Abstract
The carbazole degradative car-I gene cluster (carAaIBaIBbICIAcI) of Sphingomonas sp. strain KA1 is located on the 254-kb circular plasmid pCAR3. Carbazole conversion to anthranilate is catalyzed by carbazole 1,9a-dioxygenase (CARDO; CarAaIAcI), meta-cleavage enzyme (CarBaIBbI), and hydrolase (CarCI). CARDO is a three-component dioxygenase, and CarAaI and CarAcI are its terminal oxygenase and ferredoxin components. The car-I gene cluster lacks the gene encoding the ferredoxin reductase component of CARDO. In the present study, based on the draft sequence of pCAR3, we found multiple carbazole degradation genes dispersed in four loci on pCAR3, including a second copy of the car gene cluster (carAaIIBaIIBbIICIIAcII) and the ferredoxin/reductase genes fdxI-fdrI and fdrII. Biotransformation experiments showed that FdrI (or FdrII) could drive the electron transfer chain from NAD(P)H to CarAaI (or CarAaII) with the aid of ferredoxin (CarAcI, CarAcII, or FdxI). Because this electron transfer chain showed phylogenetic relatedness to that consisting of putidaredoxin and putidaredoxin reductase of the P450cam monooxygenase system of Pseudomonas putida, CARDO systems of KA1 can be classified in the class IIA Rieske non-heme iron oxygenase system. Reverse transcription-PCR (RT-PCR) and quantitative RT-PCR analyses revealed that two car gene clusters constituted operons, and their expression was induced when KA1 was exposed to carbazole, although the fdxI-fdrI and fdrII genes were expressed constitutively. Both terminal oxygenases of KA1 showed roughly the same substrate specificity as that from the well-characterized carbazole degrader Pseudomonas resinovorans CA10, although slight differences were observed.
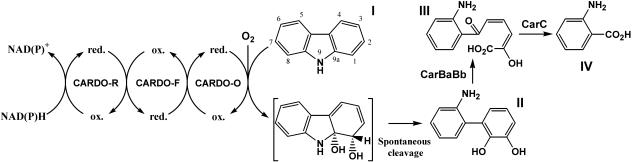
Carbazole is an N-heterocyclic aromatic compound derived from coal tar and shale oil (26) and is known to possess mutagenic and toxic activities (2, 16). To remediate carbazole-contaminated environments using biotechnological approaches, a wide variety of carbazole-degrading bacteria have been isolated and characterized (13, 14, 18, 28). Among them, the carbazole-catabolic car genes of Pseudomonas resinovorans CA10 (carCA10 genes) have been studied most extensively. Carbazole is first dioxygenated at the angular (C-9a) and adjacent (C-1) positions to yield an unstable cis-hydrodiol (Fig. 1) (28). Such initial dioxygenation is called an angular dioxygenation. Unstable cis-hydrodiol is spontaneously converted to 2′-aminobiphenyl-2,3-diol, which is further converted to anthranilate via meta-cleavage and hydrolysis (Fig. 1). Genes encoding carbazole 1,9a-dioxygenase (CARDO) were first cloned from CA10, and CARDOCA10 was found to be a three-component system, in which NAD(P)H-dependent ferredoxin reductase (CARDO-RCA10; CarAdCA10 monomer) and ferredoxin (CARDO-FCA10; CarAcCA10 monomer) transfer electrons from NAD(P)H to oxygenase (CARDO-OCA10; CarAaCA10 trimer) (Fig. 1) (25, 36). This multicomponent enzyme is a member of the Rieske non-heme iron oxygenase system (ROS). ROS members are classified into three classes and several subclasses based on the features of the electron transport chain (7). CARDO-RCA10 contains both a flavin adenine dinucleotide (FAD) and a plant-type [2Fe-2S] cluster, and CARDO-FCA10 is a ferredoxin having a Rieske-type [2Fe-2S] cluster (Rieske ferredoxin) (25). Thus, the CARDOCA10 is classified in class III.
FIG. 1.
Carbazole degradation by Car enzymes harbored by various carbazole degraders. The product of angular dioxygenation of carbazole shown in brackets is unstable and has not been detected directly. Enzyme (or protein) names: terminal oxygenase (CARDO-O), ferredoxin (CARDO-F), and reductase (CARDO-R) components of CARDO; meta-cleavage enzyme (CarBaBb); meta-cleavage compound hydrolase (CarC). ox. and red., oxidized and reduced states of the CARDO components, respectively. Compounds: I, carbazole; II, 2′-aminobiphenyl-2,3-diol; III, 2-hydroxy-6-oxo-6-(2′-aminobiphenyl)-hexa-2,4-dienoic acid; IV, anthranilic acid.
Sphingomonas sp. strain KA1 was isolated as a versatile carbazole-degrading bacterium (9) whose degradation pathway of carbazole is similar to that by CA10 (14). Previous study of the carbazole degradation (carKA1) gene cluster of KA1 (14) revealed that, unlike the carCA10 gene cluster, the carKA1 gene cluster does not contain the CARDO-R gene but does contain the genes for CARDO-O (carAa) and CARDO-F (carAc), meta-cleavage enzyme (carBaBb), and meta-cleavage compound hydrolase (carC). Interestingly, although CarAaKA1 shows high homology with CarAaCA10 (60% identity), CarAcKA1 had no relatedness with the Rieske ferredoxin, including CarAcCA10, and showed similarity to the putidaredoxin-type ferredoxins. Because CARDO-OKA1 can receive electrons from CARDO-FKA1 and catalyze angular dioxygenation of carbazole (14), CARDOKA1 consisting of CarAaKA1 and CarAcKA1 is likely to be assigned to class IIA. To confirm this consideration, we should definitely clarify all CARDO components, especially ferredoxin reductase CARDO-R, that are involved in carbazole metabolism by KA1.
