Abstract
Neoculin (NCL), a protein with sweetness approximately 500-fold that of sugar, can be utilized as a nonglycemic sweetener. It also has taste-modifying activity to convert sourness to sweetness. NCL is a heterodimer composed of an N-glycosylated acidic subunit (NAS) and a basic subunit (NBS), which are conjugated by disulfide bonds. For the production of recombinant NCL (rNCL) by Aspergillus oryzae, α-amylase with a KEX2 cleavage site, -K-R-, was fused upstream of each of NAS and NBS and the resulting fusion proteins were simultaneously expressed. For accurate and efficient cleavage of the fusion construct by KEX2-like protease, a triglycine motif was inserted after the KEX2 cleavage site. As NBS showed lower production efficiency than did NAS, a larger amount of the NBS expression plasmid than of NAS expression plasmid was introduced during cotransformation, resulting in successful production of rNCL in the culture medium. Moreover, to obtain a higher production yield of rNCL, the active form of hacA cDNA encoding a transcription factor that induces an unfolded protein response was cloned and expressed constitutively. This resulted in a 1.5-fold increase in the level of rNCL production (2.0 mg/liter). rNCL was purified by chromatography, and its NAS was found to be N-glycosylated as expected. The original sweetness and taste-modifying activity of rNCL were comparable to those of native NCL when confirmed by calcium imaging with human embryonic kidney cells expressing the human sweet taste receptor and by sensory tests.
Neoculin (NCL) is a sweet protein present in the fruits of Curculigo latifolia, a tropical plant that is native to West Malaysia. In our previous study, we characterized NCL as a heterodimeric glycoprotein composed of an N-glycosylated acidic subunit (NAS) of ca. 13 kDa and a basic subunit (NBS) of ca. 11 kDa conjugated by disulfide bonds (25). NCL per se is approximately 500 times sweeter than sugar by weight (11, 33). NCL is unique in that it has a taste-modifying activity to convert sourness to sweetness (25, 34). The molecular mechanism of the taste-modifying activity is unclear, and thus it is an intriguing topic in research on gustation. The use of NCL is expected to find practical application as a low-calorie sweetener, which is useful for people who are affected by diseases linked to the consumption of carbohydrates, such as obesity and diabetes. In addition, its unique taste-modifying activity may be valuable for the development of novel types of functional food in the food industry. However, the harvest of C. latifolia fruits is limited due to geographical and technical problems, so it is beneficial to produce recombinant NCL (rNCL) in large quantities.
Aspergillus oryzae, a filamentous fungus, has long been utilized in Japan for the production of fermented foods, such as sake, miso, and soy sauce. Due to its long history of use in the food industry, its approval level is “generally recognized as safe” by the U.S. Food and Drug Administration (3). In recent years, because of the availability of genetic engineering techniques and the determination of its genome sequence, this organism has attracted a great deal of attention as a host for the production of heterologous proteins (10, 14). In heterologous protein production by filamentous fungi, target proteins are expressed in fusion constructs with carrier proteins, such as cellobiohydrolase of Trichoderma reesei and glucoamylase of Aspergillus niger and A. oryzae (1, 6). It is possible to release the target protein from the fusion construct by inserting a KEX2 cleavage sequence, -K-R-, between the carrier and the target protein (6). In addition, the forced expression of chaperones and foldases and constitutive activation of the unfolded protein response (UPR) pathway increase the production yield of heterologous proteins by A. niger (13, 19, 22, 29).
A bacterial expression system for NCL was reported previously (27). In this system, however, the heterodimeric subunits (NAS and NBS) were expressed in independent cells, resulting in intracellular production in the form of inclusion bodies without glycosylation. Moreover, a laborious reconstitution step is required for NAS and NBS to form heterodimers with sensory activity. In contrast, the use of A. oryzae is beneficial because it has the ability to secrete heterologous proteins with complicated structures formed via posttranslational modifications, including disulfide cross-linking and glycosylation (4). To date, many examples of heterologous protein production by filamentous fungi have been reported (1, 7, 22, 24). However, with the exception of humanized antibodies (30), there have been few reports regarding the production of hetero-oligomeric proteins by filamentous fungi.
Here we report the extracellular production of rNCL with sensory activities by A. oryzae. We designed an efficient strategy to express the two heterodimeric subunits with different productivities and increased rNCL production level by constitutive induction of the UPR. Moreover, secreted rNCL conferred the original sweetness and taste-modifying activity comparable to those of native NCL. This study demonstrated that the A. oryzae production system is useful for the production of hetero-oligomeric proteins as well as for investigation of the molecular biology of this unique taste-modifying protein.
MATERIALS AND METHODS
Microbial strains.
A. oryzae NS4 strain (niaD and sC anxotroph) (32) was used as a host in expression studies. The A. oryzae strain RIB40 was used to isolate the active form of hacA cDNA. Escherichia coli DH5α was used for plasmid amplification.
Construction of rNCL expression plasmids.
All rNCL expression plasmids were constructed using a multisite gateway three-fragment vector construction kit (Invitrogen, San Diego, CA). For details, see the supplemental material.
Isolation of the active form of hacA cDNA and construction of its expression plasmid.
A gene homologous to the A. niger hacA gene was found in the A. oryzae genome sequence database (14). We isolated the active form of the hacA cDNA clone from the A. oryzae RIB40 strain under endoplasmic reticulum (ER) stress conditions. See the supplemental material for the deduced amino acid sequence of hacA and experimental procedures.
Transformation of A. oryzae.
Transformation of A. oryzae was carried out as described previously by Kitamoto (10). In cotransformation experiments, we changed the weight ratio of the NAS and NBS expression plasmids for the production of the two subunits in an equimolar ratio into the NS4 strain. The plasmid for the expression of the active form of hacA cDNA was introduced into the rNCL-producing strain.
Culture conditions of A. oryzae.
