Abstract
Kluyveromyces lactis is one of the cheese-ripening yeasts and is believed to contribute to the formation of volatile sulfur compounds (VSCs) through degradation of l-methionine. l-Methionine aminotransferase is potentially involved in the pathway that results in the production of methanethiol, a common precursor of VSCs. Even though this pathway has been studied previously, the genes involved have never been studied. In this study, on the basis of sequence homology, all the putative aminotransferase-encoding genes from K. lactis were cloned in an overproducing vector, pCXJ10, and their effects on the production of VSCs were analyzed. Two genes, KlARO8.1 and KlARO8.2, were found to be responsible for l-methionine aminotransferase activity. Transformants carrying these genes cloned in the pCXJ10 vector produced threefold-larger amounts of VSCs than the transformant containing the plasmid without any insert or other related putative aminotransferases produced.
The primary classes of compounds that contribute to cheese flavor include amino acids and their degradation products, peptides, carbonyl compounds, and fatty acids (5). The volatile fraction of cheese includes sulfur-containing compounds, such as methanethiol (MTL), methional, and dimethyl disulfide (DMDS), which contribute significantly to the aroma of cheese (16). The most common volatile sulfur compound (VSC) found in cheese is MTL, which is derived from enzymatic degradation of the amino acid l-methionine, which is present in the cheese curd (30).
The l-methionine catabolic pathways are different in bacteria and yeasts. In Brevibacterium linens, l-methionine can be converted to MTL, α-ketobutyrate, and ammonia by an l-methionine γ-lyase in a one-step reaction (2, 12). A two-step degradation pathway has also been demonstrated in several microorganisms, including yeasts (8), lactic acid bacteria (17, 32), and cheese surface bacteria (7). The latter pathway is believed to be initiated by an aminotransferase that is also called transaminase. l-Methionine degradation results in the synthesis of 4-methylthio-oxobutyric acid (MOBA) and 4-methylthiohydroxybutyric acid (MHBA), which are subsequently converted to MTL (4). This two-step metabolic sequence has been demonstrated in lactococci (17). An aminotransferase has been identified in Lactococcus lactis (32), and two aminotransferases have been partially purified from B. linens (21). In Saccharomyces cerevisiae, the genes encoding aminotransferase in the genome fall into the following three groups: (i) ScAAT1 and ScAAT2, encoding aspartate amino acid aminotransferases (11, 23, 29); (ii) ScBCA1, encoding a branched-chain amino acid aminotransferase (15); and (iii) ScARO8 and ScARO9, encoding aromatic amino acid aminotransferases (20, 28).
Despite their importance for the conversion of amino acids to aroma compounds, the aminotransferases of the microorganisms present in cheese have not been studied in detail. These aminotransferases are pyridoxal 5′-phosphate-dependent enzymes and are widely distributed in microorganisms. They are generally more or less specific for one amino acid group (e.g., aromatic amino acids or branched-chain amino acids), but they have broad overlapping substrate specificities (31). Although the products of the aminotransferase pathway have been extensively studied, the complete set of genes encoding the enzymes has not been studied in the yeasts involved in cheese ripening.
Cheese-ripening yeasts, such as Kluyveromyces lactis, develop in the early stages of cheese ripening and consume lactate, which neutralizes the pH, thus enabling growth of the surface bacteria (4). Until recently, the production of VSCs was attributed to the cheese-ripening bacteria. However, it has been shown that yeasts can also contribute to production of these compounds (3). In the present study, all the putative genes encoding an aminotransferase in the genome of K. lactis CLIB 210 (14) were identified based on homology to the S. cerevisiae genes. They were all overexpressed in K. lactis and subsequently analyzed for the production of VSCs from l-methionine.
MATERIALS AND METHODS
Strains, vector, and culture conditions.
K. lactis CLIB 640 (Collection de Levures d'Intérêt Biotechnologique [CLIB]; http://www.inra.fr/clib) was used in this study. This strain was chosen because of its ability to produce large amounts of VSCs (data not shown). Escherichia coli DH10B and the uracil-requiring strain K. lactis CBS 2359/152F (MATα met1.1 ura3) were used for cloning and propagation of the E. coli/K. lactis shuttle vector pCXJ10 (10). Precultures of K. lactis strains were grown aerobically in 100-ml Erlenmeyer flasks containing 20 ml potato dextrose broth (PDB) or yeast peptone dextrose broth at 25°C for 1 day at 150 rpm. These cultures were then used to inoculate 500-ml flasks containing 100 ml of yeast nitrogen base (YNB) medium without amino acids supplemented with phosphate buffer (50 mM), NH4Cl (5 g/liter), Casamino Acids (2 g/liter), and glucose (20 g/liter). When necessary, media were supplemented with l-methionine and uracil at final concentrations of 1 g/liter and 100 μg/ml, respectively. Luria-Bertani broth with or without 1% agar supplemented with 100 μg/ml ampicillin when necessary (e.g., for selection) was used for propagation of E. coli DH10B. All media were purchased from Difco (Detroit, MI) unless indicated otherwise.