In the genus Sphingomonas, it has been reported that catabolic genes are sometimes dispersed on the genome (4, 23). Considering that the carKA1 gene cluster is located on the 254-kb circular plasmid pCAR3 (9), it is possible that the ferredoxin reductase gene is also located on this plasmid. In the present study, using the draft sequence covering >99.9% of pCAR3, we found the multiple gene sets involved in carbazole degradation. In addition, based on the reverse transcription-PCR (RT-PCR) analysis of each CARDOKA1 component gene and reconstitution assays of CARDOKA1 components in Escherichia coli cells, we discuss the multiplicity of the carbazole degradation function of pCAR3.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. LB or 2×YT medium (35) was used for bacterial growth. To prepare the KA1 RNA, nitrogen-containing mineral medium NMM1 supplemented with carbazole or succinate was used. NMM1 has the same composition as CNFMM (29) except for the addition of 3.0 g of NH4NO3 per liter. Carbazole was added to NMM1 as described previously (29). E. coli JM109 (35) and DH5α (35) were used as hosts for pUC119 and its derivatives. Ampicillin (Ap) was added to selective media at a final concentration of 50 μg/ml. For plate cultures, the media mentioned above, solidified with 1.6% (wt/vol) agar, were used.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s)a | Reference or source |
|---|---|---|
| Bacterial strains | ||
| E. coli JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F−[traD36 proAB+lacIqlacZΔM15] | 35 |
| E. coli DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 35 |
| Sphingomonas sp. strain KA1 | Car+ | 9 |
| Plasmids | ||
| pUC119 | AprlacZ, pMB9 replicon, M13IG | 35 |
| pT7Blue(R) | AprlacZ | Novagen |
| pUKAcarAaI | Apr; pUC119 with 1.2-kb SphI-XbaI DNA fragment containing the carAaI gene of strain KA1 | This study |
| pUKAcarAaII | Apr; pUC119 with 1.2-kb SphI-XbaI DNA fragment containing the carAaII gene of strain KA1 | This study |
| pUKAcarAcI | Apr; pUC119 with 0.3-kb XbaI-KpnI DNA fragment containing the carAcI gene of strain KA1 | This study |
| pUKAcarAcII | Apr; pUC119 with 0.3-kb XbaI-KpnI DNA fragment containing the carAcII gene of strain KA1 | This study |
| pUKAfdxI | Apr; pUC119 with 0.3-kb XbaI-KpnI DNA fragment containing the fdxI gene of strain KA1 | This study |
| pUKAfdrI | Apr; pUC119 with 1.2-kb KpnI-EcoRI DNA fragment containing the fdrI gene of strain KA1 | This study |
| pUKAfdrII | Apr; pUC119 with 1.2-kb KpnI-EcoRI DNA fragment containing the fdrII gene of strain KA1 | This study |
| pUKA248 | Apr; pUC119 with 1.2-kb SphI-XbaI fragment (carAaI) from pUKAcarAaI and 0.3-kb XbaI-KpnI fragment (carAcI) from pUKAcarAcI | This study |
| pUKA249 | Apr; pUC119 with 1.2-kb SphI-XbaI fragment (carAaI) from pUKAcarAaI and 1.2-kb KpnI-EcoRI fragment (fdrII) from pUKAfdrII | This study |
| pUKA250 | Apr; pUC119 with 1.2-kb SphI-XbaI fragment (carAaI) from pUKAcarAaI and 1.2-kb KpnI-EcoRI fragment (fdrI) from pUKAfdrI | This study |
| pUKA253 | Apr; pUC119 with 1.5-kb SphI-KpnI fragment (carAaI and carAcI) from pUKA248 and 1.2-kb KpnI-EcoRI fragment (fdrII) from pUKAfdrII | This study |
| pUKA254 | Apr; pUC119 with 1.5-kb SphI-KpnI fragment (carAaI and carAcI) from pUKA248 and 1.2-kb KpnI-EcoRI fragment (fdrI) from pUKAfdrI | This study |
| pUKA255 | Apr; pUC119 with 1.2-kb SphI-XbaI fragment (carAaII) from pUKAcarAaII and 0.3-kb XbaI-KpnI fragment (carAcII) from pUKAcarAcII | This study |
| pUKA256 | Apr; pUC119 with 1.2-kb SphI-XbaI fragment (carAaII) from pUKAcarAaII and 1.2-kb KpnI-EcoRI fragment (fdrII) from pUKAfdrII | This study |
| pUKA257 | Apr; pUC119 with 1.2-kb SphI-XbaI fragment (carAaII) from pUKAcarAaII and 1.2-kb KpnI-EcoRI fragment (fdrI) from pUKAfdrI | This study |
| pUKA260 | Apr; pUC119 with 1.5-kb SphI-KpnI fragment (carAaII and carAcII) from pUKA255 and 1.2-kb KpnI-EcoRI fragment (fdrII) from pUKAfdrII | This study |
| pUKA261 | Apr; pUC119 with 1.5-kb SphI-KpnI fragment (carAaII and carAcII) from pUKA255 and 1.2-kb KpnI-EcoRI fragment (fdrI) from pUKAfdrI | This study |
| pUKA263 | Apr; pUC119 with 1.2-kb SphI-XbaI fragment (carAaI) from pUKAcarAaI, 0.3-kb XbaI-KpnI fragment (fdxI) from pUKAfdxI, and 1.2-kb KpnI-EcoRI fragment (fdrI) from pUKAfdrI | This study |
| pUKA265 | Apr; pUC119 with 1.2-kb SphI-XbaI fragment (carAaII) from pUKAcarAaII, 0.3-kb XbaI-KpnI fragment (fdxI) from pUKAfdxI, and 1.2-kb KpnI-EcoRI fragment (fdrI) from pUKAfdrI | This study |
| pUCARA | Apr; pUC119 with 5.3-kb EcoRI DNA fragment containing carAaAaAc (ORF7) carAd genes of Pseudomonas resinovorans strain CA10 | 36 |
Car+ represents an ability to grow on CAR as the sole source of carbon, nitrogen, and energy. Apr represents resistance to ampicillin.
DNA manipulations.
Total DNA of KA1 was prepared as described previously (29). Plasmids were prepared from E. coli by the alkaline lysis method (35) or with a Quantum Prep plasmid miniprep kit (Bio-Rad Laboratories, Hercules, CA). DNA fragments were extracted from agarose gels with an EZNA gel extraction kit (Omega Bio-tek, Inc., Doraville, GA). Other DNA manipulations were performed according to standard protocols (35).
Sequencing of pCAR3 and annotation.
Shotgun sequencing of pCAR3 was performed by Dragon Genomics Co. Ltd. (Shiga, Japan). Open reading frames (ORFs) were found by DNASIS-Mac, version 3.7 (Hitachi Software Engineering Co. Ltd., Yokohama, Japan). Homologous sequences were searched from the DDBJ/EMBL/GenBank DNA databases using the BLAST program (version 2.2.10) (1). The deduced amino acid sequences of ORFs were aligned using CLUSTAL W (version 1.83) (15).
RNA preparation and RT-PCR.