Small-scale culture was carried out to screen transformants producing rNCL. The transformants were grown in 20 ml of DPY medium (2% dextrin, 1% polypeptone, 0.5% yeast extract, 0.5% K2HPO4, 0.05% MgSO4 · 7H2O, pH 8.0) in 100-ml flasks incubated at 30°C and shaken at 150 rpm for 72 h. Ten transformants for each construct were examined to confirm the secretion of rNCL into the medium. For large-scale production, the strain producing rNCL was grown in 150 ml of DPY medium in 500-ml flasks with shaking. Conidia (1.5 × 106) were inoculated into each flask and incubated at 30°C with shaking at 150 rpm for 72 h. Medium samples were obtained by filtering the fungal cultures through Miracloth (Cal Biochem, La Jolla, CA).
Purification of rNCL.
The proteins were precipitated by the addition of 60% saturated (NH4)2SO4 to the culture medium. The resulting precipitate was dissolved in buffer A (3 M NaCl in 20 mM sodium acetate buffer, pH 5.0) and dialyzed against the same buffer. The dialysate was centrifuged at 20,000 × g for 30 min, filtered through a 0.22-μm filter, and chromatographed on an HIC PH-814 (Showa Denko K.K., Kawasaki, Japan) with a linear gradient of 3 to 0 M NaCl in buffer A at a flow rate of 3.0 ml/min using a Waters 600 high-pressure liquid chromatography system (Waters, Milford, MA). Fractions showing immunoreactivity with anti-NCL antibodies were pooled, dialyzed against H2O, and lyophilized. The lyophilized protein was dissolved in buffer B (50 mM sodium acetate buffer, pH 5.0) and desalted with a PD10 column (Amersham Biosciences, Piscataway, NJ). The eluate was loaded onto a Mono S cation-exchange column (Amersham Biosciences) and eluted with a linear gradient of 0 to 1 M NaCl in buffer B at a flow rate of 1.0 ml/min. Peak fractions were analyzed by immunoblotting, and the resulting positive fractions were collected, pooled, and used as a purified rNCL sample.
Purification of native NCL.
Purification of the native NCL from the fruits of C. latifolia was carried out as described previously (25). Briefly, C. latifolia fruits were lyophilized and treated with 0.05 N H2SO4 to extract the active fraction. The extract was treated with Amberlite IRC-50 (Organo, Tokyo, Japan), and the eluate was added to 60% saturated (NH4)2SO4. The resulting precipitate was desalted on a Sephadex G-25 column (Amersham Biosciences) and lyophilized to obtain native NCL.
Preparation of anti-NBS antibody.
A peptide corresponding to the C-terminal 16 residues of NBS, V122-L-Y-S-L-G-P-N-G-C-R-R-V-N-G-G137, was synthesized, and antisera were prepared by immunizing rabbits with the hemocyanin-conjugated peptide. The anti-NBS antibodies were purified by epoxy-activated Sepharose 6B (Amersham Biosciences) conjugated with the antigen peptide.
Protein characterization.
For immunoblotting, rabbit anti-NCL antibodies (kindly provided by H. Yamashita) and anti-NBS antibodies were used for the detection of rNCL. For N-terminal amino acid sequencing, protein samples transferred onto polyvinylidene difluoride membranes were analyzed by using the Procise 491 cLC protein sequencing system (Applied Biosystems, Foster City, CA). For the detection of glycoproteins, rNCL and native NCL were loaded onto sodium dodecyl sulfate (SDS)-polyacrylamide gels and then analyzed by glycoprotein staining with a ProQ-Emerald 300 glycoprotein gel stain kit (Invitrogen). Monosaccharide composition was analyzed using the ABEE labeling kit Plus S (Honen, Tokyo, Japan).
Ca2+ imaging analysis using cultured cells to detect their sweet-taste responses.
HEK293T cells were transfected with hT1R2, hT1R3, and G16/gust25 (35), which were subcloned into the pEAK10 expression vector (Edge Biosystems, Gaithersburg, MD) using Lipofectamine 2000 reagent (Invitrogen). Ca2+ imaging analysis was performed essentially as described by Ueda et al. (28). Cells were transferred onto glass coverslips approximately 24 h after transfection. After a further 24 h, cells were loaded with 10 μM of the calcium indicator dye fura-2/AM (Invitrogen) for 25 min at room temperature. The cells were washed with assay buffer (130 mM NaCl, 10 mM glucose, 5 mM KCl, 2 mM CaCl2, and 1.2 mM MgCl2 in 10 mM HEPES, pH 7.4) and subjected to Ca2+ imaging analysis. All sweet tastants were diluted with the assay buffer at the following concentrations: rNCL, 20 μM; saccharin, 10 mM; aspartame, 10 mM; and native NCL, 20 μM. As a negative control, monosodium glutamate (MSG), an umami tastant, was used at 10 mM. These tastants were applied sequentially to the cells over a period of 30 s under gravity at a flow rate of 8 ml/min. An interval of more than 3 min was taken between the application of the first sweetener and that of the second to avoid desensitizing the cells. The fura-2 fluorescence intensities resulting from excitation at 340 and 380 nm were measured at 510 nm using a computer-controlled filter changer (Lambda 10-2; Sutter, San Rafael, CA), a MicroMax cooled charge-coupled device camera (Princeton Instruments, Trenton, NJ), and an inverted fluorescence microscope (IX-70; Olympus, Tokyo, Japan). Images were recorded at 4-s intervals and analyzed using MetaFluor software (Molecular Devices, Sunnyvale, CA). Changes in intracellular free calcium ion concentration ([Ca2+]i) were determined as changes in the ratio of fluorescence emitted at the two excitation wavelengths.
Sensory evaluation of the taste-modifying activity of rNCL.
