food, and certain bacteria form symbiotic relationships with sponges. These symbiotic relationships are involved in nutrient acquisition (54), supply of photosynthate from photoautotrophic symbionts to the sponge (40, 53), stabilization of the sponge skeleton (35), metabolic waste processing (5, 52), and production of secondary metabolites (37). Sponges are a well-established source of important natural products, and some of these may be produced by the bacterial symbionts (18, 21).
Several studies have been performed on the culturable and total microbial communities of various marine sponges. The microbial diversity associated with marine sponges was reviewed by Lee et al. (28), Hentschel et al. (19), and Hill (21). Several recent reports describe the use of molecular techniques to study the diversity of sponge-associated microbial communities (14, 25, 33, 41-43, 49). Hentschel et al. (20) suggested that a uniform microbial assemblage is present in sponges from different oceans.
A major unresolved issue regarding sponge/bacterial symbiosis is the nature of the relationship between the bacteria and the sponge. In general, it is not clear whether this relationship is truly mutualistic or is in fact a commensal relationship in which the bacterium is the only party benefiting by obtaining nutrients and a specialized niche. So far, in most cases, contributions from the bacterial assemblage to the sponge have not been shown conclusively. For this reason, symbiosis is here defined as a consistent association of a microbe with the sponge host, regardless of whether a benefit has been ascribed to the microbe or the host.
An important objective has been to obtain sponge symbionts in culture to investigate the role of these bacteria in the sponge symbiosis. In a study of the Great Barrier Reef sponge Rhopaloides odorabile Thompson et al. 1987, an alphaproteobacterium (designated NW001) was the dominant bacterium in the sponge-associated culturable assemblage (48). This bacterium dominated the culturable assemblage of 44 individuals of R. odorabile independent of seasonal or spatial sampling variations and was never detected in the surrounding water. Two R. odorabile individuals that lacked strain NW001 were clearly diseased, and a related alphaproteobacterial symbiont capable of causing disease in sponges was isolated from one of these (50). Taken together, these factors provide strong circumstantial evidence that strain NW001 is a true sponge symbiont in R. odorabile.
There are two pathways whereby a developing sponge may acquire bacterial symbionts. The first is by selective absorption of specific bacteria from the large diversity of bacteria in the surrounding water column that passes through the sponge during filter feeding. The second is by vertical transmission of symbionts through the gametes of the sponge by inclusion of the bacteria in the oocytes or larvae. Transmission of symbionts via the gametes of the host has been well documented in insects, especially aphids where total numbers of the symbiont Buchnera range from 850 to over 8,000 per egg depending on the aphid species (32). Transmission of bacteria within sponge oocytes was first suggested based on electron microscope studies (17, 29). Symbiotic cyanobacteria of the sponge Chondrilla australiensis Carter 1873 are present in the sperm and are also passed through the eggs by nurse cells that transport symbionts from the surface layers of the sponge to the eggs deeper in the sponge matrix and release their contents into the egg cytoplasm (44, 45). A similar role for nurse cells was postulated in the Barents Sea sponge Leucosolenia complicata Montagu 1818 (3). Bacteria were detected in the larvae of five Oscarella species together with maternal mesohyl material (11). Furthermore, recent evidence has emerged for the vertical transmission of symbiotic bacteria of the viviparous sponge Halisarca dujardini Johnson 1842 (12).
In this study, the presence of culturable symbionts from several sponge species was investigated. A group of alphaproteobacteria, closely related to sponge symbiont strain NW001, were ubiquitous in sponges across different oceans and therefore provide a good model for studying bacterial sponge symbiosis. Seven sponge species from four locations were sampled (Table 1). The culturable bacterial communities of these sponges were studied, and total DNA was extracted from the sponges for molecular community analysis. Mycale laxissima sponges sampled from Key Largo, Fla., were found to contain larvae. This provided an ideal opportunity to study the microbiology of sponge larvae and to determine whether alphaproteobacteria and other bacterial symbionts may be vertically transmitted.
TABLE 1.
Sponge systematics, taxonomic identification, and locality and habitat dataa
| Taxon | Sample identifier
|
Locality and habitat | |
|---|---|---|---|
| BMNH | Other sources | ||
| Class Demospongiae Sollas 1885, order Poecilosclerida Topsent 1928, family Mycalidae Lundbeck 1905, Mycale (Arenochalina) laxissima (Duchassaing and Michelotti 1864) | NIWAKD 3071 | Conch Key, Key Largo, Fla. (sandy patch reef, 22 m) | |
| Family Microcionidae Carter 1875, Clathria (Microciona) prolifera Ellis and Solander 1786 | RTH | Mouth of the Hampton River, Chesapeake Bay, Md. (wharf pilings and pontoon at low tide) | |
| Family Crambeidae Lévi 1963, Monanchora unguifera (de Laubenfels 1953) | BMNH 2000.7.17.3 | MKB 2040 | Discovery Bay, Jamaica (reef slope, 40 m) |
| NIWAKD 3074 | Conch Key, Key Largo, Fla. (sandy patch reef, 16 m) | ||
| Order Halichondrida Vosmaer 1887, family Axinellidae Carter 1875, Axinella corrugata (George and Wilson 1919) | NIWAKD 3075 | Conch Key, Key Largo, Fla. (sandy patch reef, 22 m) | |
| Family Desmoxyidae Hallmann 1917, Didiscus oxeata Hechtel 1983 | BMNH 2001.7.20.1 | NIWAKD 1057 | Rio Beano, Jamaica (caves and vertical walls, 25-40 m) |
| Order Haplosclerida Topsent 1928, family Niphatidae Van Soest 1980, Niphates digitalis (Lamarck 1814) | NIWAKD 3076 | Conch Key, Key Largo, Fla. (sandy patch reef, 22 m) | |
| Family Petrosiidae Van Soest 1980, Acanthostrongylophora sp. (undescribed) | BMNH 2003.2.17.1 | NIWAKD 1107 | Manado, Sulawesi, Indonesia (silty reef base, 6-23 m) |
| BMNH 2002.5.13.2 | NIWAKD 1108 | Manado Bay, Indonesia (vertical reef slopes, 40 m) | |
Abbreviations for specimen registration numbers: BMNH, British Museum of Natural History; NIWAKD, National Institute of Water and Atmospheric Research, New Zealand (Auckland Collection); MKB, personal collection of M.K.; RTH, personal collection of R.T.H.
