Abstract
The introduction of nonnative oysters (i.e., Crassostrea ariakensis) into the Chesapeake Bay has been proposed as necessary for the restoration of the oyster industry; however, nothing is known about the public health risks related to contamination of these oysters with human pathogens. Commercial market-size C. ariakensis triploids were maintained in large marine tanks with water of low (8-ppt), medium (12-ppt), and high (20-ppt) salinities spiked with 1.0 × 105 transmissive stages of the following human pathogens: Cryptosporidium parvum oocysts, Giardia lamblia cysts, and microsporidian spores (i.e., Encephalitozoon intestinalis, Encephalitozoon hellem, and Enterocytozoon bieneusi). Viable oocysts and spores were still detected in oysters on day 33 post-water inoculation (pwi), and cysts were detected on day 14 pwi. The recovery, bioaccumulation, depuration, and inactivation rates of human waterborne pathogens by C. ariakensis triploids were driven by salinity and were optimal in medium- and high-salinity water. The concentration of human pathogens from ambient water by C. ariakensis and the retention of these pathogens without (or with minimal) inactivation and a very low depuration rate provide evidence that these oysters may present a public health threat upon entering the human food chain, if harvested from polluted water. This conclusion is reinforced by the concentration of waterborne pathogens used in the present study, which was representative of levels of infectious agents in surface waters, including the Chesapeake Bay. Aquacultures of nonnative oysters in the Chesapeake Bay will provide excellent ecological services in regard to efficient cleaning of human-infectious agents from the estuarine waters.
The Eastern oyster (Crassostrea virginica), native to the Chesapeake Bay, has been a major shellfish species in the Bay for the past 3 centuries; however, its population has been devastated over time, beyond the point of restoration, predominantly due to overharvesting and oyster diseases such as multinucleated sphere X (MSX) disease (Haplosporidium nelsoni) and Dermo disease (Perkinsus marinus) (2, 4, 23). The Asian (Suminoe) oyster (Crassostrea ariakensis), a nonnative species, holds promise for rebuilding the collapsed commercial oyster industry, as it grows quickly (reaching market size in 1 year, versus 2 years for C. virginica), tolerates MSX and Dermo diseases, has a lower natural mortality, and is indistinguishable in taste from C. virginica (2, 25). The introduction of C. ariakensis into the Bay requires the use of genetically manipulated oysters, i.e., nonreproductive triploids (4). Adverse environmental risks associated with this introduction are not known; however, reversion in the wild to the diploid reproductive stage (4, 25) could force competition with the native oysters, establish C. ariakensis as a nuisance species, or introduce new oyster epibionts and/or disease(s) (2, 4, 25).
Although the popularity of seafood is high, concerns have been voiced worldwide related to health risks from shellfish contaminated with human waterborne pathogens (32). Consumption of oysters, which are usually eaten raw, can cause outbreaks of human diseases, especially if the oysters are harvested from polluted waters (1, 32). Native oysters commercially harvested from the Chesapeake Bay have been shown to contain human enteric pathogens (8, 9, 10); however, nothing is known about the recovery efficiency, retention, and inactivation rate of such pathogens by nonnative oysters. Depuration and relaying (i.e., purification) of oysters are an effective method for purging potential pathogens from live oysters (32), but it is only effective if the oysters' level of pathogen recovery, bioaccumulation, and inactivation are well understood (which is not the case for C. ariakensis). Oyster filtration and energy assimilation rates generally depend on water salinity (2, 25). The salinity of the Chesapeake Bay water varies considerably from the upper Bay down to the Atlantic Ocean, and C. ariakensis has a much wider tolerance to variations in water salinity than the native oysters (2, 25).
Cryptosporidium parvum, Giardia lamblia, and human-infectious microsporidia (i.e., Encephalitozoon intestinalis, Encephalitozoon hellem, and Enterocytozoon bieneusi) are human enteric pathogens that inflict considerable morbidity on healthy people and can cause mortality in immunosuppressed individuals (17, 38, 39). The transmissive stages of these pathogens, i.e., oocysts, cysts, and spores, respectively, are resistant to environmental stressors and are therefore long lasting and ubiquitous in an aquatic environment (27, 33, 39). Their viability in seawater lasts from several months up to 1 year (6, 36). Cryptosporidium and Giardia are transmitted via water (19, 33), which is also involved in the transmission of microsporidian spores (3, 5, 11, 22, 35). The diameter of C. parvum oocysts does not exceed 6 μm, and G. lamblia cysts and microsporidian spores are oval and no longer than 16 and 2 μm, respectively (17, 19, 38). The identification and viability assessment of the aforementioned pathogens can be accomplished using fluorescent in situ hybridization (FISH) assays (22, 24).