The original sweetness of rNCL and its taste-modifying activity were evaluated by three panel members who had been well trained to accurately describe the intensities of sweetness. For further accuracy, they fasted for at least 30 min prior to the sensory test (25). Each panel member tasted 100 μl of the NCL solution and was asked to describe its intensity of sweetness. As the sense of original sweetness disappeared within 30 s, they were successively given 300 μl of 100 mM citrate buffer (pH 4.0) to evaluate its sweetness intensity, which was defined as the taste-modifying activity of neoculin. Sweetness scores were determined as follows: 5, very strong; 4, strong; 3, moderate; 2, weak; and 1, not sweet.
RESULTS
Expression of the two subunits of rNCL as separate constructs.
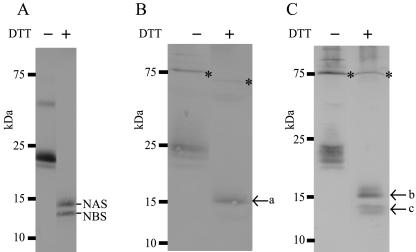
NCL is a heterodimer of NAS-NBS held together by disulfide bonds (25). On electrophoresis without dithiothreitol (DTT) treatment, the NAS-NBS heterodimer is detected as a 22-kDa band, while the two subunits (the 14-kDa band of NAS and 12-kDa band of NBS) are dissociated under reducing conditions (Fig. 1A). In the present study, we took the balanced expression of both subunits into consideration. We used α-amylase (AmyB) as a carrier protein to fuse rNCL because it is one of the most abundantly secreted proteins in A. oryzae. A KEX2 cleavage site (-K-R-) was inserted at the N terminus of each subunit to allow release of the recombinant proteins from fusion constructs (Table 1). We chose the strategy to simultaneously express the two subunits (NAS and NBS) as independent fusion constructs with AmyB. Two expression plasmids, pAAsC and pAB, were generated and cotransformed into the NS4 strain (Table 1). The A. nidulans sC gene was included in the NAS expression plasmid as a selectable marker but was not included in the NBS expression plasmid (Table 1, pAAsC and pAB rows, respectively). By immunological analysis using anti-NCL antibody, we detected a protein of approximately 15 kDa in the culture media of 10 of 12 transformants, which migrated as a 24-kDa band under nonreducing conditions, similar to the native NCL (Fig. 1B, arrow a). However, the N-terminal sequence of the 15-kDa band (NH2-A-G-S-K-R-D-S- [the N-terminal sequence of NAS is underlined]) indicated that the processing of the fusion protein occurred at an incorrect position upstream of the KEX2 cleavage site (-K-K↓A-G-S-K-R-D-S-). The protein purified from the 24-kDa band under nonreducing conditions was found to consist of only NAS and did not show any original sweetness or taste-modifying activity (data not shown). Moreover, bands of AmyB-NCL fusion proteins were detected at 75 kDa under nonreducing conditions and at 70 kDa under reducing conditions (Fig. 1B). While similar results were observed using the pAAsC-transformed transformants expressing the NAS-NAS homodimer and the AmyB-NAS fusion protein (Table 1), the transformation of the NBS expression plasmid containing the A. nidulans sC marker did not yield any products (data not shown), suggesting lower stability of NBS than NAS. It was apparent that it was necessary to improve the efficiency and accuracy of the KEX2-like protease cleavage and to increase the productivity of NBS.
FIG. 1.
Immunoblotting analysis of rNCL produced by A. oryzae transformants. Culture media of the transformants indicated below were subjected to 15% SDS-PAGE in the presence or absence of 50 mM DTT. (A) The native NCL protein. (B) The strain cotransformed with the separate NAS and NBS expression plasmids (Table 1, pAAsC and pAB rows). (C) The strain cotransformed with the modified separate NAS and NBS expression plasmids (Table 1, pA3GAsC and pA3GB rows). The NCL subunits released from the fusion protein are indicated by arrows a to c. Asterisks indicate the AmyB-NCL fusion protein.
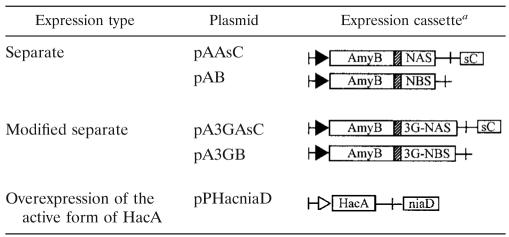
TABLE 1.
Expression plasmids used in this study
Filled arrows, amyB promoter nt 9-606 (Hara et al., 1992 [8a]); open arrow, pgkA promoter nt 7-1623 (D28484); AmyB, amyB open reading frame nt 607-2647 (Hara et al., 1992 [8a]); NAS, NAS nt 67-405 (AB167079); NBS, NBS nt 77-421 (X64110); 3G-NAS, triglycine motif attached to NAS; 3G-NBS, triglycine motif attached to NBS; HacA, active form of hacA cDNA nt 1-1038 (AB246349); hatched rectangles, KEX2 cleavage site (KR) in linker; crosses, amyB terminator nt 2044-2595 (Hara et al., 1992 [8a]); sC, A. nidulans sC gene nt 1-3627 (X82541); niaD, A. oryzae niaD gene nt 1-5140 (D49701).
Successful production of rNCL by inclusion of a triglycine motif following the KEX2 cleavage site and with the optimized weight ratio of expression plasmids during cotransformation.