MATERIALS AND METHODS
Sponge collection and taxonomic identification.
Three sponges of each species were collected by scuba divers from Jamaica in the tropical West Central Atlantic, the Keys region of Florida; the Chesapeake Bay in Maryland; and Manado, Sulawesi, Indonesia, in Southeast Asian waters. Voucher specimens were either preserved in 70% ethanol immediately upon collection or freeze-dried for chemical characterization. Freeze-dried specimens were prepared for histology by dehydration in a very weak solution of detergent for 24 h, followed by preservation in 70% ethanol. Histological sections and spicule preparations were made as in the work of Kelly-Borges and Vacelet (24). Specimens are stored for the long term in 70% ethanol. Voucher specimens representing material that was chemically characterized were registered with the Natural History Museum (formerly the British Museum of Natural History), and specimens are deposited in their sponge collections (Table 1).
Processing of sponges for microbial culture.
Disposable latex gloves were worn during collection. Sponges were removed from seawater and rinsed with sterile artificial seawater (ASW) to remove any transient bacteria from the surface and channels of the sponge. In each case, three individuals of each sponge species were processed for microbiological studies. All sponge samples were processed immediately for isolation of culturable bacteria. For each sponge, a 1-cm3 section of sponge tissue was excised with a sterile scalpel and ground into a slurry in 10 ml of sterile ASW. A dilution series was plated onto marine agar 2216 (BD Biosciences, Franklin Lakes, NJ). Additional sponge samples were stored frozen at −80°C. Larval masses from M. laxissima were carefully separated from the surrounding tissue and rinsed in sterile ASW. Triplicate water samples were collected adjacent to the sponges in every case of sponge collection, and serial dilutions were plated onto marine agar 2216. A total of eight water samples were processed in attempts to culture NW001-like alphaproteobacteria.
Identification of isolates by 16S rRNA gene sequence analysis.
Isolates were purified by streaking for single colonies that were inoculated into 25-ml volumes of marine broth 2216 (BD Biosciences) and shaken overnight at 30°C. All isolates were stored at −80°C in marine broth 2216 supplemented with 30% glycerol. DNA was extracted from isolates using the UltraClean microbial DNA isolation kit (Mo Bio Laboratories Inc., Carlsbad, CA). Isolates were identified by 16S rRNA gene sequence analysis. The 16S rRNA genes from the isolates were amplified by PCR using primers 27f and 1492r (27). Cycling conditions were a 5-min hot start at 94°C; 20 cycles of 30 s at 92°C, 2 min at 48°C, and 1.5 min at 72°C; and a final 5-min extension step at 72°C. Thermal cycling was performed in a PTC-200 cycling system (MJ Research, Waltham, Mass.). PCR products were sequenced using primers 27f and 1492r. Analysis was performed on 1,380 bp of sequence.
Characterization of isolates.
Isolates JE005, JE008, JE013, JE021, JE041, JE061, JE062, JE063 JE064, JE065, and JE066 were characterized using API 20NE strips (bioMérieux, Paris, France). Salinity requirements of strains were determined by assessing growth on the basis of visual appearance of turbidity in PYN medium (38). PYN medium was augmented with NaCl to concentrations of 0 to 8% in 1% increments.
Analysis of bacteria associated with sponge larvae.
Isolates from the larvae were processed as above. In addition, total bacterial diversity was assessed using molecular methods. DNA was extracted from larvae with the UltraClean microbial DNA kit. A high-fidelity Taq polymerase was used (Platinum Taq High Fidelity; Invitrogen Life Technologies, Carlsbad, Calif.) to amplify the 16S rRNA genes from the total larval DNA with a low-cycle-number PCR (25 cycles) in three replicate reactions. Primers 27f and 1492r were used with the following cycling conditions: a 5-min hot start at 94°C; 25 cycles of 30 s at 92°C, 2 min at 48°C, and 1.5 min at 72°C; and a final 5-min extension step at 72°C. The samples were pooled and gel purified with the QIAquick gel extraction kit (QIAGEN, Valencia, Calif.) and cloned into Escherichia coli using the TOPO XL kit (Invitrogen Life Technologies). A random selection of 40 clones was sequenced using the M13 forward primer with an ABI 377 automated sequencer using the PRISM Ready Reaction kit (Applied Biosystems, Foster City, Calif.). Clones from which >500 bp of sequence was obtained from the 5′ end of the 16S rRNA gene were included in the phylogenetic analysis.
Collection and nucleic acid extraction from seawater samples.
Water samples were collected in sterile 20-liter containers adjacent to the sponges sampled. Water samples (10 liters) were filtered through 0.22-μm-pore-size Sterivex filters (Millipore), and nucleic acids were extracted by the method of Somerville et al. (39).
Phylogenetic analysis.