The purpose of the present study was to determine the recovery efficiency of human waterborne pathogens and the bioaccumulation, depuration, and inactivation rates of these pathogens by commercial market-size C. ariakensis triploids maintained in water of various salinity levels (i.e., low, medium, and high) representing the salinity range of the Chesapeake Bay (25). The inoculum of C. parvum oocysts, G. lamblia cysts, and spores of human-infectious microsporidia simulated the concentration of these pathogens reported for surface waters, including the Chesapeake Bay (9, 33, 34, 37).
MATERIALS AND METHODS
Three 114-liter (approximately 30-gallon)-capacity marine tanks were filled with dechlorinated drinking water filtered with a Filterite 10-μm-pore-size, yarn-wound cartridge (Memtec America Corp., Baltimore, MD). Different amounts of marine salt (Marine Enterprises International, Inc., Baltimore, MD) were added to create water with 8-, 12-, and 20-ppt salinities (i.e., low, medium, and high, respectively) in these tanks. The salinity was measured daily using a salinity meter (YSI, Inc., Yellow Spring, OH) and adjusted by addition of the filtered water. Each tank was equipped with two intermittently working air stones. The room temperature was controlled at 17°C. After 10 days, 114 commercial market-size (>7.6 cm) C. ariakensis triploids were shipped overnight in a cooler from the Virginia Institute of Marine Science, Aquaculture Genetics and Breeding Technology Center, Gloucester Point, VA. The Center bred these oysters in a quarantine tank using pathogen-free water. The oysters were evenly distributed among the tanks and acclimatized for 10 days. The oysters were fed on alternate days with 2 ml/tank of concentrated shellfish diet 1800 (ReedMarine, Inc., Campbell, CA) administered according to the manufacturer's instruction.
Each tank of water was inoculated with 1.0 × 105 transmissive stages each of C. parvum oocysts, G. lamblia cysts, and spores of E. intestinalis, E. hellem, and E. bieneusi. The oocysts, cysts, and spores were enumerated using a hemocytometer. The C. parvum and G. lamblia (genotype E) stocks originated from an experimental infection of a calf, and the microsporidian spores originated from in vitro cell line infections (E. intestinalis and E. hellem) and a human fecal sample (E. bieneusi) (22). The pathogens used for water inoculation were stored at 4°C for approximately 3 weeks prior to experiments. Twenty-four hours after water inoculation (day 1 post-water inoculation [pwi]), all the water from each tank was filtered by a cellulose acetate membrane (393-mm diameter and 1.2-μm pore size) (Millipore Corp., Bedford, MA) (9). Filtered water was circulated to the respective tanks, and efforts were made to collect sediment from the bottoms of the tanks during filtration. On day 6 pwi, a set of five oysters was collected randomly from each tank, and all water from each tank was filtered as described above. The procedure of oyster and water sampling was repeated six more times at 4- to 5-day intervals, i.e., on day 10, 14, 18, 22, 27, and 33 pwi, respectively.
Filter membranes were subjected to elution overnight at 4°C using approximately 500 ml of the eluting fluid (16), the eluants were centrifuged (10,000 × g, 10 min), supernatants were discharged, and pellets were stored in 75% ethanol. The hemolymph and gills were collected from each oyster (8), and the hemolymph and gill washings from the five oysters were pooled. Pooled samples were centrifuged (10,000 × g, 10 min), supernatants discharged, and pellets stored in 75% ethanol. Alcohol was washed from the pellets by centrifugation (10,000 × g, 10 min) two times using sterile phosphate-buffered saline and evenly divided into two aliquots. One aliquot was processed for assaying C. parvum and G. lamblia with a combined FISH and direct immunofluorescent antibody (IFA) method, and the other aliquot was processed for assaying human-infective microsporidia with FISH (22, 24). FISH oligonucleotide probes were synthesized by the DNA Analysis Facility of the Johns Hopkins University, Baltimore, MD, at a 1.0 μM scale, purified by high-pressure liquid chromatography, and 5′ end labeled with a single molecule of a fluorochrome (22). A fluorescein isothiocyanate-conjugated monoclonal IFA against the cell wall antigens of Cryptosporidium and Giardia from MERIFLUOR (Cryptosporidium/Giardia test kit; Meridian Diagnostic, Inc., Cincinnati, OH) was used (22). The walls of the pathogens' transmissive stages were permeabilized (22). All combined FISH and direct IFA assay reactions were carried out in Eppendorf tubes in a total volume of 100 μl of hybridization buffer at 48oC for 1 h (22). The concentration of each oligonucleotide probe, i.e., CRY-1, GIAR-4, and GIAR-6 (22), was 1 mmol liter−1, and the IFA mixture was diluted 1:1. The FISH reactions for human-infective microsporidia were carried out in Eppendorf tubes in a total volume of 100 μl of hybridization buffer at 57oC for 3 h (22, 24). The concentration of each oligonucleotide probe, i.e., HEL 878, INT-1, and BIEN-1 (22, 24), was 1 mmol liter−1. Positive controls were prepared from the same pathogen batches used for water inoculations, and negative controls were prepared as described previously (22). After hybridization, the tubes were centrifuged twice at 4oC (2,000 × g, 5 min), and the pellets were resuspended in 100 μl of sterile phosphate-buffered saline. Five 20-μl samples were transferred into lysine-coated wells (5-mm diameter) on a Teflon-coated glass slide (Carlson Scientific, Inc., Peotone, IL) and air dried. The entire area of a well was examined with the aid of an Olympus BH2-RFL epifluorescence microscope with a dry 60× objective and a BP450-490 exciter filter and without knowledge of the samples' identities, the pathogens were enumerated, and the samples were uncoded.