DNA manipulation and PCR amplification.
Cells of K. lactis CLIB 640 were harvested from an overnight culture grown in yeast peptone dextrose broth, and DNA was extracted as described previously (19). Electrocompetent cells of strain CBS 2359/152F were prepared and transformed as described by Sanchez et al. (26). K. lactis transformants were selected on YNB agar plates without uracil.
PCR amplification was performed with a Perkin-Elmer 2400 DNA thermocycler using PyroBest Taq DNA polymerase (Cambrex Bioscience, Paris, France). The following conditions were used: 30 cycles of denaturation at 94°C for 30 s, annealing at 48°C for 30 s, and amplification at 72°C for 90 s, followed by a final amplification at 72°C for 5 min. The oligonucleotides (Eurogentec S. A., Seraing, Belgium) used in this study are listed in Table 1. They were phosphorylated and contained an artificial restriction site at the 5′ end to facilitate subsequent cloning. The oligonucleotides that were used to amplify KlARO9, KlAAT1, and KlAAT2 contained an SmaI cleavage site, the oligonucleotides that were used to amplify KlARO8.1 and KlBAT1 contained a BamHI site, and the oligonucleotides that were used to amplify KlARO8.2 contained a KpnI cleavage site. The amplified PCR products that were used as inserts were excised from a 1% agarose gel and purified using a QIAGEN extraction kit (QIAGEN S.A., France). They were subsequently cleaved overnight using the appropriate enzyme according to the manufacturer's recommendations, ethanol precipitated, and stored at −20°C until they were needed. The enzymes used in the study were purchased from New England Biolabs (St. Quentin en Yvelines, France).
TABLE 1.
Oligonucleotides used in this study and sizes of the amplified PCR products
| Gene amplified | Sequencea | PCR product size (bp) |
|---|---|---|
| KlARO8.1 | 5′-CGGGATCCCGAACCGTTACTTACCGCGATG-3′ | 2,636 |
| 5′-CGGGATCCCGCATTCCTCCCCTTCTGTGAA-3′ | ||
| KlARO8.2 | 5′-GGGGTACCCCTTCAGAATTTCGTGCTGACG-3′ | 2,126 |
| 5′-GGGGTACCCCATAGGGCAGCATGATTTTGG-3′ | ||
| KlARO9 | 5′-TCCCCCGGGGGAAAGCATGAGCGGGTCTCTAA-3′ | 2,309 |
| 5′-TCCCCCGGGGGATGTGTTGCGTGTAAGATGTGG-3′ | ||
| KlAAT1 | 5′-TCCCCGGGGGACCTATTGGCCCTTTGAGACA-3′ | 1,778 |
| 5′-TCCCCGGGGGATGGGCATTTATAATCCCTTTTG-3′ | ||
| KlAAT2 | 5′-TCCCCCGGGGGATCGGCCACACAACCATATTA-3′ | 1,931 |
| 5′-TCCCCCGGGGGAATCTGGACGTTCGGTTATCG-3′ | ||
| KlBAT1 | 5′-CGGGATCCCGCTCTCAACATGGCTTTCAC-3′ | 1,979 |
| 5′-CGGGATCCCGCATCCCATATAAGACCTAGCA-3′ | ||
| KlURA3 | 5′-TCTTGACGTTCGTTCGACTG-3′ | 558 |
| 5′-TCGACGGTTCTGTACTGCTG-3′ | ||
| ScURA3 | 5′-ATTGGATGTTCGTACCACCA-3′ | 558 |
| 5′-TCCACGGTTCTATACTGTTG-3′ |
The added sequences containing the restriction sites for cloning are underlined.
The E. coli/K. lactis shuttle vector pCXJ10 was cleaved for 3 h and linearized at the BamHI or SmaI site present in the multiple-cloning-site region. It was subsequently purified by ethanol precipitation and dephosphorylated using SAP (shrimp alkaline phosphatase; USB) for 20 min at 37°C. The KlARO8.1-containing PCR product was inserted at the BamHI restriction site, while all the other PCR fragments were inserted at the SmaI site. The procedure was completed by ligating the insert with the linearized vector at 18°C overnight with T4 DNA ligase (New England Biolabs, St. Quentin en Yvelines, France). E. coli DH10B electrocompetent cells were prepared as previously described (13). The transformants were selected on Luria-Bertani agar supplemented with ampicillin (100 μg/ml) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 μg/ml).