Sequences were compared to available databases using the Basic Local Alignment Search Tool (BLAST) (1) to determine approximate phylogenetic affiliations. Chimeric sequences were identified using the program CHECK_CHIMERA (31). Partial sequences were manually compiled and aligned using Phydit software (7). The evolutionary tree was generated using the neighbor-joining (36), Fitch-Margoliash (15), and maximum parsimony (26) algorithms in the PHYLIP package (13). Evolutionary distance matrices for the neighbor-joining and Fitch-Margoliash methods were generated as described previously by Jukes and Cantor (22). The robustness of inferred tree topologies was evaluated after 1,000 bootstrap resamplings of the neighbor-joining data.
DGGE.
DNA from alphaproteobacterial isolate JE063 from the larvae, total DNA extracted from sponge larvae, and total DNA from a seawater sample were analyzed by denaturing gradient gel electrophoresis (DGGE). Primers P2 and P3 (34) were used to amplify the 194-bp region corresponding to positions 341 and 534 in the 16S rRNA gene of E. coli. PCR amplification was performed on 100 ng of DNA with Platinum Taq (Invitrogen Life Technologies) and 25 pmol of each primer. The cycling conditions were a 5-min hot start at 94°C; 30 cycles of 1 min at 92°C, 1 min at 55°C, and 1 min at 72°C; and a final 5-min extension step at 72°C. Thermal cycling was performed in a PTC-200 cycling system (MJ Research, Waltham, Mass.). The final PCR product was loaded onto a 6% acrylamide gel with a denaturing gradient of 45 to 65%. Electrophoresis was performed using the D-Code system (Bio-Rad, Calif.) in 1× TAE (20 mM Tris acetate, 10 mM sodium acetate, 0.5 mM EDTA) at a constant temperature of 60°C and voltage of 60 V for 16 h. The gel was stained with 1× SYBR green (Molecular Probes Inc., Eugene, Oreg.) for 10 min and visualized with the Typhoon 9410 image system (Amersham Biosciences, United Kingdom).
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed on one of the alphaproteobacterial strains to investigate possible genome reduction and presence or absence of large plasmids. Intact chromosomal DNA of isolate JE063 was prepared from a 3-day-old culture grown in marine broth 2216 at 30°C with shaking. Cells were harvested and washed with cell suspension buffer (10 mM Tris-HCl [pH 7.6], 150 mM NaCl, 50 mM EDTA) and resuspended to a concentration of 5 × 109 cells/ml. Cells were mixed with InCert agarose (FMC Bioproducts, Rockland, ME) to a final concentration of 1%. The cell suspension (100 μl) was pipetted into a plug mold and chilled at −20°C for 5 min. Gel plugs were treated with lysozyme solution (10 mM Tris-HCl [pH 7.6], 50 mM NaCl, 0.2% sodium deoxycholate, 0.5% sodium lauryl sarcosine, 1 mg/ml lysozyme) at 37°C for 16 h. Plugs were transferred to proteinase K solution (100 mM EDTA [pH 8], 0.2% sodium deoxycholate, 1% sodium lauryl sarcosine, 1 mg/ml proteinase K) at 50°C for 16 h. Plugs were washed four times for 1 h in TE buffer (20 mM Tris-HCl [pH 8], 50 mM EDTA). Proteinase K was inactivated by inclusion of 1 mM phenylmethylsulfonyl fluoride in the second wash. Plugs were stored in TE buffer at 4°C. Using the CHEF-DRIII system (Bio-Rad), undigested plugs were subjected to PFGE on a 1% agarose gel with 0.5× Tris-borate-EDTA buffer to determine whether this strain contained large plasmids. The pulse program used was a ramping progression of 20 to 60 s at 200 V for 24 h. A lambda ladder and Saccharomyces cerevisiae chromosomes (Bio-Rad) were used as molecular weight markers.
FISH.
Samples for fluorescent in situ hybridization (FISH) were fixed in 70% ethanol prior to embedding and sectioning as described previously (48). Sections were hybridized with 5-carboxytetramethylrhodamine (TAMRA)-labeled probes (Invitrogen Life Technologies) eubacterial probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′) (2) and a previously described probe for strain NW001 designated NW442 (5′-AGTTAATGTCATTATCTTCACTGC-3′) (48). Negative controls used were nonsense EUB338 (5′-CTCCTACGGGAGGCAGC-3′) (2) and NW442 with a 2-bp mismatch (5′-AGTTAAGTTCATTATCTTCACTGC-3′). Probe NW442 is specific for strain NW001 (48), and strain JE061 and the other NW001-like alphaproteobacteria have identical 16S rRNA gene sequences in this target region. Hybridization was performed according to the methods described by Webster and Hill (48). Images were captured by laser scanning confocal microscopy using the Radiance 2100 system (Bio-Rad).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 16S rRNA gene sequence fragments from isolates are DQ097237 to DQ097264. The GenBank accession numbers of cloned 16S rRNA gene fragments from the uncultured assemblage are DQ098829 to DQ098850.
RESULTS
Culturable assemblage.
A morphotype with a distinctive brown, highly mucoid colony morphology was present on marine agar 2216 isolation plates from all seven sponge species. In the cases of sponges Axinella corrugata and M. laxissima (from Florida), Monanchora unguifera and Didiscus oxeata (from Jamaica), and Acanthostrongylophora sp. (from Indonesia), this was the dominant morphotype present in the culturable assemblage. Counts were obtained for sponges M. unguifera (from Jamaica and Florida), D. oxeata, and A. corrugata. In these cases, the abundances of the alphaproteobacterial morphotype expressed as a percentage of total culturable bacteria were as follows: M. unguifera (Jamaica), 65%; M. unguifera (Florida), 37%; D. oxeata, 72%; A. corrugata, 52%. In samples from sponges Microciona prolifera (from the Chesapeake Bay) and Niphates digitalis (from Florida), this morphotype was present in lower numbers as a smaller proportion of the cultural assemblage (<10%). This morphotype was never detected in the culturable assemblage from the surrounding water column at any of the sampling sites.