Statistical analysis was carried out with Statistix 7.0 (Analytical Software, St. Paul, MN). Variables were tested with the Wilk-Shapiro test and ranking plots to determine whether their distribution conformed to a normal distribution; a nonparametric test was used otherwise. Statistical significance was considered to be a P value of <0.05.
RESULTS
The combined FISH and direct IFA assays for C. parvum oocysts and G. lamblia cysts and the FISH assay for microsporidian spores revealed only viable pathogens in oysters and water samples. The oocysts were intact and had a small gap between the wall and internal structures, and the sporozoites were visible. The cysts were filled out completely with cytoplasm, without a gap between the internal structures and the wall. Spores of E. intestinalis, E. hellem, and E. bieneusi displayed typical microsporidian morphology, with more intense fluorescent staining in the polar half of the spore.
Cryptosporidium was the pathogen that was identified for the longest time period in the oysters (i.e., up to 33 days pwi), followed by the spores of human-infective microsporidia (i.e., 27 days pwi) and Giardia (i.e., 14 days pwi) (Table 1). The endpoint day of pathogen detection in oysters was influenced by the water salinity and the type of infectious agent (Table 1). All samples of oysters and water prior to the endpoint day were pathogen positive; however, irrespective of the pathogen and the water salinity, the number of pathogens in oysters progressively decreased from the values presented in Fig. 1 to a single pathogen cell (Table 1). Cryptosporidium parvum oocysts were still identified on day 33 pwi in oysters maintained in both low- and medium-salinity water and up to 27 days pwi in oysters kept in water at 20-ppt salinity (Table 1). Quite differently, in water of 20- and 12-ppt salinities, G. lamblia cysts were detected in oysters only on day 6 pwi, but they were detected on day 14 pwi in water of 8-ppt salinity (Table 1). Microsporidian spores were still identifiable on day 27 pwi in oysters maintained in both medium- and high-salinity water and on day 6 pwi in oysters kept in water at 8-ppt salinity (Table 1). Higher salinity (i.e., 12 and 20 ppt) was associated with a longer retention of microsporidian spores in oyster tissue, whereas G. lamblia cysts were retained longer at low salinity (Table 1). Water salinity had only a small effect on the endpoint detection of C. parvum oocysts (Table 1). Human waterborne pathogens were predominantly detected in oyster hemolymph, as distinct from gills (Table 1). Only microsporidian spores in oysters maintained in low-salinity water were detected in both hemolymph and gill washings (Table 1).
TABLE 1.
Endpoint day of detection of human waterborne pathogens in oysters and in seawatera
| Pathogen species | Endpoint day of detection in water with salinity ofb:
|
|||||
|---|---|---|---|---|---|---|
| 8 ppt (low)
|
12 ppt (medium)
|
20 ppt (high)
|
||||
| Oysters | Water | Oysters | Water | Oysters | Water | |
| Cryptosporidium parvum | 33 (H) | 1 | 33 (H) | ND | 27 (H) | ND |
| Giardia lamblia | 14 (H) | 1 | 6 (H) | ND | 6 (GH) | ND |
| Encephalitozoon intestinalis | 6 (H) | 33 | 27 (H) | 33 | 27 (H) | 1 |
| Encephalitozoon hellem | 6 (GH) | 33 | 27 (H) | 33 | 27 (H) | 1 |
| Enterocytozoon bieneusi | 6 (GH) | 33 | 27 (H) | 33 | 27 (H) | 1 |
Commercial market-size (>7.6-cm) triploids of Asian (Suminoe) oysters (C. ariakensis) maintained in 30-gallon-capacity marine tanks with water of various salinities were sampled (in batches of five specimens) together with the filtered water on day 6, 10, 14, 18, 22, 27 and 33 pwi with 1.0 × 105 transmissive stages of each of the pathogens. All samples collected before the endpoint day were pathogen positive.
Organisms were detected in hemolymph (H) and/or gills (G) of oysters. ND, not detected.