Southern blot hybridization.
Total DNA from the K. lactis transformants was cleaved with HindIII, loaded on a 0.8% agarose gel, and electrophoresed overnight at 40 V/cm in 1× Tris-acetate-EDTA buffer. Transfer to a GeneScreen nylon membrane (NEN, Dupont de Nemours) and hybridization were performed as described previously (9). Two DNA probes were prepared by amplification of regions that were the same size containing the ScURA3 and KlURA3 genes, which encoded the orotidine-5-phosphate decarboxylase in S. cerevisiae and K. lactis, respectively. The PCR products were excised from the gel and purified using a QIAGEN extraction kit (QIAGEN S.A., France). Equal concentrations of the products were subsequently mixed, and the compounds were radiolabeled with [α-32P]dCTP by using a MegaPrimer labeling kit (Amersham Biosciences) according to the supplier's instructions. The membranes were hybridized using Denhardt buffer at 65°C (25) and were subsequently exposed and scanned after 24 h with a Storm 860 (Amersham Biosciences) to estimate the plasmid copy number.
Plasmid maintenance.
Precultures of the transformants were grown overnight in YNB medium at 25°C at 150 rpm. Twenty milliliters of PDB supplemented with 6.7 mM l-methionine was inoculated with 200 μl of a preculture and incubated at 25°C. Samples were taken at 12 h, 24 h, and 48 h and plated on YNB medium in the presence and absence of uracil in order to determine plasmid maintenance in the nonselective medium (PDB). The optical density at 600 nm and the pH were also determined. This experiment was performed in duplicate.
Preparation of cell extracts for enzyme assays.
Precultures of the transformants were grown overnight in YNB medium at 150 rpm. One hundred milliliters of PDB supplemented with 6.7 mM l-methionine was inoculated (1%, vol/vol) and incubated for 24 h. Three individual cultures were prepared for each transformant. The cultures were centrifuged (20,000 × g, 10 min, 4°C), and the pellets were washed three times with 20 ml of 50 mM Tris HCl (pH 8.8). The biomass (200 to 250 mg) was subsequently suspended in 1 ml of 50 mM Tris HCl (pH 8.0) containing 1 mM EDTA and ground using an FP 120 FastPrep cell disrupter (Savant Instruments, Holbrook, N.Y.) in the presence of 0.6 g of glass beads (diameter, 500 μm). Then three 20-s mixing cycles (speed, 6.5 m/s) were alternated with 5 min of cooling on ice in order to minimize heat-induced denaturation of enzymes. The samples were centrifuged (20,000 × g, 5 min, 4°C), and the supernatant (cell extract) was collected for enzymatic assays. The total protein content of the cell extract was determined by the method of Bradford (8a), using bovine serum albumin as the standard.
Enzyme activity measurement.
Aminotransferase activity was assayed by measuring the formation of glutamate from l-methionine and α-ketoglutarate as the amino group acceptor. The reaction mixture contained 80 mM Tris-HCl (pH 8.0), 30 mM l-methionine, 30 mM α-ketoglutarate, 0.05 mM pyridoxal phosphate, and 20% (vol/vol) cell extract (final concentration, 1 to 1.2 mg protein/ml). The mixture was incubated for 15 min at 37°C. Once the reaction was stopped by heating at 95°C for 10 min, the amount of l-glutamic acid produced was determined colorimetrically using an assay kit (Boehringer Mannheim, R-Biopharm, Darmstadt, Germany). The absorbance at 492 nm was determined. The glutamate concentration was determined after 15 min of incubation. The aminotransferase activity was expressed as the amount of glutamate formed in nmol/g protein/s.
l-Methionine, MOBA, and MHBA analyses.
The l-methionine, MOBA, and MHBA contents of 48-h culture filtrates were determined by high-performance liquid chromatography with a Symmetry C18 column (3.5 μm; diameter, 4.6 mm; length, 100 mm; Waters) (7).
Volatile compound analysis.