Phylogenetic analysis.
Representative isolates of this morphotype were identified by 16S rRNA gene sequence analysis. Interestingly, all isolates formed a tight clade and are closely related to strain NW001 (99% identity) from the Great Barrier Reef sponge R. odorabile, previously described as an important symbiont (48). Isolates were closely related to marine isolate MBIC3368 (AB012864) (99% identity) and the marine facultative anaerobe Pseudovibrio denitrificans (AY4486423) (99% identity). A culture collection of these morphologically distinct alphaproteobacteria, containing a total of 60 isolates from the eight sponge species sampled, was assembled.
Characterization of isolates.
API 20NE profiles were identical for all JE strains tested. The strains were positive for denitrification. The strains were positive for indole production on tryptophan, glucose acidification, and gelatin hydrolysis and weakly positive for β-galactosidase. Strains were negative for oxidase, arginine dihydrolase, urease, and esculin hydrolysis and negative for assimilation of the following sugars: d-glucose, l-arabinose, d-mannose, d-mannitol, N-acetylglucosamine, d-maltose, potassium gluconate, capric acid, adipic acid, malic acid, trisodium citrate, and phenylacetic acid. All isolates had similar growth patterns as shown by salinity testing, with no growth on 0 and 8% NaCl and growth on 1 to 7% NaCl. Optimal growth was achieved with 3% NaCl. Strain characteristics are generally similar to those of P. denitrificans except for the oxidase reaction.
Isolation and phylogenetic analysis of bacteria from larvae.
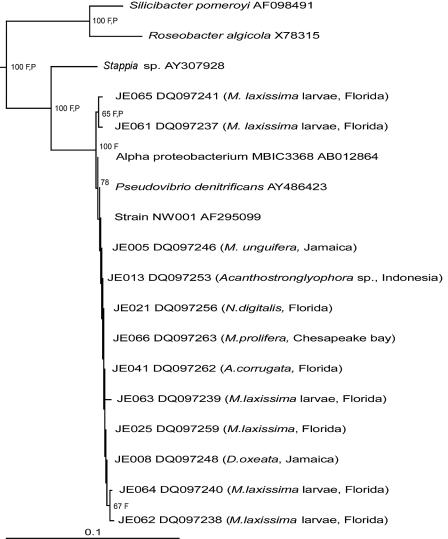
The characteristic alphaproteobacterial morphotype was the only colony type that grew on marine agar 2216 inoculated with samples from these larvae. Five isolates, designated JE061 to JE065, were selected. The relationship of 16S rRNA gene sequences of the isolates from the larvae to the other alphaproteobacterial isolates is shown in Fig. 1.
FIG. 1.
Neighbor-joining phylogenetic tree from analysis of >1,380 bp of 16S rRNA gene sequence of isolates from the different sponges studied. One representative alphaproteobacterial sequence was taken from each species. Also included are the five isolates from the larvae of M. laxissima. F and P indicate branches that were supported by the Fitch-Margoliash and maximum parsimony methods, respectively. The numbers at the roots indicate bootstrap support based on a neighbor-joining analysis of 1,000 resampled data sets and are given as percentages with only values of >50% shown. The scale bar represents 0.1 substitutions per nucleotide position. Escherichia coli was used as the outgroup.
16S rRNA gene clone library construction.
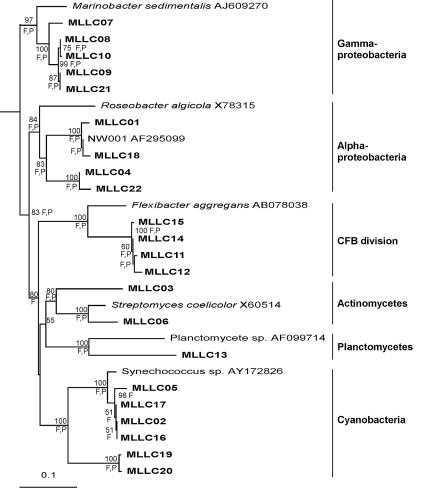
16S rRNA gene sequences from 22 larva-derived clones were used for phylogenetic analysis (Fig. 2). There was a higher diversity of bacteria within the larvae than was observed from the culture-based analysis. Six distinct groups were observed in these clones with representatives from the alphaproteobacteria (18% of clones), gammaproteobacteria (23%), actinobacteria (9%), planctomycetes (5%), and the divisions cyanobacteria (27%) and Cytophaga-Flexibacter-Bacteroides (18%). Two of the alphaproteobacterial clones had high 16S rRNA gene sequence similarity (>99%) with strain NW001.
FIG. 2.
Neighbor-joining phylogenetic tree from analysis of ca. 600 bp of 16S rRNA gene sequence of clones in the library constructed from total DNA extracted from larvae of M. laxissima. F and P indicate branches that were supported by the Fitch-Margoliash and maximum parsimony methods, respectively. The numbers at the roots indicate bootstrap support based on a neighbor-joining analysis of 1,000 resampled data sets and are given as percentages with only values of >50% shown. The scale bar represents 0.1 substitutions per nucleotide position. E. coli was used as the outgroup. CFB, Cytophaga-Flexibacter-Bacteroides.