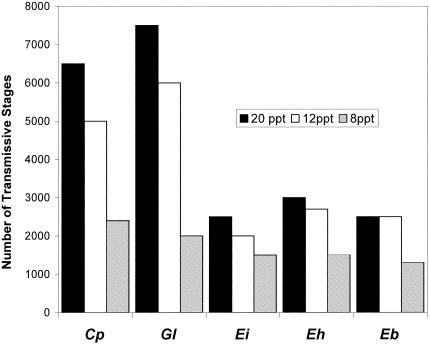
FIG. 1.
The numbers of C. parvum (Cp) oocysts, G. lamblia (Gl) cysts, and spores of E. intestinalis (Ei), E. hellem (Eh), and E. bieneusi (Eb) recovered from commercial market-size (>7.6-cm) triploids of Asian (Suminoe) oysters (Crassostrea ariakensis) maintained in 30-gallon-capacity marine tanks with various salinities (i.e., high, 20 ppt; medium, 12 ppt; and low, 8 ppt). Oysters were sampled in batches of five specimens on day 6 pwi with 1.0 × 105 transmissive stages of each of the pathogens.
Cryptosporidium parvum and G. lamblia were detected only on day 1 pwi in water filtrates from the tank with low-salinity water and were never detected in water at the two higher salinities (Table 1). Microsporidian spores were still detectable in water filtrates on day 33 pwi in water at 8- and 12-ppt salinities (Table 1). However, in the tank with high-salinity water, the spores were detected only on day 1 pwi (Table 1). Microsporidian spores were filtered out most efficiently by oysters kept in high-salinity water, as these pathogens were detected only on day 1 pwi (Table 1). The numbers of spores in the tank with low-salinity water were constantly high (mean, 88.0 ± 9.9 spores/liter), significantly higher than in water kept at 12-ppt salinity (mean, 19.1 ± 3.4 spores/liter) (Wilcoxon rank sum test; t = 8.2, P < 0.01), indicating that the filtration efficiency of oysters in 8-ppt-salinity water was low.
The results presented in Table 1 demonstrate that the recovery efficiency was, in general, higher for C. parvum oocysts and G. lamblia cysts than for microsporidian spores, because most of the water samples (89%) were oocyst and cyst negative while still positive for spores (83%). This indicates that the oysters did not efficiently filter out the microsporidian spores or did not retain the spores in their tissues at the same level as they retained oocysts and cysts. Overall, oysters maintained in high-salinity water had efficient recovery and retention of all pathogens (total of 83% of pathogen negative water samples) compared with oysters kept in low- and medium-salinity water, where all water samples were positive at all time points for microsporidian spores (Table 1). Irrespective of the pathogen species, the recovery efficiency was highest in oysters kept in high-salinity water, followed by oysters kept in medium- and low-salinity water (Fig. 1). The differences in the numbers of pathogens recovered by oysters in high-, medium-, and low-salinity water were significant (Kruskal-Wallis one-way analysis of variance; F = 8.7, P < 0.01). Overall, the cumulative number of oocysts and cysts filtered out by oysters was significantly higher than the cumulative number of microsporidian spores of all three species (Wilcoxon rank sum test, t = 1.90; P < 0.05) (Fig. 1).
In general, oysters depurated microsporidian spores very slowly and at a rate similar to that for C. parvum oocysts. In contrast, oysters depurated G. lamblia cysts quite quickly; however, the observation that water samples were negative for the cysts (Table 1) indicates that the cysts were most likely digested by the oysters. The most inefficient depuration of microsporidian spores and C. parvum oocysts was observed in oysters maintained in low- and medium-salinity water, and the most inefficient depuration of G. lamblia cysts was observed in oysters kept in low-salinity water (Table 1).
Trends in pathogen inactivation in oyster tissue were similar for C. parvum oocysts and microsporidian spores but different for G. lamblia cysts (Table 1). Giardia cysts were inactivated on average two times faster than the C. parvum oocysts and microsporidian spores. In oysters maintained in high- and medium-salinity water, the cysts were detected only on day 6 pwi, and in oysters kept in 8-ppt-salinity water, the cysts were detected on day 14 pwi (Table 1).