After the precultivation of KA1 in 5 ml of NMM1 supplemented with 10 mM succinate at 30°C, cells were gathered by centrifugation at 5,000 × g and then washed twice using CFMM (17). The washed cells were suspended in 500 μl of CFMM. Fifty microliters of the resultant cell suspension was added to 5 ml of NMM1 supplemented with 10 mM succinate, 10 mM each succinate and carbazole, or 10 mM carbazole (all media contained 1% [vol/vol] dimethyl sulfoxide [DMSO]). After a 2-h incubation with reciprocal shaking (300 strokes/min) at 30°C, the cells were harvested and used for extraction of total RNA by a NucleoSpin RNA II (Macherey-Nagel & Co., Düren, Germany) combined with RQ1 RNase-free DNase (Promega, Madison, WI). For RT-PCR, a One Step RNA PCR kit (AMV) (Takara Bio Inc.) was used. In RT-PCR, 100 ng of total RNA was used as a template. Detailed information on the RT-PCR primer sets and the conditions employed for respective gene amplifications are provided in Table S1 in the supplemental material. Control experiments without the addition of reverse transcriptase were also performed.
qRT-PCR.
The primer sets used in quantitative RT-PCR (qRT-PCR) and RT conditions are provided in Table S1 in the supplemental material. To synthesize each cDNA, ThermoScript reverse transcriptase (Invitrogen Corp., Carlsbad, CA) and 10 ng of total RNA (as a template) were used. After the RT reaction, quantitative real-time PCR with SYBR GREEN PCR Master Mix (Applied Biosystems, Foster City, CA) was performed using the synthesized cDNA as a template on the ABI7700 sequence detection system (Applied Biosystems). The copy number of each mRNA was determined by a standard curve using a series of known concentrations of the target sequence according to the method of Habe et al. (10). For normalization, 16S rRNA of KA1 was used as an internal standard. For each sample, the mean value from triplicate real-time PCRs was used to calculate the transcript abundance. The mRNA levels of each gene in the control sample (NMM1 supplemented with succinate) were set at 1.0.
Construction of expression plasmids for car genes.
Each of the carAaI, carAaII, carAcI, carAcII, fdxI, fdrI, and fdrII genes was separately amplified by PCR using the respective primer sets shown in Table S2 in the supplemental material, which were designed to introduce appropriate restriction sites and the effective Shine-Dalgarno sequence for the E. coli transcription system. In PCR amplification, total DNA of KA1 was used as a template. The amplified products were digested at the introduced restriction sites and were ligated into the corresponding sites of pUC119 to produce plasmids for the expression of single CARDOKA1 components. After their nucleotide sequences were confirmed to be identical to those designed, their insert fragments were used for the construction of plasmids to direct the expression of each of the CARDOKA1 components. For example, pUKA248 was constructed by cloning the 0.3-kb XbaI-KpnI fragment from pUKAcarAcI into the corresponding site of pUKAcarAaI. For the construction of pUKA249 and pUKA253, the 1.2-kb KpnI-EcoRI fragment from pUKAfdrII was ligated into the corresponding sites of pUKAcarAaI and pUKA248, respectively. Other plasmids were constructed similarly.
Biotransformation of carbazole by E. coli cells expressing putative CARDO components.
E. coli JM109 harboring appropriate plasmids was cultivated in 5 ml of LB supplemented with Ap at 37°C with reciprocal shaking (300 strokes min−1), and then 100 μl of the culture was transferred to 100 ml of the same medium. After cultivation at 30°C with shaking at 120 rpm until the optical density at 600 nm (OD600) reached 0.4 to 0.5, isopropyl-1-thio-β-d-galactopyranoside (IPTG) was added to 0.5 mM and the cells further cultivated at 30°C for 15 h. Then the cells were harvested by centrifugation (5,000 × g, 10 min, 4°C), washed twice with CNF buffer (2.2 g Na2HPO4, 0.8 g KH2PO4 per liter), and resuspended in the same buffer to an OD600 of 12 to 13. Carbazole was dissolved at 100 mM in DMSO and 50 μl added to 5-ml cell suspensions in the reaction tube. After incubation on a reciprocal shaker (300 strokes min−1) at 30°C for 20 h, the mixtures were extracted with an equal volume of ethyl acetate. The extracts were analyzed by gas chromatography-mass spectrometry (GC-MS) after derivatization with N-methyl-N-trimethylsilyltrifluoroacetamide (MSTFA) as described previously (27, 39). To define the conversion ratio, we used the following formula: conversion ratio (%) = 100 × (peak area for the TIC of the product)/(peak area for the TIC of the carbazole + peak area for the TIC of the product), where TIC is the total ion current. All experiments were conducted independently at least three times.
Biotransformation analysis to determine substrate specificity.
E. coli JM109 harboring pUKA253 or pUKA260 was cultivated as described above, and then 5 ml of the culture was transferred to 1 liter of the same medium. A resting cell suspension (OD600, 17 to 18) was prepared as described above except for the incubation temperature (25°C) and induction time after the addition of IPTG (12 h). Anthracene was dissolved in N,N-dimethylformamide at 10 mg/ml (wt/vol), and the other putative substrates shown in Table S3 in the supplemental material were dissolved in DMSO at the same concentration. Addition of substrate, subsequent incubation, and GC-MS analysis were performed as described above.
Nucleotide sequence accession numbers.
The nucleotide sequences of the car-I, car-II, fdxIfdrI, and fdrII loci were deposited in the DDBJ DNA database under accession numbers AB095953, AB220949, AB220950, and AB220951, respectively.
RESULTS
pCAR3 loci encompassing the carbazole degradation genes.
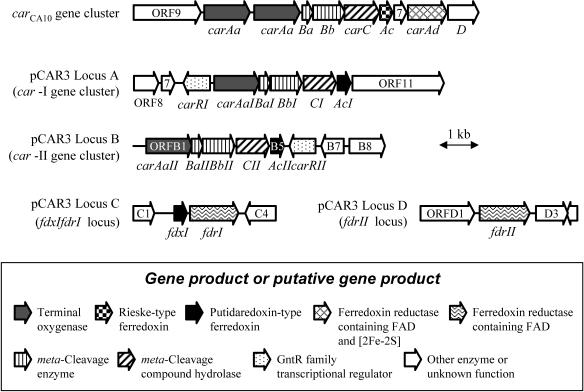
The draft sequence of pCAR3 having a single gap (estimated to be <80 bp) revealed the loci encompassing the genes that could be involved in the transformation of carbazole to anthranilate. In addition to the previously characterized car gene cluster (14), probable carbazole degradation genes were found in three other distinct loci of pCAR3 (Fig. 2 and Table 2).
FIG. 2.
Genetic organization of the car-I gene cluster (locus A), car-II gene cluster (locus B), fdxIfdrI locus (locus C), and fdrII locus (locus D) encompassing the pCAR3 genes involved in the degradation of carbazole by Sphingomonas sp. strain KA1. The genetic organization of the car gene cluster responsible for carbazole degradation in P. resinovorans CA10 is also shown. The carAa gene of CA10 is tandemly duplicated (36). The arrows in the physical maps indicate the size, location, and direction of transcription of the ORFs derived from the nucleotide sequence data.