DGGE.
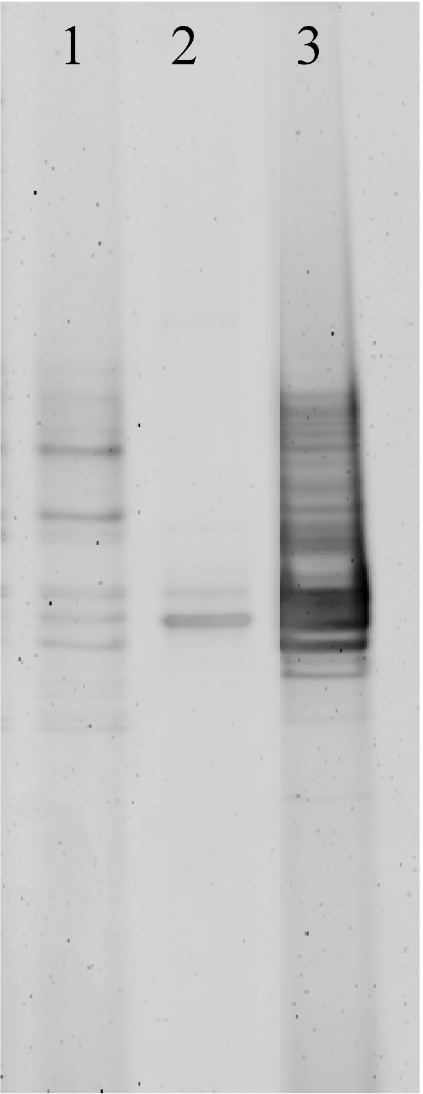
The DGGE profiles from the larvae, isolate JE063, and seawater were compared (Fig. 3). Isolate JE063 generated two bands that corresponded in position to bands in the larvae, suggesting the presence of this isolate in the larval assemblage. Unfortunately, this could not be confirmed by sequence analysis of bands excised from DGGE gels as this approach was not successful in yielding usable sequence data. The banding pattern from total DNA from the larvae was more complex than that for isolate JE063 alone, corroborating the clone library analysis in indicating the presence of an assemblage of bacteria in addition to the NW001-like alphaproteobacteria.
FIG. 3.
DGGE analysis of DNA extracted from M. laxissima larvae and from alphaproteobacterial isolates obtained from these larvae. Lane 1, larval DNA; lane 2, isolate JE063; lane 3, DNA from seawater microbial communities.
The banding pattern from the seawater sample indicated a much higher diversity than that from the larvae, having fewer clear dominant bands. A high number of bands was present in the region corresponding to the bands from isolate JE063. Sequence analysis of excised DNA from this region was not successful. DGGE was therefore not conclusive in indicating the presence or absence of NW001-like alphaproteobacteria in the water sample.
PFGE.
Pulsed-field electrophoresis was performed on cells of isolate JE063 from the larva samples. Undigested samples revealed the presence of two large plasmids of approximately 540 kb and 430 kb.
FISH.
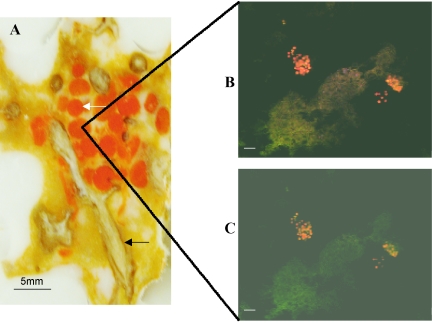
Figure 4A shows a cross-section of M. laxissima tissue containing a group of the orange larvae. FISH was performed on the larvae, focusing on the central parts of sectioned larvae. Eubacterial probe EUB338 (Fig. 4B) revealed clusters of bacteria that were densely associated with larvae rather than being evenly distributed throughout the mesohyl. The NW001-specific probe NW442 (Fig. 4C) showed the presence of the NW001-like alphaproteobacteria within these clusters. The sections shown in Fig. 4B and Fig. 4C are consecutive. Analysis of FISH images allowed us to estimate that NW001-like alphaproteobacteria represented approximately 50% of the total bacteria present in the larvae.
FIG. 4.
(A) Cross-section of the mesohyl of M. laxissima showing bright orange larvae among paler orange choanosome and tan skeletal fibers. The section is embedded in solid OCT (Tissue-Tek; Sakura, Inc., Torrance, Calif.). The white arrow indicates larvae; the black arrow indicates sponge fibers. (B and C) FISH images of the inside of the larvae, obtained by scanning confocal microscopy. The larvae were hybridized with TAMRA-labeled universal bacterial probe EUB338 (B) and TAMRA-labeled probe NW442, specific for sponge-associated alpha-proteobacteria (C). Scale bars, 10 μm.
DISCUSSION
This is the first report of a culturable bacterium that is vertically transmitted through sponge larvae. The presence of closely related alphaproteobacteria in the culturable communities of eight sponge species (48) from different locations in the Atlantic and Southeast Asian waters indicates a close relationship between these bacteria and sponges. The presence of closely related bacteria in sponges of different species is consistent with the findings of Hentschel et al. (20). The high numbers of the alphaproteobacteria that were detected and the fact that these bacteria were not cultured from the water column samples suggests that they are not present merely as food particles. However, it is important to note that although these bacteria dominate the culturable assemblage from several sponges, they are not dominant in the total bacterial assemblage within these sponges. A previous 16S rRNA gene community analysis of A. corrugata showed that the NW001-like alphaproteobacteria dominated the culturable assemblage in this sponge but in the molecular community analysis only 1 clone in 50 contained the NW001-like alphaproteobacterial 16S rRNA gene sequence (9). This confirms a previous study in which the alphaproteobacterium NW001 dominated the culturable assemblage of R. odorabile but was not detected in a 16S rRNA gene library (48).