DISCUSSION
The introduction of C. ariakensis into the Chesapeake Bay, although a debatable option, has been proposed as a necessity for restoration of the oyster industry, with economic and ecological benefits (4, 25). The present study demonstrated that C. ariakensis triploids can retain viable human waterborne pathogens in their tissue, similar to oysters native to the Chesapeake Bay (C. virginica) (8, 9, 10); thus, the nonnative oysters could be involved in the epidemiology of food-borne diseases whenever they are consumed in a raw form. However, viable C. parvum oocysts persisted in C. ariakensis (i.e., up to 33 days) almost five times longer than in C. virginica (7 days) even when a sixfold-smaller C. parvum inoculum was used for water contamination (7). In a study that utilized the European oyster (Ostrea edulis), viable C. parvum oocysts were identified on day 31 postexposure (12). The present study has demonstrated for the first time that oysters can bioaccumulate spores of human-infectious microsporidia and supports previous findings that oysters depurate Cryptosporidium very inefficiently (13). The concentration of human pathogens from ambient water by C. ariakensis triploids and retention of these pathogens without (or with minimal) inactivation and a very low depuration rate provide evidence that these oysters will present a public health threat to the human food chain if they are harvested from polluted water. This conclusion is reinforced by the concentration of waterborne pathogens (25 cells/liter/day) used in the present study, which was representative of pathogen levels observed in surface waters, including the Chesapeake Bay (9, 21, 33, 34, 37). The concentrations of waterborne C. parvum oocysts measured in the Nanticoke, Patuxent, and Potomac Rivers and the Tangier Sound were 79, 31, 10, and 8 oocysts/liter, respectively (9). Crassostrea virginica oysters have been demonstrated to efficiently remove another human pathogen, Toxoplasma gondii oocysts, which are present in coastal waters (28, 29, 30).
The introduction of C. ariakensis into the Chesapeake Bay is based on large-scale aquaculture operations that utilize standard containments (i.e., floats) holding hundreds of oysters (4, 25). This type of aquaculture, which was prompted by the thin shells of C. ariakensis (much thinner than C. virginica), does not ensure physical restoration of indigenous oyster reefs (2, 4, 25). The present study indicates that such containment of nonnative oysters provides excellent ecological services in regard to efficient cleaning of human-infectious agents from surrounding water. However, these environmental benefits are associated with public heath risks from contaminated oysters intended for human consumption.
The ultimate fate of human pathogens taken up by oysters is not clear. The progressively decreasing pathogen levels in oyster tissue over time and the fact that only viable pathogens were detected indicate that nonviable cells disintegrate rapidly, most likely because they are being digested by the hemocytes (18). The best example in the present study was provided by G. lamblia cysts, which were detected no longer than 2 weeks after exposure. Giardia cysts were never found in the native oysters commercially harvested from the Chesapeake Bay (8, 9, 10, 21); however, it is difficult to assume that the water was free of cysts, given the very broad anthropozoonotic range of this parasite within the Chesapeake Bay and its watershed (19). The different patterns of oyster interactions with waterborne pathogens in the present study were most likely related to the size of the pathogen. Giardia lamblia cysts and C. parvum oocysts are significantly bigger than microsporidian spores (16, 6,and 2 μm, respectively) and were filtered out more efficiently and bioaccumulated better, while the spores were recovered less efficiently and were most likely released back into the water with oyster pseudofeces or feces.
Field trials in the Chesapeake Bay have demonstrated that C. ariakensis considerably outperforms the native oysters, displaying significant growth and meat weight gain, and is associated with the lowest mortality rates in medium-salinity (i.e., 14- to 16-ppt) and high-salinity (i.e., 29- to 31-ppt) sites (2, 25). In support, the present study demonstrated that the recovery, bioaccumulation, depuration, and inactivation of human pathogens by C. ariakensis were driven by salinity and were optimal in medium- and high-salinity water.
Crassostrea ariakensis is a suspension feeder with a clearance rate (i.e., number of cleared particles per unit of time) proportionally related to its size (2, 25), so that larger oysters filter more particles than smaller ones do. The pathogens used in the present study do not multiply in the environment; their origin in the Chesapeake Bay is from fecal contamination (21), and their size falls within the range of particles filtered by oysters. Because C. ariakensis grows faster than C. virginica under similar ecological conditions in the Chesapeake Bay (2, 25), this species filters out more particulate matter suspended in the water, indicating that it may remove more waterborne pathogens than C. virginica. However, the results of the present study pertain to C. ariakensis triploids, and it remains unknown whether diploids will be equally efficient in the removal of waterborne pathogens. It is important to note that reversion to diploids is a progressive process that more frequently occurs in low- and medium-salinity sites within the Chesapeake Bay (4, 25).