TABLE 2.
ORFs found in four pCAR3 loci, which contain the genes probably involved in carbazole conversion to anthranilate
| Locus and accession no. | Position (bp) in sequence (directiona) | Name of gene | GC content (%) | No. of amino acids | Probable function | Homologous protein
|
|||
|---|---|---|---|---|---|---|---|---|---|
| Identity (%)b | Protein or ORF name | Source | Accession no. | ||||||
| Locus A | |||||||||
| AB095953 | 3162-3785 (n) | ORF8 | 57.2 | 207 | Conserved hypothetical protein similar to SMa10599 (truncated) | 27 | SalaDRAFT_3075 | Sphingopyxis alaskensis RB2256 | ZP_00577426 |
| AB095953 | 3862-4188 (n) | ORF7 | 52.6 | 108 | Putative RNA-binding protein (truncated) | 68 | Atlg32790 | Arabidopsis thaliana | NP_174556 |
| AB095953 | 4409-5089 (c) | carRI (ORF6) | 60.8 | 226 | Transcriptional regulator of car operon | 51 | CarRII | Sphingomonas sp. strain KA1 | |
| 36 | CarR | Janthinobacterium sp. strain J3 | BAC56739 | ||||||
| AB095953 | 5192-6328 (n) | carAaI (ORF1) | 57.1 | 378 | Terminal oxygenase component of CARDO | 75 | CarAaII | Sphingomonas sp. strain KA1 | |
| 59 | CarAa | P. resinovorans CA10 | BAC41545 | ||||||
| AB095953 | 6328-6609 (n) | carBaI (ORF2) | 55.3 | 93 | Subunit of meta-cleavage enzyme | 66 | CarBaII | Sphingomonas sp. strain KA1 | |
| 36 | CarBa | P. resinovorans CA10 | BAC41546 | ||||||
| AB095953 | 6602-7405 (n) | carBbI (ORF3) | 59.5 | 267 | Subunit of meta-cleavage enzyme | 72 | CarBbII | Sphingomonas sp. strain KA1 | |
| AB095953 | 7448-8272 (n) | carCI (ORF4) | 59.6 | 274 | meta-Cleavage compound hydrolase | 67 | CarCII | Sphingomonas sp. strain KA1 | |
| 57 | CarC | P. resinovorans CA10 | BAC41548 | ||||||
| AB095953 | 8313-8642 (n) | carAcI (ORF5) | 62.7 | 109 | Ferredoxin component of CARDO | 56 | CarAcII | Sphingomonas sp. strain KA1 | |
| 42 | CamB | Pseudomonas putida | P00259 | ||||||
| AB095953 | 8684-10999 (n) | ORF11 | 58.1 | 771 | Outer membrane receptor | 44 | Saro02002648 | Novosphingobium aromaticivorans DSM12444 | ZP_00302784 |
| Locus B | |||||||||
| AB220949 | 344-1495 (n) | carAaII (ORFB1) | 67.4 | 383 | Terminal oxygenase component of CARDO | 75 | CarAaI | Sphingomonas sp. strain KA1 | BAC56759 |
| 55 | CarAa | P. resinovorans CA10 | BAC41545 | ||||||
| AB220949 | 1492-1773 (n) | carBaII (ORFB2) | 68.5 | 93 | Subunit of meta-cleavage enzyme | 66 | CarBaI | Sphingomonas sp. strain KA1 | BAC56760 |
| 32 | CarBa | P. resinovorans CA10 | BAC41546 | ||||||
| AB220949 | 1766-2575 (n) | carBbII (ORFB3) | 69.8 | 269 | Subunit of meta-cleavage enzyme | 72 | CarBbI | Sphingomonas sp. strain KA1 | BAC56761 |
| 44 | CarBb | P. resinovorans CA10 | BAC41547 | ||||||
| AB220949 | 2626-3444 (n) | carCII (ORFB4) | 67.5 | 272 | meta-Cleavage compound hydrolase | 67 | CarCI | Sphingomonas sp. strain KA1 | BAC56762 |
| 59 | CarC | P. resinovorans CA10 | BAC41548 | ||||||
| AB220949 | 3499-3828 (n) | carAcII (ORFB5) | 70.6 | 109 | Ferredoxin component of CARDO | 56 | CarAcI | Sphingomonas sp. strain KA1 | BAC56763 |
| 45 | CamB | P. putida | P00259 | ||||||
| AB220949 | 3968-4630 (c) | carRII (ORFB6) | 71.3 | 220 | Transcriptional regulator of car operon | 51 | CarRI | Sphingomonas sp. strain KA1 | BAC56758 |
| 37 | CarR | Janthiobacterium sp. strain J3 | BAC56739 | ||||||
| AB220949 | 4771-5340 (c) | tnpR (ORFB7) | 59.4 | 189 | Resolvase | 95 | TnpR | Flavobacterium sp. strain ATCC 27551(pPDL2) | CAD13184 |
| AB220949 | 5481-6383 (n) | tnpA (ORFB8) | 61.8 | 300 | Transposase (truncated) | 58 | TnpA | Flavobacterium sp. strain ATCC 27551(pPDL2) | CAD13183 |
| Locus C | |||||||||
| AB220950 | 1-534 (n) | ORFC1 | 58.0 | 177 | Resolvase/integrase (truncated) | 76 | mlr9024 | Mesorhizobium loti MAFF303099(pMLa) | NP_085611 |
| AB220950 | 1038-1355 (n) | fdxI (ORFC2) | 56.8 | 105 | Putidaredoxin-type [2Fe-2S] ferredoxin | 66 | ELI2682 | Erythrobacter litoralis HTCC2594 | ZP_00377441 |
| 40 | CarAcI | Sphingomonas sp. strain KA1 | BAC56763 | ||||||
| 38 | CarAcII | Sphingomonas sp. strain KA1 | |||||||
| 32 | CamB | P. putida | P00259 | ||||||
| AB220950 | 1432-2655 (n) | fdrI (ORFC3) | 60.8 | 407 | Ferredoxin reductase component of CARDO | 64 | Saro02000237 | N. aromaticivorans DSM12444 | ZP_00304456 |
|
|
|
|
|
|
59
|
FdrII
|
Sphingomonas sp. strain KA1
|
|
|
| 59 | RedA2 | S. wittichii RW1 | CAA05635 | ||||||
| 38 | CamA | P. putida | P16640 | ||||||
| AB220950 | 2831-3607 (c) | ORFC4 | 62.1 | 258 | Transcriptional regulator (IcIR family) | 28 | Chlo02004436 | Chloroflexus aurantiacus | ZP_00356137 |
| Locus D | |||||||||
| AB220951 | 1-1368 (n) | ORFD1 | 63.9 | 455 | Pyruvate/2-oxoglutarate dehydrogenase complex, dehydrogenase (E1) component, β subunit | 80 | Saro02001437 | N. aromaticivorans DSM12444 | ZP_00303572 |
| AB220951 | 1504-2766 (n) | fdrII (ORFD2) | 62.4 | 420 | Ferredoxin reductase component of CARDO | 76 | RedA2 | S. wittichii RW1 | CAA05635 |
| 59 | FdrI | Sphingomonas sp. strain KA1 | |||||||
| 41 | CamA | P. putida | P16640 | ||||||
| AB220951 | 2923-3729 (n) | ORFD3 | 58.0 | 268 | Transglycosylase | 49 | ORF925 | N. aromaticivorans F199(pNL1) | AAD03969 |
| AB220951 | 3696-3971 (c) | ORFD4 | 60.5 | 91 | Hypothetical protein | 62 | ORF921 | N. aromaticivorans F199(pNL1) | AAD03968 |
n, normal; c, complementary.