Five milliliters of culture was analyzed using a headspace analyzer (Hewlett-Packard 7695A purge and trap concentrator; Hewlett-Packard, Palo Alto, CA) coupled to a mass spectrophotometer detector (Hewlett-Packard 6890A quadruple mass spectrometer; Hewlett-Packard), as previously described (22). The samples were heated for 10 min at 60°C prior to a 15-min purge with helium at a flow rate of 30 ml/min. Volatile compounds were then adsorbed on a Tenax porous polymer adsorbent trap column (60 to 80 mesh; 0.25 g; 30 by 0.32 cm; Tekmar, Cincinnati, Ohio) at room temperature. Desorption was accomplished by heating the column for 2 min at 225°C, followed by cryofocusing at 150°C and heating the interface at 180°C for 1 min. Gas chromatography began after injection of the volatile compounds into a nonpolar capillary column (HP-5 MS; 30 m by 0.25 mm; film thickness, 0.25 μm; Hewlett Packard). The compounds, adsorbed on the column, were eluted using helium as the carrier gas at a flow rate of 1.6 ml/min. The oven temperature was maintained at 5°C for 8 min. Subsequently, the temperature was raised at a rate of 3°C/min to 20°C and then at a rate of 10°C/min to a final temperature of 150°C. Mass spectrometry was used to detect the separated compounds. Data were collected in the range from 29 to 300 atomic mass units at a rate of 3.4 scans/s. Volatile compounds were identified on the basis of their ion chromatograms.
Sequence analysis.
BLAST was used to screen sequence databases for homology (1). Sequence alignments were generated using CLUSTAL W (18) and CLUSTAL X (27) and were manually adjusted in Genedoc (http://www.psc.edu/biomed/genedoc). Phylogenetic trees were generated by the Phylip maximum parsimony method (http://evolution.genetics.washington.edu/phylip.html). Trees were visualized with Njplot (24).
Data analyses.
All analyses were performed in triplicate. Data were expressed as means ± standard deviations, and they were analyzed using SPSS 11.5 for Windows software. One-way analysis of variance was performed. Scheffe multiple-range tests (α ≤ 0.05) were applied to the individual variables to compare means and to assess whether a difference was significant. Different letters (e.g., a, b, c, d, and e) were assigned to significantly different groups, while for those results situated between two groups, a combination of the two corresponding letters was used (e.g., ab).
RESULTS
Identification of the K. lactis aminotransferase genes.
Genes encoding putative aminotransferases were detected in the completely sequenced genome (14) of K. lactis CLIB 210 (http://www.cbi-labri.fr/genolevures/) by using BLASTP and the S. cerevisiae aminotransferase genes as queries. Three groups of orthologous genes were found; one group consisted of two orthologues of ScAAT1 and ScAAT2, one group consisted of the branched-chain aminotransferase gene ScBAT1, and the third group consisted of two ScARO8 aromatic aminotransferase orthologues and one ScARO9 aromatic aminotransferase orthologue.
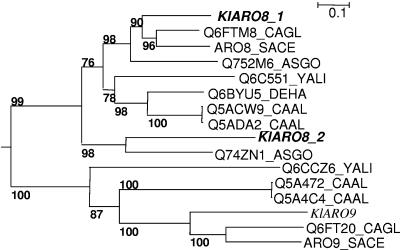
The KlAAT1 (KLLA0F13640g) and KlAAT2 (KLLA0F17754g) genes exhibited 63% and 65% identity at the protein sequence level with ScAAT1 and ScAAT2, respectively. They were located on the F chromosome of K. lactis. KlBAT1 (KLLA0A10307g) on the A chromosome of K. lactis exhibited 76% identity with the ScBAT1 gene. The KlARO8.1 (KLLA0F10021g) and KlARO8.2 (KLLA0A04906g) genes were present on the F and A chromosomes of K. lactis, respectively; these two orthologues exhibited 65% and 55% identity to ScARO8, respectively. The last aromatic amino acid aminotransferase gene, KlARO9 (KLLA0D01110g), was located on the A chromosome and exhibited 61% identity with ScARO9. The phylogenetic relationships of the K. lactis aromatic aminotransferases with homologues in other yeasts species are shown in Fig. 1. This figure shows that both K. lactis and Ashbya gossypii possess two copies of the ARO8 gene (KlARO8.1 and KlARO8.2; Q752M6_ASGO and Q74ZN1_ASGO), whereas S. cerevisiae, Candida glabrata, Debaryomyces hansenii, and Yarrowia lipolytica have only one gene. The phylogenetic relationships between ARO8 homologues show that KlARO8.2 and AgARO8.2 branched before speciation of all hemiascomycetous yeasts and therefore cannot be considered orthologues of ARO8.1. This result can be explained by early duplication of the ancestral ARO8 gene before the divergence of the yeasts, followed by the loss of one copy in all species except A. gossypii and K. lactis, or by acquisition (lateral gene transfer) of a gene close to ARO8.1 in the A. gossypii/K. lactis branch.
FIG. 1.
Phylogenetic relationships of several yeast aminotransferases. The phylogeny was derived from a maximum likelihood analysis of various orthologues of the ScAro8 and ScAro9 proteins and was rooted with Ralstonia solanacearum Q8Y2V3. Bootstrap values based on 100 replicates are indicated at the nodes. Kl, Kluyveromyces lactis; ASGO, Ashbya gossypii; CAGL, Candida glabrata; SACE, Saccharomyces cerevisiae; YALI, Yarrowia lipolytica; DEHA, Debaryomyces hansenii; CAAL, Candida albicans.