The extraction of human pathogens from oysters is a complex process, but identifying specific pathogen species and assessing viability are even more challenging. The FISH method meets both of these challenges, as it utilizes a fluorescence-labeled oligonucleotide probe targeted to species-specific sequences of the 18S rRNA of these pathogens (22, 24). Because rRNA is present only in large copy numbers in viable organisms (22, 24), FISH facilitates the enumeration of viable pathogens. The main advantage of FISH is the assessment of viability of even a single pathogen cell (22, 24). Such resolution is not available or is impractical with any other technique. For example, using mouse bioassay and highly sensitive reverse transcription-PCR, the lowest number of C. parvum oocysts that can be assessed for viability is 103 (26). Additionally, as demonstrated previously (22) and by the present study, FISH assays can be multiplexed for identification of human waterborne pathogens in molluscan shellfish. Implementation of FISH assays into monitoring programs and seafood product testing will enable shellfish producers to better assess the product safety. However, irrespective of the identification technique, pathogen extraction from oyster tissue is not 100% efficient. A previous study that utilized Chesapeake Bay molluscan shellfish spiked with known numbers of human pathogens demonstrated approximately 50% extraction efficiency (20). Recently, a range of 48 to 70% extraction efficiency of C. parvum oocysts from oysters has been reported (31). Thus, we conclude that the numbers of viable pathogens reported in the present study were potentially underestimated.
The present study provides realistic and useful information for the risk assessment of food-borne infections related to the consumption of contaminated triploids of C. ariakensis. However, although there is no doubt that C. ariakensis can be contaminated with infectious agents that cause serious diseases in people, the presence of viable pathogens in oysters is not equivalent to the cause of infections in people consuming such oysters. A set of host-related, environment-derived, and pathogen-specific factors interact in the epidemiology of human food-borne diseases caused by consumption of contaminated shellfish (14, 15). Also, under natural exposure situations, the pathogens enter the gastrointestinal tract of a person while they are embedded in oyster tissue, which could prevent, inhibit, or delay the arrival of a set of stimuli triggering pathogen excystation and subsequent infection. The pathogens used in the present study have life-threatening consequences in people with various immune deficiencies, and it is thought that in this population a very low number of pathogens can cause infection (17, 38, 39). However, it is also believed that people with immunological impairments are usually aware of the potential health hazards associated with consumption of raw food items, particularly molluscan shellfish, and they therefore avoid this type of food and potential infections (33).
Acknowledgments
We thank J. King, NOAA Chesapeake Bay Office, Annapolis, MD; S. K. Allen and N. Geyerhahn, VIMS, Gloucester Point, VA; and K. Gibson, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, for facilitating this study.
The study was supported by the NOAA Chesapeake Bay Office (grant no. NA04NMF4570426), by a NATO (Brussels, Belgium) Collaborative Linkage Grant (no. 979765), by the Johns Hopkins Center in Urban Environmental Health (grant no. P30 ES03819), and by the Alternatives Research and Development Foundation.
REFERENCES
- 1.Bean, N. H., S. J. Goulding, C. Lao, and E. J. Lao. 1996. Surveillance for foodborne-disease outbreaks—United States, 1988-1992. Morb. Mortal. Wkly. Rep. 45:1-66. [PubMed] [Google Scholar]
- 2.Calvo, G. W., M. W. Luckenbach, S. K. Allen, and E. M. Burreson. 2001. Comparative field study of Crassostrea ariakensis and Crassostrea virginica in relation to salinity in Virginia. J. Shellfish Res. 20:221-229. [Google Scholar]
- 3.Cotte, L., M. Rabodonirina, F. Chapuis, F. Bailly, F. Bissuel, C. Raynal, P. Gelas, F. Persat, M. A. Piens, and C. Treppo. 1999. Waterborne outbreak of intestinal microsporidiosis in persons with and without human immunodeficiency virus infection. J. Infect. Dis. 180:2003-2008. [DOI] [PubMed] [Google Scholar]
- 4.Dew, J. R., J. Berkson, E. M. Hallerman, and S. K. Allen, Jr. 2003. A model for assessing the likelihood of self-sustaining populations resulting from commercial production of triploid Suminoe oysters (Crassostrea ariakensis) in Chesapeake Bay. Fish Bull. 101:758-768. [Google Scholar]