Percentage of amino acids that are identical when sequences are aligned with sequences listed in the DDBJ/EMBL/GenBank databases by using the CLUSTAL W multiple sequence alignment program (version 1.83) (15).
The deduced amino acid sequences of six ORFs (ORFB1 to ORFB6) in locus B exhibited 51 to 75% identity with those of the previous car genes on pCAR3, and we designated this cluster the car-II gene cluster, carAaIIBaIIBbIICIIAcII-carRII. The previous car gene cluster was renamed the car-I gene cluster (Fig. 2, locus A). The amino acid sequence of CarAaII displayed 75 and 59% identity with those of CarAaI and CarAaCA10, respectively, and contained both the Rieske-type [2Fe-2S] cluster consensus and Fe(II)-binding motifs (31). The amino acid sequence of CarAcII showed similarity to that of several putidaredoxin-type ferredoxins and 55% identity with that of CarAcI. CarAcII contained four Cys residues (CX5CX2CXnC), typical of putidaredoxin-type [2Fe-2S] cluster ligands (3, 5, 38). ORFC2 (designated fdxI) at locus C (Fig. 2) showed similarity to CarAcI and CarAcII. The deduced amino acid sequences of ORFC3 (designated fdrI), which may constitute an operon with the fdxI gene in locus C, and ORFD2 (designated fdrII) at locus D (Fig. 2) displayed 59 and 76% identities with RedA2, the ferredoxin reductase component of the dioxin dioxygenase of Sphingomonas wittichii RW1 (6). FdrI and FdrII shared 38 to 41% identity with putidaredoxin reductase (20, 32) and rhodocoxin reductase (24), and also had a FAD-binding motif and two ADP-binding motifs (for FAD and NADH) (40).
As well as CarBaI, CarBbI, and CarCI in the car-I gene cluster, CarBaII, CarBbII, and CarCII shared moderate homology with the CarBa, CarBb, and CarC from CA10 as summarized in Table 2. Their functions were predicted as follows: CarBaII and CarBbII, structural and catalytic subunits, respectively, of the meta-cleavage enzyme; CarCII, meta-cleavage compound hydrolase.
Transcriptional analyses of the putative CARDO component genes.
The primer set for the carAaIBaIBbICIAcI, carAaIIBaIIBbII, carBbIICII, carCIIAcII, fdxIfdrI, or fdrII genes could amplify DNA fragments with the expected sizes from the RNA of carbazole-grown KA1 cells (data not shown). Therefore, it was revealed that all seven genes (carAaI, carAaII, carAcI, carAcII, fdxI, fdrI, and fdrII) are expressed in carbazole-grown KA1 cells, suggesting that the gene products could be involved in CARDO system formation. Also, these results indicated that the carAaIBaIBbICIAcI and fdxIfdrI genes are transcribed as single transcriptional units. Furthermore, although we could not detect the RT-PCR product spanning the region from carAaII to carAcII (data not shown), the amplifications of three segmental fragments covering the entire car-II gene cluster (carAaIIBaIIBbII, carBbIICII, and carCIIAcII regions) were detected, and we concluded that the car-II gene cluster was also cotranscribed.
To analyze the inducibilities of the putative CARDO component genes, the mRNA levels of each gene after 2 h in carbazole-exposed or nonexposed KA1 were investigated by qRT-PCR. The mRNAs of carAaI, carAcI, carAaII, and carAcII in carbazole-exposed cells were about 13-, 10-, 15-, and 11-fold more abundant than those in nonexposed cells. Together with the results of RT-PCR analyses, these findings clearly indicated that transcription of the car-I and car-II gene clusters was induced when KA1 cells were exposed to carbazole. In contrast, the expression levels of ferredoxin and ferredoxin reductase genes were not elevated in response to carbazole exposure (1.0- to 1.2-fold induction).
Functional analyses of putative CARDO.
Biotransformation experiments were performed with carbazole using E. coli cells expressing putative CARDO components in various combinations. Although we could not detect their expression by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown), considering that the same promoter and Shine-Dalgarno sequence were used, the expression level of each gene was assumed to be similar. The production ratios of 2′-aminobiphenyl-2,3-diol from carbazole detected for cells harboring pUKA254 (carAaI, carAcI, and fdrI) and pUKA253 (carAaI, carAcI, and fdrII) were much higher than those seen with cells harboring any other recombinant plasmids containing carAaI (Table 3). These results clearly indicated that CarAaI can catalyze angular dioxygenation of carbazole efficiently in conjunction with both CarAcI and FdrI (or FdrII). Similarly, as shown in Table 3, E. coli cells expressing CarAaII exhibited a much higher conversion ratio when both CarAcII and FdrI (or FdrII) were coexpressed, indicating that CarAaII could function as a terminal oxygenase of CARDO with the aid of both CarAcII and FdrI (or FdrII). In addition, the expression of FdxI with CarAaI/FdrI or CarAaII/FdrI also increased the conversion ratio (see pUKA263 versus pUKA250 and pUKA265 versus pUKA257 in Table 3), although it was markedly lower than the expression observed when CarAcI or CarAcII was supplied as ferredoxin. These results suggested that FdxI could also transfer electrons from FdrI to CarAaI/CarAaII, but the efficiency of electron transfer was markedly lower than those of CarAcI and CarAcII.
TABLE 3.