Effect of amplification of genes encoding aminotransferases on conversion of l-methionine to VSCs.
The DNA regions containing the genes of interest were PCR amplified from CLIB 640 and inserted in the pCXJ10 plasmid. Ligation mixtures were used to transform E. coli DH10B. Several transformants were selected and subjected to PCR on colonies and restriction analysis. Subsequently, one transformant containing each individual gene was picked, the DNA was extracted, and K. lactis CBS 2359/152F cells were transformed by electroporation. The cells were selected on YNB medium based on the ability to grow without added uracil.
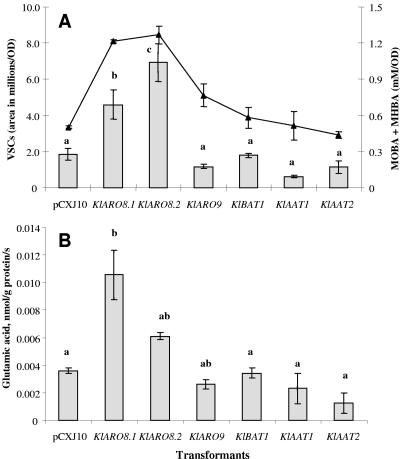
The production of MOBA and MHBA by the various transformants is shown in Fig. 2A. We compared the transformants containing the putative genes and the plasmid without any insert, pCXJ10. The average total production for the plasmid without any insert, pCXJ10, ranged from 0.2 mM/optical density unit to 0.5 mM/optical density unit at 12 h (data not shown) and 24 h (Fig. 2A). The transformants containing the KlARO8.1 and KlARO8.2 genes produced the most MOBA and MHBA after 24 h, ∼1.2 mM/optical density unit in both cases. This value was 2.5 times greater than the value for the control with no insert, pCXJ10, showing that these genes are involved in the l-methionine catabolic pathway. After 24 h, 60 to 70% of the initial amount of l-methionine was consumed by all transformants (Table 2). Also, growth at 24 h (as estimated from the optical density at 600 nm) was measured for the transformants (Table 3).
FIG. 2.
Production of VSCs, MOBA, and MHBA and aminotransferase activities of various CBS 2359/152F transformants after 24 h of incubation in PDB in the presence of 6.7 mM l-methionine. The results are expressed as means ± standard deviations (error bars) for three repetitions. Letters indicate statistical groups. (A) Total production of VSCs (bars) and MOBA and MHBA (triangles). (B) Aminotransferase activity.
TABLE 2.
Consumption of l-methionine by each transformant after 12 h and 24 h of incubation in the presence of 6.7 mM l-methionine
| Transformant |
l-Methionine consumption (mM/optical density unit)
|
|
|---|---|---|
| 12 h | 24 h | |
| pCXJ10 | 2.6 ± 0.8 (39)a | 3.6 ± 0.5 (54) |
| KlARO8.1 | 3.7 ± 0.1 (54) | 4.2 ± 0.2 (63) |
| KlARO8.2 | 3.6 ± 0.4 (54) | 4.6 ± 0.3 (68) |
| KlARO9 | 4.5 ± 0.2 (67) | 4.7 ± 0.1 (71) |
| KlBAT1 | 2.7 ± 0.7 (40) | 4.0 ± 0.5 (60) |
| KlAAT1 | 3.8 ± 0.2 (56) | 4.4 ± 0.1 (65) |
| KlAAT2 | 4.2 ± 0.0 (63) | 4.6 ± 0.2 (70) |
The numbers in parentheses are the percentages of the initial amount of methionine consumed.
TABLE 3.
Estimated copy number of plasmid, plasmid maintenance, and growth for each transformant grown in PDB after 24 h
| Transformant | Copies/cell | Plasmid maintenance (%)a | Growth (OD600)b |
|---|---|---|---|
| pCXJ10 | 5 | 30 | 0.78 |
| KlARO8.1 | 2 | 8 | 0.37 |
| KlARO8.2 | 7 | 48 | 0.40 |
| KlARO9 | 6 | 31 | 0.38 |
| KlBAT1 | 5 | 35 | 0.84 |
| KlAAT1 | 6 | 31 | 0.32 |
| KlAAT2 | 6 | 44 | 0.63 |
Plasmid maintenance was calculated by dividing the number of CFU/ml for cells grown on YNB without uracil by the number of CFU/ml for cells grown on YNB medium with uracil after 48 h of incubation. The values are means from duplicate experiments.
The optical densities at 600 nm (OD600) are means from duplicate experiments.