The NW001-like alphaproteobacteria found in this and previous studies (9, 48) are closely related to Pseudovibrio denitrificans isolated from shallow water in Taiwan (38) with 99% identity in a 16S rRNA gene sequence over >1,380 bp. Although it is possible that P. denitrificans is genomically distinct from the sponge-derived alphaproteobacteria since our phylogenetic analysis is based on only 16S rRNA gene sequences, this seems unlikely because P. denitrificans clusters within the sponge-derived alphaproteobacteria and is also similar on the basis of biochemical characterization. Detection of P. denitrificans in the water column could mean that this organism and the closely related NW001-like alphaproteobacteria discussed in this study may be present in low numbers in the water column. Another possibility is that the shallow water sample from Taiwan contained alphaproteobacterial cells from nearby sponges that had expelled this organism due to disease, damage, or death.
The analysis of larvae from M. laxissima has provided evidence that these alphaproteobacteria are transferred from parent sponges to their offspring via the germ line. This is the first report of the successful culture of a bacterial symbiont of a sponge that is transferred through the gametes. It is clear that at least in the case of M. laxissima and possibly also in other sponges, these symbionts are most likely transmitted from one generation to the next rather than being acquired by the sponges from the surrounding water column. The developmental process associated with oocyte formation was reviewed by Ereskovskii (10). The nurse cells surrounding the oocyte serve as a source of yolk-type inclusions and are likely to contain symbiotic bacteria.
The possibility that the NW001-like alphaproteobacterial symbionts are acquired from the water column rather than through vertical transmission cannot be totally excluded. A 1-kg sponge can filter 24,000 liters per day (47). As pointed out by Hill (21), if a specific bacterium is present at a density of only 2.4 cells per 10,000 liters, 1 cell of this potential symbiont would still be acquired by the sponge each day. This is several orders of magnitude below the detection limit of most culture-based and molecular techniques, which generally depend on sample sizes in the range of milliliters to 10 liters. Although we convincingly demonstrate association of the NW001-like alphaproteobacterial symbionts with sponge larvae, we do not have data on the age of these larvae, and gametes were not directly sampled. It is therefore conceivable that larvae are colonized from the surrounding water early in their development. Future studies should focus on examination of gametes and further investigations of larvae in M. laxissima and other sponges in which we have demonstrated the occurrence of the NW001-like alphaproteobacterial symbionts.
In our study, alphaproteobacteria were the only bacteria cultured from the larval masses. However, molecular methods showed that there is a more complex assemblage of bacteria associated with the larvae, as revealed by DGGE and supported by the 16S rRNA gene clone library analysis of the larvae. The clone library analysis indicated the presence of alphaproteobacteria other than the NW001-like alphaproteobacteria, gammaproteobacteria, cyanobacteria, actinomycetes, and Flexibacter-like strains. Since these bacteria are presumably transmitted vertically between sponge generations they are likely to be important symbionts for the sponge. These bacteria were not grown with the straightforward culturing methods used in this study. FISH studies revealed that the NW001-like alphaproteobacteria comprised approximately 50% of the total bacteria observed within the larvae. This supports the presence of other types of bacteria in addition to alphaproteobacteria. The numbers of the NW001-like alphaproteobacteria detected in the 16S rRNA gene library analysis of the larvae were considerably less than 50%, suggesting that the NW001-like alphaproteobacteria may be underrepresented in 16S rRNA gene clone libraries.
The presence of the NW001-like alphaproteobacteria in such a wide variety of sponge species from various locations and detection within sponge larvae indicate that these alphaproteobacteria are true sponge symbionts. It is not yet clear whether the sponge benefits from the presence of these bacteria or what the nature of this relationship is. Webster et al. (48, 50) suggested that the NW001-like alphaproteobacterium was involved in sponge health as it was absent in two diseased individuals of R. odorabile.
It is striking that both phylogenetic analysis and biochemical profiling demonstrate that the alphaproteobacterial isolates are nearly identical despite being isolated from different sponge species collected from different oceans. The fact that no apparent species-specific coevolution has occurred within each host may indicate that the relationship was established relatively recently. It is possible that coevolution may be revealed by more detailed genomic analysis. However, in the case of Photobacterium leiognathi, which forms a bioluminescent symbiosis with various species of leiognathid fish, genomic profiling using PFGE demonstrated substantial heterogeneity with the same genomotype rarely observed in different individuals of the same host species (8). The related symbiont ‘Photobacterium kishitanii’ is a facultative symbiont that was also not cospeciating with its different species of host fish (4).
Unconventional genome organization is common in the alpha subgroup of the Proteobacteria (23). In alphaproteobacteria such as Rhizobium spp. (6) and Agrobacterium tumefaciens (16), large plasmids are often implicated in symbiotic processes. For these reasons, isolate JE063 was analyzed by PFGE to determine chromosome organization and screen for the presence of large plasmids. There was a single chromosome and two large plasmids of 430 and 540 kb. The possible role of these plasmids in symbiosis and/or sponge colonization by isolate JE063 will be an interesting area of study. This readily culturable alphaproteobacterial symbiont provides an ideal model system for future studies of the relationship between sponges and their bacterial symbionts.
Acknowledgments
Funding for this study was provided by the Microbial Observatories Program, National Science Foundation (MCB-0238515), and this support is gratefully acknowledged.