The addition of a triglycine motif to the C terminus of the KEX2 cleavage site has been reported to improve cleavage accuracy and efficiency in A. niger (26, 30). Therefore, we constructed modified expression plasmids with insertion of this motif into the NH2 termini of NAS and NBS (Table 1, pA3GAsC and pA3GB rows, respectively). As it was suggested that NBS showed lower stability, we introduced NAS and NBS expression plasmids in various weight ratios during cotransformation. In 2 of 12 strains obtained by cotransformation of 2 μg of pA3GAsC and 10 μg of pA3GAB, the two bands of approximately 15 and 13 kDa that were seen under reducing conditions were detected in the culture medium by immunological analysis (Fig. 1C, arrows b and c, respectively). The band shown in Fig. 1C, arrow c, is a doublet. This may be due to processing and glycosylation. The two bands were approximately 24 kDa under nonreducing conditions, which suggested that these strains secreted NAS-NBS heterodimers conjugated by disulfide bonds into the culture medium. With other weight ratios of the two expression plasmids during cotransformation (e.g., 2 μg of pA3GAsC and 2 μg of pA3GAB), only NAS-NAS homodimers were produced and NBS was not detected (data not shown). After purification (see below), the N-terminal sequence of the 15-kDa protein was determined as NH2-G-G-G-D-S-V-L-L-S- (the N terminus of NAS is underlined) and that of the 13-kDa protein was determined as NH2-G-G-G-D-N-V-L-L-S- (the N terminus of NBS is underlined), confirming that both of the subunits (NAS and NBS) were produced successfully in the culture medium. Furthermore, this result demonstrated that the appropriate cleavages occurred between the KEX2 cleavage site and the triglycine motif (-K-K-A-G-S-K-R↓G-G-G-D-S- and -K-K-A-G-S-K-R↓G-G-G-D-N-) in both the AmyB-NAS and AmyB-NBS fusion proteins, respectively. Neverthless, AmyB-NCL fusion proteins were observed at a molecular weight of approximately 75 kDa (Fig. 1C). The transformant producing the highest level of rNCL was designated as AB2-2.
Improvement of rNCL production level by overexpression of the active form of hacA cDNA.
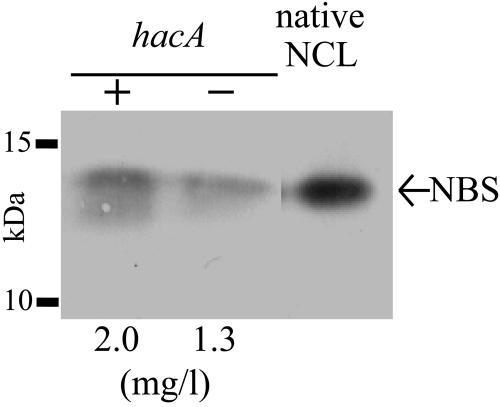
The HacA protein of A. niger is a transcription factor that induces the expression of chaperones and foldases in response to ER stress (20). Under conditions of ER stress, the unconventional 20-nucleotide (nt) intron in the hacA mRNA is spliced out, resulting in the translation of an active transcription factor (20). Although forced expression of the active form of hacA cDNA increases the production yield of heterologous proteins by A. niger (29), the effects of constitutive activation of UPR on the production of hetero-oligomeric heterologous proteins have not been reported. To improve the rNCL production level, a plasmid expressing the active form of the hacA cDNA under the control of the pgkA promoter (for constitutive expression) was prepared and introduced into the AB2-2 strain. The A. oryzae niaD gene (18) was used as a selectable marker. Immunological analysis using anti-NBS antibody was carried out with the culture medium of the transformant. We compared eight different transformants of the hacA-introduced AB2-2 strain and the control and found that the average amount of rNCL produced in the former case was 1.5 times higher than that in the control (Fig. 2). This indicated that the constitutive expression of the active form of hacA cDNA enhanced the level of production of a heterodimeric heterologous protein, rNCL.
FIG. 2.
Improvement of the level of rNCL production by overexpression of the active form of hacA cDNA. Immunological analysis with anti-NBS antibody was carried out using the culture media of the strain overexpressing the active form of hacA cDNA (+) and the niaD marker-transformed strain (−). The amounts of rNCL produced in the culture medium were calculated by measuring the intensities of the bands and comparing with those of native NBS. The average value of the rNCL production level is shown below the panel.
Purification and characterization of rNCL.
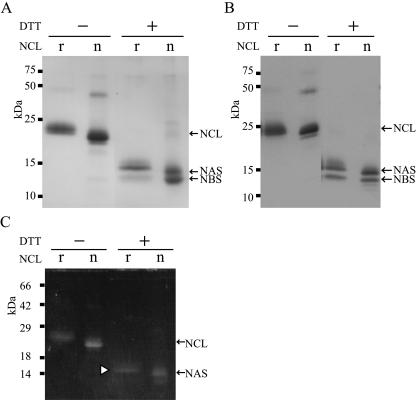
We next purified rNCL from the culture supernatant of the AB2-2 strain as described in Materials and Methods. Purification was performed by ammonium sulfate precipitation and phenyl-hydrophobic interaction and ion-exchange chromatography. The purified rNCL was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and analyzed by Coomassie brilliant blue (CBB) staining. Under nonreducing conditions, rNCL was observed as a band of approximately 24 kDa near the 22-kDa band of native NCL (Fig. 3A). Under reducing conditions, two bands appeared at approximately 15 and 13 kDa, which were almost in accordance with the bands of native NAS (14 kDa) and NBS (12 kDa), respectively. The difference in mobility on SDS-PAGE is due to the addition of GGG at the N terminus and also to the heterogeneity of the sugar chains added. Although the band of the recombinant NBS showed less staining than that of NAS by CBB (Fig. 3A), the intensities of the two bands under immunoblotting analysis were similar to those of the respective native proteins (Fig. 3B). These results suggested that the recombinant NAS and NBS were produced in an almost equimolar ratio. Next, we performed glycoprotein staining for rNCL, and the results indicated that both rNCL and native NCL responded positively under nonreducing conditions (Fig. 3C). Under reducing conditions, a positive band appeared at the position of NAS in both recombinant (Fig. 3C) and native NCL. Monosaccharide composition analysis of rNCL demonstrated the presence of mannose, N-acetylglucosamine, and galactose. The range of electrophoretic mobility of the recombinant NAS was within approximately 2 kDa, probably due to the heterogeneity of N-glycans in size (data not shown). It was inferred that the N-glycan of the recombinant NAS was of the high-mannose type, which is known to be common in aspergilli (2).
FIG. 3.