Even though the transformants containing the KlAAT1 and KlAAT2 genes consumed large amounts of l-methionine, they converted only 11% and 10% of the l-methionine consumed to MOBA and MHBA, respectively, after 24 h of incubation. The pCXJ10 control converted only 14% of the initial l-methionine to MOBA and MHBA, while the transformants that contained the KlARO8.1 and KlARO8.2 genes converted the largest amounts of l-methionine to MOBA and MHBA. The transformants containing the KlARO8.1 and KlARO8.2 genes converted 29% and 27% of the l-methionine to the intermediate products, respectively, resulting in production of significant amounts of MOBA and MHBA. The transformant containing the KlARO9 gene converted 16% of l-methionine, while the transformant containing the KlBAT1 gene converted 15% of the initial amount of l-methionine and the final amounts of MOBA and MHBA were lower than those produced by the KlARO8.1 and KlARO8.2 transformants.
Whether the intermediate products of the pathway were subsequently chemically and/or enzymatically converted to MTL, the common precursor of many VSCs (7, 30), was examined by gas chromatography-mass spectrometry. DMDS, dimethyl trisulfide, and S-methylthioacetate were the dominant VSCs produced by K. lactis (data not shown). The total amounts of VSCs (DMDS, dimethyl trisulfide, and S-methylthioacetate) produced are shown in Fig. 2A. The KlARO8.1 and KlARO8.2 transformants produced the largest amounts of VSCs after 24 h; they produced two and three times more VSCs than the control (a transformant containing the plasmid without an insert), respectively. In the transformants carrying the other putative aminotransferase genes VSC production was maintained at lower levels.
The l-methionine transaminase activities of the different transformants were assayed (Fig. 2B). Even though the amounts detected were small, two transformants, the KlARO8.1 and KlARO8.2 transformants, produced about 2 to 3 times more glutamate than the transformant with no insert produced (Fig. 2B), indicating that there was higher aminotransferase activity and, as a result, larger amounts of VSCs (Fig. 2A).
Southern blot hybridization analysis.
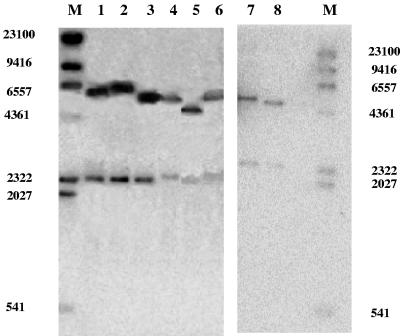
As the experiments described above were performed with cells grown in PDB with no selective pressure for the maintenance of the various plasmids, we wondered whether the copy number of the plasmids carried by the transformants could affect the results. The vector used in this study is a high-copy number plasmid, and it was shown previously that cells grown in a nonselective medium were able to retain the plasmid for approximately 30 generations (10). We therefore performed a Southern blot analysis to determine the copy numbers of the plasmid in the various transformants after 24 h (Fig. 3). Assuming that the quantities of radiolabeled KlURA3 and ScURA3 genes were the same, the copy number of the plasmid was deduced from the ratio of the intensities of the KlURA3 and ScURA3 bands. The estimated copy number of each transformant is shown in Table 3. As the average copy numbers were similar for all transformants except the KlARO8.1 transformant, we concluded that the copy number did not affect the measurements of MOBA/MHBA, VSCs, or enzymatic activity. In addition, if the levels of VSCs and MOBA/MHBA were calculated per copy number, the KlARO8.1 transformant produced 5 times more MOBA/MHBA than the control plasmid without an insert produced. This result was confirmed by measuring plasmid maintenance in cells grown with and without uracil. The KlARO8.1 transformant had the lowest plasmid maintenance, with two copies/cell, while the KlARO8.2 transformant showed the highest plasmid maintenance, with seven copies/cell in the nonselective medium PDB after 24 h of incubation (Table 3). In the latter case, 50% of the plasmid was maintained in the cells after 24 h of incubation; this transformant was also the main producer of VSCs.
FIG. 3.
Hybridization of total DNA from various CBS 2359/152F transformants with URA genes. The membranes were hybridized with ScURA (upper band) and KlURA (lower band). Lanes M, HindIII-digested λ DNA; lane 1, KlARO8.2; lane 2, KlAAT2; lane 3, KlAAT1; lane 4, KlARO8.1; lane 5, pCXJ10; lane 6, KlBAT1; lane 7, KlARO9; lane 8, pCXJ10. The sizes (in bp) of the HindIII-digested λ DNA fragments are indicated on the left and on the right.