We thank Steven Miller, Director, and Otto Rutten, Science Manager, for providing sampling opportunities at Key Largo through the UNC—Wilmington National Undersea Research Center. George Burbanck and Robert Jordan at Hampton University are thanked for assistance in Chesapeake Bay collections. Mark Hamann of the University of Mississippi is thanked for facilitating collections at the Discovery Bay Marine Laboratories in Jamaica. Mark Hamann and Reinhart Garang are thanked for assistance in sample collection in Indonesia at the Nusantara Diving Center, Molas Beach, Manado. We thank Joy E. M. Watts for helpful comments on the manuscript.
Footnotes
Contribution no. 05-118 from the Center of Marine Biotechnology.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anakina, R. P., and A. L. Drozdov. 2001. Gamete structure and fertilization in the Barents Sea sponge Leucosolenia complicata. Russ. J. Mar. Biol. 27:143-150. [Google Scholar]
- 4.Ast, J. C., and P. V. Dunlap. 2005. Phylogenetic resolution and habitat specificity of members of the Photobacterium phosphoreum species group. Environ. Microbiol. 7:1641-1654. [DOI] [PubMed] [Google Scholar]
- 5.Beer, S., and M. Ilan. 1998. In situ measurements of photosynthetic irradiance responses of two Red Sea sponges growing under dim light conditions. Mar. Biol. 131:613-617. [Google Scholar]
- 6.Brom, S., L. Girard, A. Garcia-de los Santos, J. M. Sanjuan-Pinilla, J. Olivares, and J. Sanjuan. 2002. Conservation of plasmid-encoded traits among bean-nodulating Rhizobium species. Appl. Environ. Microbiol. 68:2555-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun, J. 1995. Computer-assisted classification and identification of actinomycetes. Ph.D. thesis. University of Newcastle upon Tyne, Newcastle upon Tyne, United Kingdom.
- 8.Dunlap, P. V., A. Jiemjit, J. C. Ast, M. M. Pearce, R. R. Marques, and C. R. Lavilla-Pitogo. 2004. Genomic polymorphism in symbiotic populations of Photobacterium leiognathi. Environ. Microbiol. 6:145-158. [DOI] [PubMed] [Google Scholar]
- 9.Enticknap, J. J., R. Thompson, O. Peraud, J. E. Lohr, M. T. Hamann, and R. T. Hill. 2004. Molecular analysis of a Florida Keys sponge: implications for natural products discovery. Mar. Biotechnol. 6:S288-S293. [Google Scholar]
- 10.Ereskovskii, A. V. 1999. Development of sponges of the order Haplosclerida. Russ. J. Mar. Biol. 25:361-371. [Google Scholar]
- 11.Ereskovsky, A. V., and N. Bouryesnault. 2002. Cleavage pattern in Oscarella species (Porifera, Demospongiae, Homoscleromorpha): transmission of maternal cells and symbiotic bacteria. J. Nat. Hist. 36:1761-1775. [Google Scholar]
- 12.Ereskovsky, A. V., E. Gonobobleva, and A. Vishnyakov. 2005. Morphological evidence for vertical transmission of symbiotic bacteria in the viviparous sponge Halisarca dujardini Johnston (Porifera, Demospongiae, Halisarcida). Mar. Biol. 146:869-875. [Google Scholar]
- 13.Felsenstein, J. 2004. PHYLIP (Phylogenetic Inference Package), version 3.6. Department of Genetics, University of Washington, Seattle.
- 14.Fieseler, L., M. Horn, M. Wagner, and U. Hentschel. 2004. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 70:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitch, W. M., and E. Margoliash. 1967. Construction of phylogenetic trees: a method based on mutation distances as estimated from cytochrome c sequences is of general applicability. Science 155:279-284. [DOI] [PubMed] [Google Scholar]
- 16.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 17.Gallisian, M. F., and J. Vacelet. 1976. Ultrastructure des quelques stades de l'ovognese de spongiares du genre Verongia (Dictyoceratida). Ann. Sci. Nat. Zool. Biol. Anim. 18:381-404. [Google Scholar]
- 18.Haygood, M. G., E. W. Schmidt, S. K. Davidson, and D. J. Faulkner. 1999. Microbial symbionts of marine invertebrates: opportunities for microbial biotechnology. J. Mol. Microbiol. Biotechnol. 1:33-43. [PubMed] [Google Scholar]
- 19.Hentschel, U., L. Fieseler, M. Wehrl, C. Gernert, M. Steinert, J. Hacker, and M. Horn. 2003. Microbial diversity of marine sponges, p. 59-88. In W. E. G. Müller (ed.), Sponges (Porifera). Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 20.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, R. T. 2004. Microbes from marine sponges: a treasure trove of biodiversity for natural products discovery, p. 177-190. In A. T. Bull (ed.), Microbial diversity and bioprospecting. ASM Press, Washington, D.C.
- 22.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 23.Jumas-Bilak, E., S. Michaux-Charachon, G. Bourg, M. Ramuz, and A. Allardet-Servent. 1998. Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J. Bacteriol. 180:2749-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly-Borges, M., and J. Vacelet. 1995. A revision of Diacarnus Burton and Negombata de Laubenfels (Demospongiae; Latrunculiidae) with descriptions of new species from the West Central Pacific and the Red Sea. Mem. Queensl. Mus. 38:477-503. [Google Scholar]
- 25.Kim, T. K., M. J. Garson, and J. A. Fuerst. 2005. Marine actinomycetes related to the “Salinospora” group from the Great Barrier Reef sponge Pseudoceratina clavata. Environ. Microbiol. 7:509-518. [DOI] [PubMed] [Google Scholar]
- 26.Kluge, A. G., and F. S. Farris. 1969. Quantitative phyletics and the evolution of annurans. Syst. Zool. 18:1-32. [Google Scholar]
- 27.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Hoboken, N.J.