Carbazole conversion by E. coli expressing a putative CARDO component(s)
| Plasmid used | Protein(s) expressed in E. coli JM109
|
Conversion ratio (%)a | ||
|---|---|---|---|---|
| Oxygenase | Ferredoxin | Reductase | ||
| pUKAcarAaI | CarAaI | 0.090 (±0.030) | ||
| pUKA248 | CarAaI | CarAcI | 0.34 (±0.21) | |
| pUKA250 | CarAaI | FdrI | 0.19 (±0.089) | |
| pUKA254 | CarAaI | CarAcI | FdrI | 96 (±1.4) |
| pUKA263 | CarAaI | FdxI | FdrI | 11.7 (±6.9) |
| pUKA249 | CarAaI | FdrII | 0.19 (±0.067) | |
| pUKA253 | CarAaI | CarAcI | FdrII | 95 (±4.4) |
| pUKAcarAaII | CarAaII | NDb | ||
| pUKA255 | CarAaII | CarAcII | 0.24 (±0.026) | |
| pUKA257 | CarAaII | FdrI | ND | |
| pUKA261 | CarAaII | CarAcII | FdrI | 40 (±13) |
| pUKA265 | CarAaII | FdxI | FdrI | 0.18 (±0.052) |
| pUKA256 | CarAaII | FdrII | ND | |
| pUKA260 | CarAaII | CarAcII | FdrII | 61 (±1.7) |
Conversion ratio (%) = 100 × (peak area for TIC of 2′-aminobiphenyl-2,3-diol)/(peak area for TIC of carbazole + peak area for TIC of 2′-aminobiphenyl-2,3-diol). Values are means ± standard deviations calculated from at least triplicate assays.
ND, no product was produced from carbazole.
We further examined whether CarAaI and CarAaII could receive electrons from ferredoxins not encoded on the same car gene clusters. When CarAcII was expressed with CarAaI/FdrI (pUKA268) or CarAaI/FdrII (pUKA267), 77% ± 3.3% and 87% ± 5.2% conversion occurred, respectively. Both of these figures were only a little lower than those seen with CarAcI (pUKA254 and pUKA253) (Table 3). These results suggested that CarAaI could receive electrons from CarAcII and CarAcI with similar efficiencies. However, the conversion ratio of carbazole by E. coli expressing CarAaII/CarAcI/FdrI (or FdrII) was markedly lower than that detected when CarAcII was replaced with CarAcI (8.3% ± 2.0% and 6.2% ± 1.2% conversion were observed for E. coli harboring pUKA270 [CarAaII/CarAcI/FdrI] and pUKA269 [CarAaII/CarAcI/FdrII], respectively). Therefore, we concluded that CarAcI could transfer electrons to CarAaII but that the electron transferability to CarAaII is substantially lower than that to CarAcII.
Based on these results, although all CARDO components found on pCAR3 can theoretically function in various combinations, we concluded that two CARDO systems, composed of CarAaI/CarAcI/(FdrI or FdrII) (designated CARDOKA1-1) and CarAaII/CarAcII/(FdrI or FdrII) (designated CARDOKA1-2) primarily function as initial oxygenases for carbazole in KA1.
Substrate ranges of two CARDOs of KA1.
The substrate ranges of CARDOKA1-1 and CARDOKA1-2 were determined by biotransformation analyses using recombinant E. coli. (Detailed data are provided in Table S3 in the supplemental material). Both CARDOs catalyzed angular dioxygenation (28) of carbazole, dibenzofuran, dibenzo-p-dioxin (DD), and phenoxathiin. The ratio of angular dioxygenation of 9-fluorenone was markedly poorer in comparison with carbazole, dibenzofuran, DD, or phenoxathiin. Neither CARDOKA1-1 nor CARDOKA1-2 could catalyze any oxygenation reactions for dibenzothiophene sulfone. With fluorene, both CARDOs catalyzed the monooxygenation of the methylene carbon and probable lateral dioxygenation (28) at unidentified positions. For biphenyl, naphthalene, and anthracene, both CARDOs catalyzed lateral dioxygenations as was the case with CARDOCA10 (28).
In conclusion, CARDOKA1-1 and CARDOKA1-2 have a wide substrate range, which is similar to that of CARDOCA10. However, there are two noteworthy differences (see Table S3 in the supplemental material). (i) CARDOKA1-1 preferably oxygenates anthracene, probably at C-2 and C-3, whereas CARDOKA1-2 and CARDOCA10 preferably oxygenate at C-1 and C-2 to yield cis-1,2-dihydroxy-1,2-dihydroanthracene. (ii) It is likely that the angular dioxygenation activities of CARDOKA1-2 for DD or phenoxathiin are relatively lower than those in the other two CARDO systems.
DISCUSSION
Two car gene clusters were present on plasmid pCAR3, and two CARDOs, composed of CarAaI/CarAcI/(FdrI or FdrII) (CARDOKA1-1) and CarAaII/CarAcII/(FdrI or FdrII) (CARDOKA1-2), function in KA1. Plural isofunctional degradation genes on a single bacterial genome have been reported. For example, multiple gene sets for biphenyl/polychlorinated biphenyl degradation are known to be dispersed on the chromosome and large linear plasmids in Rhodococcus sp. strain RHA1 (11, 19, 22, 42). Duplication of degradation genes has also been reported in several degradation plasmids, such as the 2,4-dichlorophenoxyacetic acid-degrading plasmids pJP4 (21) and pEST4011 (41) and the toluene/xylene-degrading plasmid pWW53 (30). It would be of great interest to determine the physiological roles of the two copies of the car gene cluster in carbazole metabolism by KA1. As described above, the substrate specificities of the two CARDOs are nearly identical, although slight differences were observed. These facts suggest that duplication does not broaden the degradation (or growth) substrate range of KA1 but raises its degradation rate of carbazole. Estimation of the in vivo concentration of each CARDO component and generation of knockouts of either car-I or car-II (or either fdrI or fdrII) followed by quantitative determination of the carbazole catabolic capacities will yield information to reveal the importance of multiple degradation genes.
The draft sequence of pCAR3 showed that the fdxIfdrI cistron is located 50 and 85 kb downstream of the car-I and car-II gene clusters, respectively (data not shown). The fdrII gene is located about 80 and 115 kb downstream of the car-I and car-II gene clusters, respectively (data not shown). The dispersion of the ROS component genes in pCAR3 contrasts with the well-organized degradation operon in pseudomonads, where the four or three genes are assembled, or at least cotranscribed, as is observed with the carCA10 gene cluster (Fig. 2) (36). Because similar dispersions of ROS component genes have been reported many times in sphingomonads (4, 23, 33, 34), it is possible that gene dispersion is one of the characteristics of sphingomonads. It is possible that constitutive expression of the fdrI and fdrII genes provides enough reductase to transfer electrons from NAD(P)H to ferredoxin. If this is the case, a well-organized operon is not necessary to achieve the appropriate carbazole degradation capacity. Another possibility is that the organization of car gene clusters of pCAR3 has not yet fully evolved and been optimized and that there is room for further evolution of the carbazole catabolic operon when KA1 is exposed to some selective pressure. On the other hand, the ferredoxin reductases, FdrI and FdrII, may be shared with other redox systems, possibly to maximize the catabolic potential while limiting its genetic burden. In this case, carbazole-dependent control of their expression would be disadvantageous for KA1.