DISCUSSION
K. lactis is generally recognized as safe for industrial use. During cheese ripening, this yeast can produce significant amounts of esters, imparting fruity flavors. Moreover, it has been shown that by providing an exogenous source of MTL in K. lactis, the amounts and variety of VSCs are significantly increased, which contributes to the aroma of the cheese (3). However, the mechanisms resulting in the production of VSCs are still controversial.
In the present study, all genes that putatively encode aminotransferase on the basis of sequence homology in the cheese-ripening yeast K. lactis were found and cloned in the overproducing plasmid pCXJ10. The KlBAT1, KlAAT1, KlAAT2, KlARO8.1, KlARO8.2, and KlARO9 genes were closely related to the S. cerevisiae BAT1, AAT1, AAT2, ARO8, and ARO9 genes encoding several aminotransferases. According to the results of this study, the KlARO8.1 and KlARO8.2 genes seem to be functional orthologues of the ScARO8 gene and to have an important role in the l-methionine pathway. The transformants with plasmids containing these genes produced the largest amounts of MOBA and MHBA and subsequently converted these compounds to VSCs. This was confirmed by the fact that after 24 h the transformant that produced the largest amount of MOBA was also the transformant that produced the largest amount of VSCs. The KlBAT1 gene and the KlAAT1 and KlAAT2 genes had no effect on the conversion of l-methionine to VSCs. The transformants containing these genes did not produce significant amounts of DMDS or any other VSC compared to the control transformant (plasmid without any insert). The transformant containing the KlARO9 gene produced consistent amounts of MOBA and MHBA that were not subsequently entirely converted to VSCs. Why the transformant that produced the largest amount of MOBA but not the largest amount of MHBA is the transformant that produced the largest amount of VSCs has not been determined. Although the aminotransferase pathway is active in K. lactis, the subsequent conversion of MOBA and/or MHBA to MTL and other VSCs still remains to be elucidated. A MOBA demethiolating activity has been found previously in K. lactis (3), but the genes involved and the mechanism have yet to be studied.
No studies have been conducted with any other yeast except Y. lipolytica, in which, unlike with K. lactis, the orthologue of a branched-chain aminotransferase-encoding gene was shown to be involved in the l-methionine pathway (6). As no other gene has been studied, it is possible that an aromatic aminotransferase is also involved in the pathway in Y. lipolytica, as it is in K. lactis. No information is available for other cheese-ripening yeasts, and it would be worthwhile to determine how the pathway proceeds in these organisms and whether they all convert l-methionine to MTL with the involvement of the same genes.
This is the first study in which all the possible aminotransferase-encoding genes from a yeast were studied in relation to the catabolism l-methionine. We elucidated the genes involved in the l-methionine catabolic pathway in K. lactis and showed that it is a complex pathway involving two ScARO8 orthologues. We show here for the first time that the role of these genes in the production and conversion of l-methionine to methanethiol is significant and that the two genes contribute at comparable rates.
Acknowledgments
D.-M. Kagkli is grateful to the EU for a Marie Curie Fellowship (YETI program QLK3_CT_2000_60055).
We thank Micheline Wésolowski-Louvel for providing strain CBS 2359/152F and Bao Weiguo for providing the pCXJ10 vector used in this study, as well as Roselyne Tâche and Armelle Delile for excellent technical assistance.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amarita, F., M. Yvon, M. Nardi, E. Chambellon, J. Delettre, and P. Bonnarme. 2004. Identification and functional analysis of the gene encoding methionine-gamma-lyase in Brevibacterium linens. Appl. Environ. Microbiol. 70:7348-7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arfi, K., H. E. Spinnler, R. Tâche, and P. Bonnarme. 2002. Production of volatile compounds by cheese-ripening yeasts: requirement for a methanethiol donor for S-methyl thioacetate synthesis by Kluyveromyces lactis. Appl. Microbiol. Biotechnol. 58:503-510. [DOI] [PubMed] [Google Scholar]
- 4.Arfi, K., R. Tâche, H. E. Spinnler, and P. Bonnarme. 2003. Dual influence of the carbon source and l-methionine on the synthesis of sulfur compounds in the cheese-ripening yeast Geotrichum candidum. Appl. Microbiol. Biotechnol. 61:359-365. [DOI] [PubMed] [Google Scholar]
- 5.Aston, J. W., and L. K. Creamer. 1986. Contribution of the components of the water-soluble fraction to the flavor of cheddar cheese. N. Z. J. Dairy Sci. Technol. 21:229-248. [Google Scholar]