- 28.Lee, Y. K., J.-H. Lee, and H. K. Lee. 2001. Microbial symbiosis in marine sponges. J. Microbiol. 39:254-264. [Google Scholar]
- 29.Levi, C., and P. Levi. 1976. Embryogenese de Chondrosia reinformis (Nardo), demosponge ovipare, et transmission des bacteries symbiotiques. Ann. Sci. Nat. Zool. Biol. Anim. 18:367-380. [Google Scholar]
- 30.Levi, C., and P. Levi. 1965. Populations bacteriennes dans les eponges. J. Microsc. (Paris) 4:60. [Google Scholar]
- 31.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mira, A., and N. A. Moran. 2002. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb. Ecol. 44:137-143. [DOI] [PubMed] [Google Scholar]
- 33.Montalvo, N. F., N. M. Mohamed, J. J. Enticknap, and R. T. Hill. 2005. Novel actinobacteria from marine sponges. Antonie Leeuwenhoek 87:29-36. [DOI] [PubMed] [Google Scholar]
- 34.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rützler, K. 1985. Associations between Caribbean sponges and photosynthetic organisms, p. 455-466. In K. Rützler (ed.), New perspectives in sponge biology. Smithsonian Institution Press, Washington, D.C.
- 36.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt, E. W., A. Y. Obraztsova, S. K. Davidson, D. J. Faulkner, and M. G. Haygood. 2000. Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel δ-proteobacterium, “Candidatus Entotheonella palauensis.” Mar. Biol. 136:969-977. [Google Scholar]
- 38.Shieh, W. Y., Y. T. Lin, and W. D. Jean. 2004. Pseudovibrio denitrificans gen. nov., sp. nov., a marine, facultatively anaerobic, fermentative bacterium capable of denitrification. Int. J. Syst. Evol. Microbiol. 54:2307-2312. [DOI] [PubMed] [Google Scholar]
- 39.Somerville, C. C., I. T. Knight, W. L. Straube, and R. R. Colwell. 1989. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl. Environ. Microbiol. 55:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steindler, L., S. Beer, and M. Ilan. 2002. Photosymbiosis in intertidal and subtidal tropical sponges. Symbiosis 33:263-273. [Google Scholar]
- 41.Taylor, M. W., P. J. Schupp, I. Dahllof, S. Kjelleberg, and P. D. Steinberg. 2004. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6:121-130. [DOI] [PubMed] [Google Scholar]
- 42.Taylor, M. W., P. J. Schupp, R. de Nys, S. Kjelleberg, and P. D. Steinberg. 2005. Biogeography of bacteria associated with the marine sponge Cymbastela concentrica. Environ. Microbiol. 7:419-433. [DOI] [PubMed] [Google Scholar]
- 43.Usher, K. M., J. Fromont, D. C. Sutton, and S. Toze. 2004. The biogeography and phylogeny of unicellular cyanobacterial symbionts in sponges from Australia and the Mediterranean. Microb. Ecol. 48:167-177. [DOI] [PubMed] [Google Scholar]
- 44.Usher, K. M., J. Kuo, J. Fromont, and D. C. Sutton. 2001. Vertical transmission of cyanobacterial symbionts in the marine sponge Chondrilla australiensis (Demospongiae). Hydrobiologia 461:9-13. [Google Scholar]
- 45.Usher, K. M., D. C. Sutton, S. Toze, J. Kuo, and J. Fromont. 2005. Inter-generational transmission of microbial symbionts in the marine sponge Chondrilla australiensis (Demospongiae). Mar. Freshw. Res. 56:125-131. [Google Scholar]
- 46.Vacelet, J., and C. Donadey. 1977. Electron microscope study of the association between some sponges and bacteria. J. Exp. Mar. Biol. Ecol. 30:301-314. [Google Scholar]
- 47.Vogel, S. 1977. Current-induced flow through living sponges in nature. Proc. Natl. Acad. Sci. USA 74:2069-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webster, N. S., and R. T. Hill. 2001. The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an α-Proteobacterium. Mar. Biol. 138:843-851. [Google Scholar]
- 49.Webster, N. S., A. P. Negri, M. M. Munro, and C. N. Battershill. 2004. Diverse microbial communities inhabit Antarctic sponges. Environ. Microbiol. 6:288-300. [DOI] [PubMed] [Google Scholar]
- 50.Webster, N. S., A. P. Negri, R. I. Webb, and R. T. Hill. 2002. A spongin-boring α-proteobacterium is the etiological agent of disease in the Great Barrier Reef sponge Rhopaloeides odorabile. Mar. Ecol. Prog. Ser. 232:305-309. [Google Scholar]
- 51.Webster, N. S., K. J. Wilson, L. L. Blackall, and R. T. Hill. 2001. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 67:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilkinson, C. R. 1978. Microbial associations in sponges. I. Ecology, physiology and microbial populations of coral reef sponges. Mar. Biol. 49:161-167. [Google Scholar]
- 53.Wilkinson, C. R. 1983. Net primary productivity in coral reef sponges. Science 219:410-412. [DOI] [PubMed] [Google Scholar]
- 54.Wilkinson, C. R., and R. Garrone. 1980. Nutrition of marine sponges. Involvement of symbiotic bacteria in the uptake of dissolved carbon, p. 157-161. In D. C. Smith and Y. Tiffon (ed.), Nutrition in the lower metazoa. Pergamon Press, Oxford, United Kingdom.