According to the classification of Batie et al. (7), both CARDOs of KA1 are classified in class IIA. Class IIA ROS is a three-component oxygenase, in which the electron transfer components comprise a simple flavoprotein and a putidaredoxin-type ferredoxin. Until now, almost all ROSs have been assigned to class IIB or III. As for class IIA ROSs, only a few examples have been reported, such as the pyrazon dioxygenase of an unidentified bacterium (37), the dioxin dioxygenase of S. wittichii RW1 (6), and the dicamba O-demethylase of Pseudomonas maltophilia DI-6 (8, 12). Whereas CARDOKA1-1/2 belongs to class IIA, according to the properties of ferredoxin and ferredoxin reductase, CARDOCA10 is a class III ROS. Considering that the ferredoxins contained in CARDOKA1-1/2 and CARDOCA10 are different from one another, it is likely that the respective oxygenases have different ferredoxin selectivity, although markedly high amino acid sequence homology was observed between the two oxygenases (CarAaI and CarAaII) and CarAaCA10 (Table 2). To confirm this, ferredoxin interchangeability between CARDO-OKA1-1/2 and CARDO-OCA10 should be determined.
Supplementary Material
Acknowledgments
This work was supported by the program Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN) and a grant-in-aid (Hazardous Chemicals) from the Ministry of Agriculture, Forestry, and Fisheries of Japan (HC-06-2325-1).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein databases search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcos, J. C., and M. F. Argus. 1968. Molecular geometry and carcinogenic activity of aromatic compounds. New perspectives. Adv. Cancer Res. 11:305-471. [DOI] [PubMed] [Google Scholar]
- 3.Armengaud, J., J. Gaillard, and K. N. Timmis. 2000. A second [2Fe-2S] ferredoxin from Sphingomonas sp. strain RW1 can function as an electron donor for the dioxin dioxygenase. J. Bacteriol. 182:2238-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armengaud, J., B. Happe, and K. N. Timmis. 1998. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J. Bacteriol. 180:3954-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armengaud, J., and K. N. Timmis. 1997. Molecular characterization of Fdx1, a putidaredoxin-type [2Fe-2S] ferredoxin able to transfer electrons to the dioxin dioxygenase of Sphingomonas sp. RW1. Eur. J. Biochem. 247:833-842. [DOI] [PubMed] [Google Scholar]
- 6.Armengaud, J., and K. N. Timmis. 1998. The reductase RedA2 of the multi-component dioxin dioxygenase system of Sphingomonas sp. RW1 is related to class-I cytochrome P450-type reductase. Eur. J. Biochem. 253:437-444. [DOI] [PubMed] [Google Scholar]
- 7.Batie, C. J., D. P. Ballou, and C. C. Correll. 1991. Phthalate dioxygenase reductase and related flavin-iron-sulfur containing electron transferases, p. 543-556. In F. Muller (ed.), Chemistry and biochemistry of flavoenzymes, vol. 3. CRC Press, Boca Raton, Fla. [Google Scholar]
- 8.Chacraborty, S., M. Behrens, P. L. Herman, A. F. Arendsen, W. R. Hagen, D. L. Carlson, X.-Z. Wang, and D. P. Weeks. 2005. A three-component dicamba O-demethylase from Pseudomonas maltophilia, strain DI-6: purification and characterization. Arch. Biochem. Biophys. 437:20-28. [DOI] [PubMed] [Google Scholar]
- 9.Habe, H., Y. Ashikawa, Y. Saiki, T. Yoshida, H. Nojiri, and T. Omori. 2002. Sphingomonas sp. strain KA1, carrying a carbazole dioxygenase gene homologue, degrades chlorinated dibenzo-p-dioxins in soil. FEMS Microbiol. Lett. 211:43-49. [DOI] [PubMed] [Google Scholar]
- 10.Habe, H., J.-S. Chung, A. Ishida, K. Kasuga, K. Ide, T. Takemura, H. Nojiri, H. Yamane, and T. Omori. 2005. The fluorene catabolic linear plasmid in Terrabacter sp. strain DBF63 carries the β-ketoadipate pathway genes, pcaRHGBDCFIJ, also found in proteobacteria. Microbiology 151:3713-3722. [DOI] [PubMed] [Google Scholar]
- 11.Hauschild, J. E., E. Masai, K. Sugiyama, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1996. Identification of an alternative 2,3-dihydroxybiphenyl 1,2-dioxygenase in Rhodococcus sp. strain RHA1 and cloning of the gene. Appl. Environ. Microbiol. 62:2940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herman, P. L., M. Behrens, S. Chacraborty, B. M. Chrastil, J. Barycki, and D. P. Weeks. 2005. A three-component dicamba O-demethylase from Pseudomonas maltophilia, strain DI-6. J. Biol. Chem. 280:24759-24767. [DOI] [PubMed] [Google Scholar]
- 13.Inoue, K., H. Habe, H. Yamane, T. Omori, and H. Nojiri. 2005. Diversity of carbazole-degrading bacteria having the car gene cluster: isolation of a novel gram-positive carbazole-degrading bacterium. FEMS Microbiol. Lett. 245:145-153. [DOI] [PubMed] [Google Scholar]
- 14.Inoue, K., J. Widada, S. Nakai, T. Endoh, M. Urata, Y. Ashikawa, M. Shintani, Y. Saiki, T. Yoshida, H. Habe, T. Omori, and H. Nojiri. 2004. Divergent structures of carbazole degradative car operons isolated from gram-negative bacteria. Biosci. Biotechnol. Biochem. 68:1467-1480. [DOI] [PubMed] [Google Scholar]
- 15.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 16.Jha, A. M., and M. K. Bharti. 2002. Mutagenic profiles of carbazole in the male germ cells of Swiss albino mice. Mutat. Res. 500:97-101. [DOI] [PubMed] [Google Scholar]
- 17.Kasuga, K., H. Habe, J.-S. Chung, T. Yoshida, H. Nojiri, H. Yamane, and T. Omori. 2001. Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63. Biochem. Biophys. Res. Commun. 283:195-204. [DOI] [PubMed] [Google Scholar]
- 18.Kilbane, J. J., II, A. Daram, J. Abbasian, and K. J. Kayser. 2002. Isolation and characterization of Sphingomonas sp. GTIN11 capable of carbazole metabolism in petroleum. Biochem. Biophys. Res. Commun. 297:242-248. [DOI] [PubMed] [Google Scholar]