- 5.Dowd, S. E., C. P. Gerba, and I. L. Pepper. 1998. Confirmation of the human-pathogenic microsporidia Enterocytozoon bieneusi, Encephalitozoon intestinalis, and Vittaforma corneae in water. Appl. Environ. Microbiol. 64:3332-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fayer, R. 2004. Infectivity of microsporidia spores stored in seawater at environmental temperatures. J. Parasitol. 90:654-657. [DOI] [PubMed] [Google Scholar]
- 7.Fayer, R., C. A. Farley, E. J. Lewis, J. M. Trout, and T. K. Graczyk. 1997. Potential role of the Eastern oyster, Crassostrea virginica, in the epidemiology of Cryptosporidium parvum. Appl. Environ. Microbiol. 63:2086-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fayer, R., T. K. Graczyk, E. J. Lewis, J. M. Trout, and C. A. Farley. 1998. Survival of infectious Cryptosporidium parvum oocysts in seawater and Eastern oysters (Crassostrea virginica) in the Chesapeake Bay. Appl. Environ. Microbiol. 64:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fayer, R., E. J. Lewis, J. M. Trout, T. K. Graczyk, M. C. Jenkins, J. Higgins, L. Xiao, and A. A. Lal. 1999. Cryptosporidium parvum in oysters from commercial harvesting sites in the Chesapeake Bay. Emerg. Infect. Dis. 5:706-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fayer, R., J. M. Trout, E. J. Lewis, L. Xiao, A. Lal, M. C. Jenkins, and T. K. Graczyk. 2002. Temporal variability of Cryptosporidium in the Chesapeake Bay. Parasitol. Res. 88:998-1003. [DOI] [PubMed] [Google Scholar]
- 11.Fournier, S., O. Liguory, M. Santillana-Hayat, E. Guillot, C. Sarfari, N. Dumoutier, J. Molina, and F. Derouin. 2000. Detection of microsporidia in surface water: a one-year follow-up study. FEMS Immunol. Med. Microbiol. 29:95-100. [DOI] [PubMed] [Google Scholar]
- 12.Freire-Santos, F., A. M. Oteiza-Lopez, C. A. Vergara-Castiblanco, E. Ares-Mazas, E. Alvarez-Suraex, and O. Garcia-Martin. 2000. Detection of Cryptosporidium oocysts in bivalve molluscs destined for human consumption. J. Parasitol. 86:853-854. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Couso, H., F. Freire-Santos, J. Martinez-Urataza, O. Garcia-Martin, and M. E. Ares-Mazas. 2003. Contamination of bivalve molluscs by Cryptosporidium oocysts: the need for new quality control standards. Int. J. Food Microbiol. 87:97-105. [DOI] [PubMed] [Google Scholar]
- 14.Graczyk, T. K. 2003. Human waterborne parasites in molluscan shellfish. J. Parasitol. 89:557-561. [Google Scholar]
- 15.Graczyk, T. K., and K. J. Schwab. 2000. Foodborne infections vectored by molluscan shellfish. Curr. Gastroenterol. Rep. 2:305-309. [DOI] [PubMed] [Google Scholar]
- 16.Graczyk, T. K., M. R. Cranfield, and R. Fayer. 1997. Recovery of waterborne oocysts of Cryptosporidium parvum from water samples by the membrane-filter dissolution method. Parasitol. Res. 83:121-125. [DOI] [PubMed] [Google Scholar]
- 17.Graczyk, T. K., R. Fayer, and M. R. Cranfield. 1997. Zoonotic potential of Cryptosporidium parvum: implications for waterborne cryptosporidiosis. Parasitol. Today 13:348-351. [DOI] [PubMed] [Google Scholar]
- 18.Graczyk, T. K., R. Fayer, E. J. Lewis, C. A. Farley, and J. M. Trout. 1997. In vitro interactions between hemocytes of the Eastern oyster, Crassostrea virginica Gmelin, 1791 and Cryptosporidium parvum oocysts. J. Parasitol. 83:949-952. [PubMed] [Google Scholar]
- 19.Graczyk, T. K., R. C. A. Thompson, R. Fayer, P. Adams, U. M. Morgan, and E. J. Lewis. 1999. Giardia duodenalis of genotype A recovered from clams in the Chesapeake Bay subestuary, Rhode River. Am. J. Trop. Med. Hyg. 61:526-529. [DOI] [PubMed] [Google Scholar]
- 20.Graczyk, T. K., R. Fayer, E. J. Lewis, J. M. Trout, and C. A. Farley. 1999. Cryptosporidium oocysts in Bent mussels (Ischadium recurvum) in the Chesapeake Bay. Parasitol. Res. 85:518-521. [DOI] [PubMed] [Google Scholar]
- 21.Graczyk, T. K., R. Fayer, M. C. Jenkins, J. M. Trout, J. Higgins, E. J. Lewis, and C. A. Farley. 2000. Susceptibility of the Chesapeake Bay to environmental contamination with Cryptosporidium parvum. Environ. Res. 82:106-112. [DOI] [PubMed] [Google Scholar]
- 22.Graczyk, T. K., D. B. Conn, F. Lucy, D. Minchin, L. Tamang, L. N. S. Moura, and A. J. DaSilva. 2004. Human waterborne parasites in zebra mussels (Dreissena polymorpha) from the Shannon River drainage, Ireland. Parasitol. Res. 93:389-391. [DOI] [PubMed] [Google Scholar]
- 23.Hallerman, E., M. Leffler, S. Mills, and S. K. Allen. 2002. Aquaculture of triploid C. ariakensis in Chesapeake Bay: a symposium report. Maryland Publication UM-SG-TS- 2002-01, Virginia Publication VSG-02-03. http://www.mdsg.umd.edu/oysters/exotic/ariakensis01/ariakensis_oct01.pdf.