Purification and characterization of rNCL produced by A. oryzae. (A) SDS-PAGE analysis. The purified rNCL (see Materials and Methods) and native NCL (8 μg of each protein) were subjected to 15% SDS-PAGE in the presence or absence of 50 mM DTT and detected by CBB staining. (B) Immunoblotting analysis. Proteins (100 ng of the purified rNCL and native NCL) were transferred onto polyvinylidene difluoride membranes and probed with anti-NCL antibody. (C) Glycoprotein staining. The purified rNCL and native NCL (8 μg of each) were subjected to 15% SDS-PAGE in the presence or absence of 50 mM DTT. Glycoproteins were detected using a ProQ Emerald glycoprotein stain kit (Invitrogen). The white arrowhead indicates recombinant NAS. Lanes r, purified rNCL; lanes n, native NCL.
Evaluation of the sweetness of rNCL.
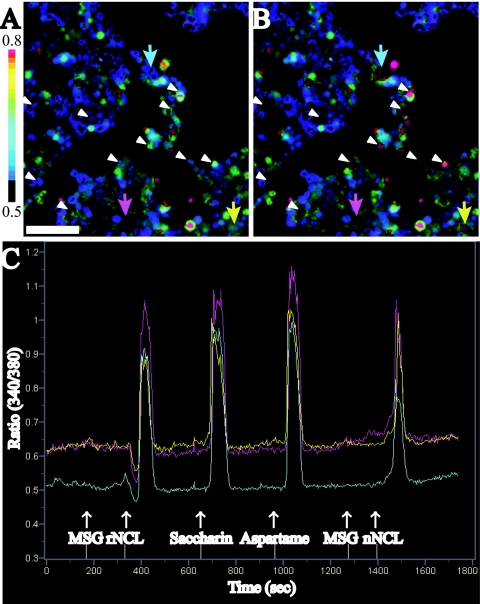
To evaluate the sweetness of the recombinant product, we examined the responses of the human sweet taste receptor to rNCL by calcium imaging analysis. T1R2 and T1R3, both of which belong to the G protein-coupled receptor family, were reported to associate with each other and to function together as a sweet taste receptor (5, 12, 21, 35). Human T1R2/T1R3 responds to low-molecular-weight sweeteners (sucrose, saccharin, several d-amino acids, and sweet peptide aspartame) and sweet proteins (thaumatin, monellin, and brazzein) (9, 12, 21, 31, 35). First, cDNAs for both hT1R2 and hT1R3 were transiently introduced into cultured HEK293T cells together with the chimeric Gα protein, G16/gust25, a promiscuous phospholipase C-linked G protein (28). The receptor activation of hT1R2/T1R3 can be detected by using fura-2/AM calcium indicator dye, which allows monitoring of the intracellular calcium increase caused by phospholipase C activation (9, 21, 35). An increase in calcium in response to rNCL was detected in HEK293T cells expressing hT1R2/T1R3 (Fig. 4A and B). These cells also responded to other sweet substances, such as saccharin, aspartame, and native NCL (Fig. 4C). Conversely, 88% (107/122 cells) and 97% (96/99 cells) of cells that responded to aspartame and to saccharin also responded to rNCL, respectively. These results indicated that the frequencies of cell responses to rNCL were similar to those to the other sweeteners, saccharin and aspartame. Furthermore, the intensities of cell responses to 20 μM rNCL were the same as those of an equal concentration of the native NCL (Fig. 4C). These results indicated that rNCL conferred the original sweetness at the same level as native NCL did.
FIG. 4.
Sweet-tasting effect assay. (A and B) Sample images of HEK293T cells expressing hT1R2/T1R3 before (A) and after (B) addition of rNCL (20 μM). The color scale indicates the F340/F380 ratio, where F is fluorescence intensity. White arrowheads and colored arrows show NCL-responding cells (scale bar, 100 μm). (C) Line traces show the ratiometric value changes for representative cells. White arrows indicate the time of application of 20 μM NCL, 10 mM saccharin, 10 mM aspartame, 20 μM native NCL (nNCL), and 10 mM MSG (negative control). Each trace was derived from cells indicated by colored arrows (A and B). Note that MSG, an umami tastant (12), did not trigger any cell responses.
Taste-modifying activity of rNCL compared to that of native NCL.
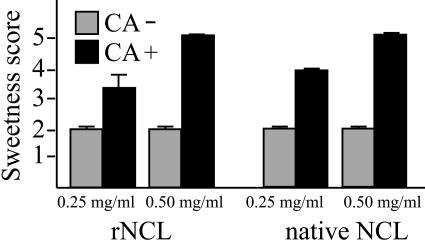
NCL per se has sweetness but this taste disappears in a short time (referred to as “original sweetness”) (25, 33). When an acid solution is placed on the tongue shortly after the disappearance of the original sweetness of NCL, the sourness of the acid is restrained and a stronger sweetness is elicited (25, 33). This taste-modifying activity of rNCL was evaluated by sensory tests. The activity was represented by sweetness intensities (5 ranks) determined by a panel who tasted 100 mM citrate buffer (pH 4.0) shortly after the disappearance of the original sweetness of NCL. While the average score of the original sweetness in 0.25 mg/ml rNCL was 2.0, stronger sweetness (score, 3.5) was elicited when citrate buffer was added after the disappearance of the original sweetness of rNCL (Fig. 5; left). Likewise, when 0.5 mg/ml rNCL was used, stronger sweetness (score, 5.0) was observed in the presence of the acid than the original sweetness (score 2.0). Similar scores were obtained when the native NCL was evaluated at the same concentrations as rNCL (Fig. 5, right). These results indicated that the taste-modifying activity of rNCL was comparable to that of native NCL.
FIG. 5.
Taste-modifying activity assay. Gray columns show intensities of the original sweetness of rNCL (left) and native NCL (right). Black columns show the sweetness intensities when added with 100 mM citrate buffer (pH 4.0) (CA) shortly after the disappearance of the original sweetness of NCL. The sweetness scores are shown as means ± standard deviations (error bars) (n = 3). (Sweetness scores were determined as follows: 5, very strong; 4, strong; 3, moderate; 2, weak; and 1, not sweet).