- 6.Bondar Cernat, D., J. M. Beckerich, and P. Bonnarme. 2005. Involvement of a branched-chain aminotransferase in production of volatile sulfur compounds in Yarrowia lipolytica. Appl. Environ. Microbiol. 71:4585-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnarme, P., C. Lapadatescu, M. Yvon, and H. E. Spinnler. 2001. l-Methionine degradation potentialities of cheese-ripening microorganisms. J. Dairy Res. 68:663-674. [DOI] [PubMed] [Google Scholar]
- 8.Bonnarme, P., K. Arfi, C. Dury, S. Helinck, M. Yvon, and H. E. Spinnler. 2001. Sulfur compound production by Geotrichum candidum from l-methionine: importance of the transamination step. FEMS Microbiol. Lett. 205:247-252. [DOI] [PubMed] [Google Scholar]
- 8a.Bradford M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Brun, C., P. Surdej, and R. Miassod. 1993. Relationship between scaffold-attached regions, sequences replicating autonomously in yeast, and a chromosomal replication origin in the Drosophila rDNA. Exp. Cell Res. 208:104-114. [DOI] [PubMed] [Google Scholar]
- 10.Chen, X. J. 1996. Low- and high-copy number shuttle vectors for replication in the budding yeast Kluyveromyces lactis. Gene 172:131-136. [DOI] [PubMed] [Google Scholar]
- 11.Cronin, V. B., B. Maras, D. Barra, and S. Doonan. 1991. The amino acid sequence of the aspartate aminotransferase from baker's yeast (Saccharomyces cerevisiae). Biochem. J. 277:335-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dias, B., and B. Weimer. 1998. Purification and characterization of l-methionine gamma-lyase from Brevibacterium linens BL2. Appl. Environ. Microbiol. 64:3327-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dower, W., J. F. Miller, and C. W. Ragsale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44.15229592 [Google Scholar]
- 15.Eden, A., G. Simchen, and N. Benvenisty. 1996. Two yeast homologs of ECA39, a target for c-Myc regulation, code for cytosolic and mitochondrial branched-chain amino acid aminotransferases. J. Biol. Chem. 271:20242-20245. [DOI] [PubMed] [Google Scholar]
- 16.Engels, W. J. M., and J. Visser. 1994. Isolation and comparative characterization of components that contribute to the flavor of different types of cheese. Neth. Milk Dairy J. 48:127-140. [Google Scholar]
- 17.Gao, S., E. S. Mooberry, and J. L. Steele. 1998. Use of 13C nuclear magnetic resonance and gas chromatography to examine methionine catabolism by lactococci. Appl. Environ. Microbiol. 64:4670-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 20.Iraqui, I., S. Vissers, M. Cartiaux, and A. Urrestarazu. 1998. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 257:238-248. [DOI] [PubMed] [Google Scholar]
- 21.Lee, C. W., and M. J. Desmazeaud. 1985. Partial purification and some properties of an aromatic amino acid and an aspartate aminotransferase in Brevibacterium linens. J. Gen. Microbiol. 131:459-467. [Google Scholar]
- 22.Martin, N., C. Berger, C. Le Du, and H. E. Spinnler. 2001. Aroma compound production in cheese curd by coculturing with selected yeast and bacteria. J. Dairy Sci. 84:2125-2135. [DOI] [PubMed] [Google Scholar]
- 23.Morin, P. J., G. S. Subramanian, and T. D. Gilmore. 1992. AAT1, a gene encoding a mitochondrial aspartate aminotransferase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1171:211-214. [DOI] [PubMed] [Google Scholar]
- 24.Perrière, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Sanchez, M., F. J. Iglesias, C. Santamaria, and A. Dominguez. 1993. Transformation of Kluyveromyces lactis by electroporation. Appl. Environ. Microbiol. 59:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urrestarazu, A., S. Vissers, I. Iraqui, and M. Grenson. 1998. Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Mol. Gen. Genet. 257:230-237. [DOI] [PubMed] [Google Scholar]
- 29.Verleur, N., Y. Elgersma, C. W. Van Roermund, H. F. Tabak, and R. J. Wanders. 1997. Cytosolic aspartate aminotransferase encoded by the AAT2 gene is targeted to the peroxisomes in oleate-grown Saccharomyces cerevisiae. Eur. J. Biochem. 247:972-980. [DOI] [PubMed] [Google Scholar]
- 30.Weimer, B., K. Seefeldt, and B. Dias. 1999. Sulfur metabolism in bacteria associated with cheese. Antonie Leeuwenhoek 76:247-261. [PubMed] [Google Scholar]
- 31.Yvon, M., and L. Rijnen. 2001. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 11:185-201. [Google Scholar]
- 32.Yvon, M., S. Thirouin, L. Rijnen, D. Fromentier, and J. C. Gripon. 1997. An aminotransferase from Lactococcus lactis initiates conversion of amino acids to cheese flavor compounds. Appl. Environ. Microbiol. 63:414-419. [DOI] [PMC free article] [PubMed] [Google Scholar]