- 24.Hester, F. D., H. D. A. Linquist, A. M. Bobst, and F. W. Schaffer. 2000. Fluorescent in situ detection of Encephalitozoon hellem spores with a 6-carboxyfluorescein-labeled ribosomal RNA-targeted oligonucleotide probe. J. Eukaryot. Microbiol. 47:299-308. [DOI] [PubMed] [Google Scholar]
- 25.Hudson, K. L., A. Kozlowski, and S. K. Allen. 2004. Biosecurity and comparison field trials of triploid Crassostrea ariakensis with C. virginica. p. 103-114. NOAA Chesapeake Bay Office Integrated Research Program Symposium Report. http://www.vims.edu/vsc/.
- 26.Jenkins, M. C., J. Trout, M. S. Abrahamsen, J. Higgins, and R. Fayer. 2000. Estimating viability of Cryptosporidium parvum oocysts using reverse transcriptase-polymerase chain reaction (RT-PCR) directed at mRNA encoding amyloglucosidase. J. Microbiol. Methods 34:97-106. [DOI] [PubMed] [Google Scholar]
- 27.Kucerova-Pospisilova, Z., D. Carr, G. Leicht, M. Scanlon, and G. S. Visvesvara. 1999. Environmental resistance of Encephalitozoon spores. J. Eukaryot. Microbiol. 46:11S-13S. [PubMed] [Google Scholar]
- 28.Lindsay, D. S. 2001. Toxoplasma gondii research: summary of the seventh international workshop on opportunistic protists. J. Eukaryot. Microbiol. (Suppl.) 190S. [DOI] [PubMed]
- 29.Lindsay, D. S., K. K. Phelps, S. A. Smith, G. Flick, S. S. Sumner, and J. P. Dubey. 2001. Removal of Toxoplasma gondii oocysts from sea water by Eastern oysters (Crassostrea virginica). J. Eukaryot. Microbiol. (Suppl.) 197S-198S. [DOI] [PubMed]
- 30.Lindsay, D. S., M. V. Collins, S. M. Mitchell, C. N. Wetch, A. C. Rosypal, G. J. Flick, A. M. Zajac, A. Lindquist, and J. P. Dubey. 2004. Survival of Toxoplasma gondii in Eastern oysters (Crassostrea virginica). J. Parasitol. 90:1054-1057. [DOI] [PubMed] [Google Scholar]
- 31.MacRae, M., C. Hamilton, N. J. Strachan, S. Wright, and I. D. Ogden. 2005. The detection of Cryptosporidium parvum and Escherichia coli O157 in UK bivalve shellfish. J. Microbiol. Methods 60:395-401. [DOI] [PubMed] [Google Scholar]
- 32.Rippey, S. R. 1994. Infectious diseases associated with molluscan shellfish consumption. Clin. Microbiol. Rev. 7:419-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose, J. B., J. T. Lisle, and M. LeChevallier. 1997. Waterborne cryptosporidiosis: incidence, outbreaks, and treatment strategies, p. 93-110. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC, Boca Raton, Fla.
- 34.Solo-Gabriele, H. M., A. LeRoy, L. J. Fitzerald, J. M. Dubon, S. M. Neumeister, M. K. Baum, and C. J. Palmer. 1998. Occurrence of Cryptosporidium oocysts and Giardia cysts in water supplies of San Pedro Sula, Honduras. Rev. Panam. Salud Publica 4:398-400. [DOI] [PubMed] [Google Scholar]
- 35.Sparfel, J. M., C. Sarfati, O. Liquory, B. Caroff, N. Dumoutier, B. Gueglio, E. Billaud, F. Raffi, L. M. Molina, M. Miegeville, and F. Derouin. 1997. Detection of microsporidia and identification of Enterocytozoon bieneusi in surface water by filtration followed by specific PCR. J. Eukaryot. Microbiol. 44:78S. [DOI] [PubMed] [Google Scholar]
- 36.Tamburrini, A., and E. Pozio. 1999. Long-term survival of Cryptosporidium parvum oocysts in seawater and in experimentally infected mussels (Mytilus galloprovincialis). Int. J. Parasitol. 29:711-715. [DOI] [PubMed] [Google Scholar]
- 37.Thurston-Enriquez, J. A., P. Watt, S. E. Dowd, R. Enriquez, I. L. Pepper, and C. P. Gerba. 2002. Detection of protozoan parasites and microsporidia in irrigation water used for crop production. J. Food Prot. 65:378-382. [DOI] [PubMed] [Google Scholar]
- 38.Weber, R., R. T. Bryan, D. A. Schwartz, and R. L. Owen. 1994. Human microsporidial infections. Clin. Microbiol. Rev. 7:426-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfe, M. S. 1992. Giardiasis. Clin. Microbiol. Rev. 5:93-100. [DOI] [PMC free article] [PubMed] [Google Scholar]