DISCUSSION
In the present study, we succeeded in obtaining extracellular production of a heterodimeric protein, NCL, by A. oryzae (Fig. 1C). Moreover, the purified rNCL showed an original sweetness and taste-modifying activity equivalent to those of the native NCL (Fig. 4 and 5).
Although humanized antibodies consisting of heavy and light chains conjugated with disulfide bonds were produced by A. niger (30), no other examples of production of such hetero-oligomeric proteins by filamentous fungi have been reported. As NCL is a heterodimer composed of two subunits (NAS and NBS) linked to each other via disulfide bonds, it was important to establish a production system capable of expressing the two subunits in an equimolar ratio with the correct dimeric conformation. Initially, we attempted to produce rNCL as tandem constructs of the two subunits (NAS-NBS and NBS-NAS) but failed in all cases (data not shown). Recently, our group has completed X-ray crystallographic analysis of native NCL, and the results demonstrated that NAS and NBS are conformationally in a parallel orientation (A. Shimizu-Ibuka, Y. Morita, T. Terada, T. Asakura, K. Nakajima, S. Iwata, T. Misaka, H. Sorimachi, S. Arai and K. Abe, submitted). Therefore, the joining of NAS and NBS in tandem may have caused an improper conformation, which may have resulted in degradation by ER quality control. We then tried cotransformation of NAS and NBS. In this case, we first investigated whether the two transformants introducing the NAS or NBS plasmid independently express equimolar amounts of NAS and NBS proteins. However, it resulted that only NAS protein was efficiently produced, and there was no expression of NBS protein. This may be due to the fact that, although NAS and NBS share 92% identity at the nucleotide sequence level (25), either or both the genome integration efficiency and the mRNA stability are different between the two genes. Also, some differences in stabilities between the NAS and NBS proteins can be considered as a reason for the observed differences in their expression efficiencies. This may be because it acts as a good substrate of KEX2-like protease, as it contains two KEX2 recognition sequences, -R69-R70- and R132-R133-. Another reason may be that NAS attaches an N-linked glycan (-N103-G104-T105-), which is likely preferable for secretion (4), while NBS lacks N-glycosylation sites. It is thus suggested that NBS can be secreted only when it forms a heterodimer with NAS with the attachment of an N-linked glycan.
It is necessary to take into consideration the amounts of expression plasmids introduced during cotransformation. There have been no previous reports of changing the weight ratio between two expression plasmids in cotransformation in an attempt to achieve the production of heterologous protein by filamentous fungi. In the present study, we succeeded in the production of rNCL only when a larger amount of the NBS expression plasmid than that of the NAS expression plasmid was introduced during cotransformation. The copy numbers of the introduced genes were investigated by PCR using NAS- and NBS-specific primers. We carried out transformation with the NAS and NBS expression plasmids at 1:1 (molar ratio) and with them at 1:5. The NAS band intensities were almost similar in both cases, while in the latter a much stronger NBS band intensity resulted, suggesting that the latter (1:5) had higher copy numbers of NBS than that of the former (1:1) (data not shown). Thus, our success in efficiently producing the NAS-NBS heterodimer is probably due to the fact that the sufficient integration of the NBS plasmid has no selectable marker. Increasing the NBS copy number can lead to enhanced expression of the NBS protein.
In heterologous protein production by filamentous fungi, the target protein is released from the fusion construct by the insertion of a KEX2 cleavage site (6). In the present study, the AmyB-NAS fusion protein was processed incorrectly at the sequence upstream of the KEX2 cleavage site (see Results). Other groups reported that aberrant processing occurred around the expected cleavage site when glucoamylase was used as a carrier (7, 8, 15-17, 23), which suggests that the use of α-amylase as a carrier did not cause the inaccurate cleavage. The insertion of a triglycine motif at the C terminus of the KEX2 cleavage site led to correct processing after the KEX2 cleavage site (see Results), which was consistent with the results reported previously (26, 30). Nevertheless, AmyB-NCL fusion proteins were observed at a molecular weight of approximately 75 kDa (Fig. 1C). Low efficiency of processing at the KEX2 cleavage site was also observed in the case of the production of humanized antibodies (30). In the production of human lysozyme by A. oryzae, however, the target protein was released completely from the α-amylase-fused construct without insertion of a triglycine motif (data not shown), suggesting that low cleavage efficiency at the KEX2 cleavage site was not due to the use of α-amylase as a carrier. To our knowledge, in the case of production of heterologous proteins other than humanized antibodies (30) using glucoamylase as a carrier, the processing of the fusion construct occurred efficiently with insertion of the KEX2 cleavage site (7, 8, 16, 17, 23). Taking these results together, we hypothesized that the low processing efficiency at the KEX2 cleavage site was caused by steric hindrance of oligomeric proteins, such as NCL and humanized antibodies, with complex structures, regardless of the use of α-amylase or glucoamylase as a carrier.
Finally, we performed a sweetness assay by using HEK cells transiently expressing human sweet taste receptors, and the results indicated that the cell response to rNCL produced by A. oryzae was comparable to that of the native NCL (Fig. 4). Sensory tests also confirmed that rNCL had taste-modifying activity like that of the native NCL (Fig. 5). Therefore, it was suggested that the overall structure of rNCL was very similar to that of the native NCL, which was verified by circular dichroism spectroscopy (data not shown). Although rNAS has an N-glycan consisting of mannose, N-acetylglucosamine, and galactose (clearly different from that of native NCL, which consisted of mannose, N-acetylglucosamine, fucose, and xylose [25]), these results indicated that the structural differences in the N-glycan of rNCL did not affect its sensory activities. The rNCL production system used in the present study will facilitate mutation analysis on NCL, which will contribute to the evaluation of the critical regions or amino acid residues involved in taste-modifying activity.