- 19.Kitagawa, W., A. Suzuki, T. Hoaki, E. Masai, and M. Fukuda. 2001. Multiplicity of aromatic ring hydroxylation dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1, demonstrated by denaturing gradient gel electrophoresis. Biosci. Biotechnol. Biochem. 65:1907-1911. [DOI] [PubMed] [Google Scholar]
- 20.Koga, H., E. Yamaguchi, L. Matsunaga, H. Aramaki, and T. Horiuchi. 1989. Cloning and nucleotide sequences of NADH-putidaredoxin reductase gene (camA) and putidaredoxin gene (camB) involved in cytochrome P-450cam hydroxylase of Pseudomonas putida. J. Biochem. 106:831-836. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, C. M., J. H. Leveau, A. J. Zehnder, and J. R. van der Meer. 2000. Characterization of a second tfd gene cluster for chlorophenol and chlorocatechol metabolism on plasmid pJP4 in Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 182:1465-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masai, E., K. Sugiyama, N. Iwashita, S. Shimizu, J. E. Hauschild, T. Hatta, K. Kimbara, K. Yano, and M. Fukuda. 1997. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphABC genes in Rhodococcus sp. strain RHA1. Gene 187:141-149. [DOI] [PubMed] [Google Scholar]
- 23.Nagata, Y., K. Miyauchi, and M. Takagi. 1999. Complete analysis of genes and enzymes for gamma-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. J. Ind. Microbiol. Biotechnol. 23:380-390. [DOI] [PubMed] [Google Scholar]
- 24.Nagy, I., G. Schoofs, F. Compernolle, P. Proost, J. Vanderleyden, and R. de Mot. 1995. Degradation of the thiocarbamate herbicide EPTC (S-ethyl dipropylcarbamothiolate) and biosafening by Rhodococcus sp. strain NI86/21 involve an inducible cytochrome P-450 system and aldehyde dehydrogenase. J. Bacteriol. 177:676-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nam, J.-W., H. Nojiri, H. Noguchi, H. Uchimura, T. Yoshida, H. Habe, H. Yamane, and T. Omori. 2002. Purification and characterization of carbazole 1,9a-dioxygenase, a three-component dioxygenase system of Pseudomonas resinovorans strain CA10. Appl. Environ. Microbiol. 68:5882-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nestler, F. H. M. 1974. Characterization of wood-preserving coal-tar creosote by gas-liquid chromatography. Anal. Chem. 46:46-53. [Google Scholar]
- 27.Nojiri, H., J.-W. Nam, M. Kosaka, K. Morii, T. Takemura, K. Furihata, H. Yamane, and T. Omori. 1999. Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase from Pseudomonas sp. strain CA10. J. Bacteriol. 181:3105-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nojiri, H., and T. Omori. 2002. Molecular bases of aerobic bacterial degradation of dioxins: involvement of angular dioxygenation. Biosci. Biotechnol. Biochem. 66:2001-2016. [DOI] [PubMed] [Google Scholar]
- 29.Nojiri, H., H. Sekiguchi, K. Maeda, M. Urata, S. Nakai, T. Yoshida, H. Habe, and T. Omori. 2001. Genetic characterization and evolutionary implications of a car gene cluster in the carbazole degrader Pseudomonas sp. strain CA10. J. Bacteriol. 183:3663-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osborne, D. J., R. W. Pickup, and P. A. Williams. 1988. The presence of two complete homologous meta pathway operons on TOL plasmid pWW53. J. Gen. Microbiol. 134:2965-2975. [DOI] [PubMed] [Google Scholar]
- 31.Parales, R. E. 2003. The role of active-site residues in naphthalene dioxygenase. J. Ind. Microbiol. Biotechnol. 30:271-278. [DOI] [PubMed] [Google Scholar]
- 32.Peterson, J. A., M. C. Lorence, and B. Amarneh. 1990. Putidaredoxin reductase and putidaredoxin. Cloning, sequence determination and heterologous expression of the proteins. J. Biol. Chem. 265:6066-6073. [PubMed] [Google Scholar]
- 33.Pinyakong, O., H. Habe, T. Yoshida, H. Nojiri, and T. Omori. 2003. Identification of three novel salicylate 1-hydroxylases involved in the phenanthrene degradation of Sphingobium sp. strain P2. Biochem. Biophys. Res. Commun. 301:350-357. [DOI] [PubMed] [Google Scholar]
- 34.Romine, M. F., L. C. Stillwell, K. K. Wong, S. J. Thurston, E. C. Sisk, C. Sensen, T. Gaasterland, J. K. Fredrickson, and J. D. Saffer. 1999. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J. Bacteriol. 181:1585-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sato, S., J.-W. Nam, K. Kasuga, H. Nojiri, H. Yamane, and T. Omori. 1997. Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA10. J. Bacteriol. 179:4850-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauber, K., C. Fröhner, G. Rosenberg, J. Eberspächer, and F. Lingens. 1977. Purification and properties of pyrazon dioxygenase from pyrazon-degrading bacteria. Eur. J. Biochem. 74:89-97. [DOI] [PubMed] [Google Scholar]
- 38.Sevrioukova, I. F., C. Garcia, H. Li, B. Bhaskar, and T. L. Poulos. 2003. Crystal structure of putidaredoxin, the [2Fe-2S] component of the P450cam monooxygenase system from Pseudomonas putida. J. Mol. Biol. 333:377-392. [DOI] [PubMed] [Google Scholar]
- 39.Takagi, T., H. Nojiri, T. Yoshida, H. Habe, and T. Omori. 2002. Detailed comparison between the substrate specificities of two angular dioxygenases, dibenzofuran 4,4a-dioxygenase from Terrabacter sp. and carbazole 1,9a-dioxygenase from Pseudomonas resinovorans. Biotechnol. Lett. 24:2099-2106. [Google Scholar]
- 40.Tan, H. M., H. Y. Tang, C. L. Joanou, N. H. Abdel-Wahab, and J. R. Mason. 1993. The Pseudomonas putida ML2 plasmid-encoded genes for benzene dioxygenase are unusual in codon usage and low G+C content. Gene 130:33-39. [DOI] [PubMed] [Google Scholar]
- 41.Vedler, E., M. Vahter, and A. Heinaru. 2004. The complete sequenced plasmid pEST4011 contains a novel IncP1 backbone and a catabolic transposon harboring tfd genes for 2,4-dichlorophenoxyacetic acid degradation. J. Bacteriol. 186:7161-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada, A., H. Kishi, K. Sugiyama, T. Hatta, K. Nakamura, E. Masai, and M. Fukuda. 1998. Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 64:2006-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.