The results of the present study indicated that A. oryzae is a host capable of producing hetero-oligomeric proteins that undergo complex posttranslational modifications. rNCL produced in this study is expected to be applicable for use in both clinical settings and in the food industry because of the utilization of A. oryzae, a safe microorganism, which has long been used in fermentation processes in food.
Supplementary Material
Acknowledgments
We thank Haruyuki Yamashita for valuable comments and for providing the anti-NCL antibody. We are also grateful to Toru Akita for valuable advice and gifts of C. latifolia fruits. We thank Charles S. Zuker and Nicholas J. P. Ryba for providing hT1R2 and hT1R3 cDNAs.
This study was supported by grants-in-aid 15380058 (to K.K.) and 16108004 (to K.A.) from the Ministry of Education, Culture, Sports, Science and Technology in Japan.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Archer, D. B. 2000. Filamentous fungi as microbial cell factories for food use. Curr. Opin. Biotechnol. 11:478-483. [DOI] [PubMed] [Google Scholar]
- 2.Archer, D. B., and J. F. Peberdy. 1997. The molecular biology of secreted enzyme production by fungi. Crit. Rev. Biotechnol. 17:273-306. [DOI] [PubMed] [Google Scholar]
- 3.Barbesgaard, P., H. P. Heldt-Hansen, and B. Diderichsen. 1992. On the safety of Aspergillus oryzae: a review. Appl. Microbiol. Biotechnol. 36:569-572. [DOI] [PubMed] [Google Scholar]
- 4.Conesa, A., P. J. Punt, N. van Luijk, and C. A. van den Hondel. 2001. The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33:155-171. [DOI] [PubMed] [Google Scholar]
- 5.Damak, S., M. Rong, K. Yasumatsu, Z. Kokrashvili, V. Varadarajan, S. Zou, P. Jiang, Y. Ninomiya, and R. F. Margolskee. 2003. Detection of sweet and umami taste in the absence of taste receptor T1r3. Science 301:850-853. [DOI] [PubMed] [Google Scholar]
- 6.Gouka, R. J., P. J. Punt, and C. A. van den Hondel. 1997. Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl. Microbiol. Biotechnol. 47:1-11. [DOI] [PubMed] [Google Scholar]
- 7.Gouka, R. J., P. J. Punt, and C. A. van den Hondel. 1997. Glucoamylase gene fusions alleviate limitations for protein production in Aspergillus awamori at the transcriptional and (post)translational levels. Appl. Environ. Microbiol. 63:488-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gouka, R. J., M. van der Heiden, T. Swarthoff, and C. T. Verrips. 2001. Cloning of a phenol oxidase gene from Acremonium murorum and its expression in Aspergillus awamori. Appl. Environ. Microbiol. 67:2610-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Hara, S., K. Kitamoto, and K. Gomi. 1992. New developments in fermented beverages and foods with Aspergillus. Biotechnology 23:133-153. [PubMed] [Google Scholar]
- 9.Jiang, P., Q. Ji, Z. Liu, L. A. Snyder, L. M. Benard, R. F. Margolskee, and M. Max. 2004. The cysteine-rich region of T1R3 determines responses to intensely sweet proteins. J. Biol. Chem. 279:45068-45075. [DOI] [PubMed] [Google Scholar]
- 10.Kitamoto, K. 2002. Molecular biology of the Koji molds. Adv. Appl. Microbiol. 51:129-153. [DOI] [PubMed] [Google Scholar]
- 11.Kurihara, Y. 1992. Characteristics of antisweet substances, sweet proteins, and sweetness-inducing proteins. Crit. Rev. Food Sci. Nutr. 32:231-252. [DOI] [PubMed] [Google Scholar]
- 12.Li, X., L. Staszewski, H. Xu, K. Durick, M. Zoller, and E. Adler. 2002. Human receptors for sweet and umami taste. Proc. Natl. Acad. Sci. USA 99:4692-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lombrana, M., F. J. Moralejo, R. Pinto, and J. F. Martin. 2004. Modulation of Aspergillus awamori thaumatin secretion by modification of bipA gene expression. Appl. Environ. Microbiol. 70:5145-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machida, M., K. Asai, M. Sano, T. Tanaka, T. Kumagai, G. Terai, K. Kusumoto, T. Arima, O. Akita, Y. Kashiwagi, K. Abe, K. Gomi, H. Horiuchi, K. Kitamoto, T. Kobayashi, M. Takeuchi, D. W. Denning, J. E. Galagan, W. C. Nierman, J. Yu, D. B. Archer, J. W. Bennett, D. Bhatnagar, T. E. Cleveland, N. D. Fedorova, O. Gotoh, H. Horikawa, A. Hosoyama, M. Ichinomiya, R. Igarashi, K. Iwashita, P. R. Juvvadi, M. Kato, Y. Kato, T. Kin, A. Kokubun, H. Maeda, N. Maeyama, J. Maruyama, H. Nagasaki, T. Nakajima, K. Oda, K. Okada, I. Paulsen, K. Sakamoto, T. Sawano, M. Takahashi, K. Takase, Y. Terabayashi, J. R. Wortman, O. Yamada, Y. Yamagata, H. Anazawa, Y. Hata, Y. Koide, T. Komori, Y. Koyama, T. Minetoki, S. Suharnan, A. Tanaka, K. Isono, S. Kuhara, N. Ogasawara, and H. Kikuchi. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157-1161. [DOI] [PubMed] [Google Scholar]
- 15.MacKenzie, D. A., J. A. Kraunsoe, J. A. Chesshyre, G. Lowe, T. Komiyama, R. S. Fuller, and D. B. Archer. 1998. Aberrant processing of wild-type and mutant bovine pancreatic trypsin inhibitor secreted by Aspergillus niger. J. Biotechnol. 63:137-146. [DOI] [PubMed] [Google Scholar]
- 16.Martin, J. A., R. A. Murphy, and R. F. Power. 2003. Cloning and expression of fungal phytases in genetically modified strains of Aspergillus awamori. J. Ind. Microbiol. Biotechnol. 30:568-576. [DOI] [PubMed] [Google Scholar]
- 17.Mikosch, T., P. Klemm, H. G. Gassen, C. A. van den Hondel, and M. Kemme. 1996. Secretion of active human mucus proteinase inhibitor by Aspergillus niger after KEX2-like processing of a glucoamylase-inhibitor fusion protein. J. Biotechnol. 52:97-106. [DOI] [PubMed] [Google Scholar]
- 18.Minetoki, T., Y. Nunokawa, K. Gomi, K. Kitamoto, C. Kumagai, and G. Tamura. 1996. Deletion analysis of promoter elements of the Aspergillus oryzae agdA gene encoding α-glucosidase. Curr. Genet. 30:432-438. [DOI] [PubMed] [Google Scholar]
- 19.Moralejo, F. J., A. J. Watson, D. J. Jeenes, D. B. Archer, and J. F. Martin. 2001. A defined level of protein disulfide isomerase expression is required for optimal secretion of thaumatin by Aspergillus awamori. Mol. Genet. Genomics 266:246-253. [DOI] [PubMed] [Google Scholar]
- 20.Mulder, H. J., M. Saloheimo, M. Penttila, and S. M. Madrid. 2004. The transcription factor HACA mediates the unfolded protein response in Aspergillus niger, and up-regulates its own transcription. Mol. Genet. Genomics 271:130-140. [DOI] [PubMed] [Google Scholar]
- 21.Nelson, G., M. A. Hoon, J. Chandrashekar, Y. Zhang, N. J. Ryba, and C. S. Zuker. 2001. Mammalian sweet taste receptors. Cell 106:381-390. [DOI] [PubMed] [Google Scholar]
- 22.Nevalainen, K. M., V. S. Te'o, and P. L. Bergquist. 2005. Heterologous protein expression in filamentous fungi. Trends Biotechnol. 23:468-474. [DOI] [PubMed] [Google Scholar]
- 23.Punt, P. J., A. Drint-Kuijvenhoven, B. C. Lokman, J. A. Spencer, D. Jeenes, D. A. Archer, and C. A. van den Hondel. 2003. The role of the Aspergillus niger furin-type protease gene in processing of fungal proproteins and fusion proteins. Evidence for alternative processing of recombinant (fusion-) proteins. J. Biotechnol. 106:23-32. [DOI] [PubMed] [Google Scholar]
- 24.Punt, P. J., N. van Biezen, A. Conesa, A. Albers, J. Mangnus, and C. van den Hondel. 2002. Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 20:200-206. [DOI] [PubMed] [Google Scholar]
- 25.Shirasuka, Y., K. Nakajima, T. Asakura, H. Yamashita, A. Yamamoto, S. Hata, S. Nagata, M. Abo, H. Sorimachi, and K. Abe. 2004. Neoculin as a new taste-modifying protein occurring in the fruit of Curculigo latifolia. Biosci. Biotechnol. Biochem. 68:1403-1407. [DOI] [PubMed] [Google Scholar]
- 26.Spencer, J. A., D. J. Jeenes, D. A. MacKenzie, D. T. Haynie, and D. B. Archer. 1998. Determinants of the fidelity of processing glucoamylase-lysozyme fusions by Aspergillus niger. Eur. J. Biochem. 258:107-112. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki, M., E. Kurimoto, S. Nirasawa, Y. Masuda, K. Hori, Y. Kurihara, N. Shimba, M. Kawai, E. Suzuki, and K. Kato. 2004. Recombinant curculin heterodimer exhibits taste-modifying and sweet-tasting activities. FEBS Lett. 573:135-138. [DOI] [PubMed] [Google Scholar]
- 28.Ueda, T., S. Ugawa, H. Yamamura, Y. Imaizumi, and S. Shimada. 2003. Functional interaction between T2R taste receptors and G-protein α subunits expressed in taste receptor cells. J. Neurosci. 23:7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valkonen, M., M. Ward, H. Wang, M. Penttila, and M. Saloheimo. 2003. Improvement of foreign-protein production in Aspergillus niger var. awamori by constitutive induction of the unfolded-protein response. Appl. Environ. Microbiol. 69:6979-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward, M., C. Lin, D. C. Victoria, B. P. Fox, J. A. Fox, D. L. Wong, H. J. Meerman, J. P. Pucci, R. B. Fong, M. H. Heng, N. Tsurushita, C. Gieswein, M. Park, and H. Wang. 2004. Characterization of humanized antibodies secreted by Aspergillus niger. Appl. Environ. Microbiol. 70:2567-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, H., L. Staszewski, H. Tang, E. Adler, M. Zoller, and X. Li. 2004. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc. Natl. Acad. Sci. USA 101:14258-14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada, O., B. R. Lee, and K. Gomi. 1997. Transformation system for aspergillus oryzae with double auxotrophic mutations, niaD and sC. Biosci. Biotechnol. Biochem. 61:1367-1369. [Google Scholar]
- 33.Yamashita, H., T. Akabane, and Y. Kurihara. 1995. Activity and stability of a new sweet protein with taste-modifying action, curculin. Chem. Senses 20:239-243. [DOI] [PubMed] [Google Scholar]
- 34.Yamashita, H., S. Theerasilp, T. Aiuchi, K. Nakaya, Y. Nakamura, and Y. Kurihara. 1990. Purification and complete amino acid sequence of a new type of sweet protein taste-modifying activity, curculin. J. Biol. Chem. 265:15770-15775. [PubMed] [Google Scholar]
- 35.Zhao, G. Q., Y. Zhang, M. A. Hoon, J. Chandrashekar, I. Erlenbach, N. J. Ryba, and C. S. Zuker. 2003. The receptors for mammalian sweet and umami taste. Cell 115:255-266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.