Abstract
Stable isotope probing (SIP) of benzene-degrading bacteria in gasoline-contaminated groundwater was coupled to denaturing gradient gel electrophoresis (DGGE) of DNA fragments amplified by reverse transcription-PCR from community 16S rRNA molecules. Supplementation of the groundwater with [13C6]benzene together with an electron acceptor (nitrate, sulfate, or oxygen) showed that a phylotype affiliated with the genus Azoarcus specifically appeared in the 13C-RNA fraction only when nitrate was supplemented. This phylotype was also observed as the major band in DGGE analysis of bacterial 16S rRNA gene fragments amplified by PCR from the gasoline-contaminated groundwater. In order to isolate the Azoarcus strains, the groundwater sample was streaked on agar plates containing nonselective diluted CGY medium, and the DGGE analysis was used to screen colonies formed on the plates. This procedure identified five bacterial isolates (from 60 colonies) that corresponded to the SIP-identified Azoarcus phylotype, among which two strains (designated DN11 and AN9) degraded benzene under denitrifying conditions. Incubation of these strains with [14C]benzene showed that the labeled carbon was mostly incorporated into 14CO2 within 14 days. These results indicate that the Azoarcus population was involved in benzene degradation in the gasoline-contaminated groundwater under denitrifying conditions. We suggest that RNA-based SIP identification coupled to phylogenetic screening of nonselective isolates facilitates the isolation of enrichment/isolation-resistant microorganisms with a specific function.
Contamination of groundwater with gasoline is a serious environmental problem, since it may affect drinking water resources and has impacts on the oligotrophic environment. Benzene, toluene, and xylenes (BTX) are the major components of gasoline-derived contaminants and are of great concern because they are toxic (15) and soluble in water (16). Among them, benzene is of particular health concern due to its carcinogenicity (2, 53).
Benzene is known to be biodegraded readily under aerobic conditions. However, contamination of subsurface aquifers with gasoline often results in the development of anaerobic zones (5, 13, 30), where benzene is particularly persistent. Many studies have therefore investigated anaerobic benzene degradation in the environment, showing that degradation occurs, albeit slowly, under nitrate-reducing (10, 11, 12, 14, 36, 51, 52), sulfate-reducing (17, 31), iron-reducing (6, 7, 23, 32, 33, 44), perchlorate-reducing (12, 14), and methanogenic conditions (21, 27, 52, 57). Although these studies have identified microbial populations occurring in these enrichments by using molecular techniques (40, 44, 52), no studies have succeeded in isolating benzene-degrading organisms from them. So far, only two bacterial isolates, both affiliated with the genus Dechloromonas, have been shown to anaerobically degrade benzene in axenic cultures (11, 12, 14), although they were isolated after enrichment on different substrates, such as 4-chlorobenzoate.
We have been studying a gasoline-contaminated subsurface aquifer undergoing monitored natural attenuation. Geochemical analyses have suggested that intrinsic anaerobic BTX biodegradation has occurred in the aquifer ∼50 m downstream of the gasoline source area, where electron acceptors such as oxygen, nitrate, and sulfate were depleted, with corresponding reductions in BTX concentrations (49). Molecular ecological analyses have revealed that the community structures in the biodegradation zone are different from those in surrounding uncontaminated zones. Several phylotypes affiliated with the genus Azoarcus were detected as the major populations in the biodegradation zone (49).
The present study was carried out to identify microorganisms degrading benzene in the gasoline-contaminated aquifer. Recently, scientists have developed culture-independent approaches for linking microbial community function to the phylogenetic identities of key organisms (9, 29, 37, 39). Stable isotope probing (SIP) is one such method, enabling the identification of members in a microbial community responsible for specific activities based on the incorporation of stable isotopes (e.g., 13C) into cellular components (34, 35, 37). This study used RNA-based SIP (RNA-SIP) to label and identify [13C6]benzene-degrading organisms. RNA-SIP exploits the relatively efficient 13C incorporation into RNA compared to that into DNA (20, 37), which is particularly useful when substrate degradation and growth rates are likely to be slow, as in the context of anaerobic benzene degradation. In addition, SIP information was confirmed by isolating RNA-SIP-identified bacteria and analyzing their benzene-degrading ability under denitrifying conditions.
MATERIALS AND METHODS
Groundwater sample.
The gasoline-contaminated groundwater used in this study was obtained from a BTX-contaminated subsurface aquifer situated at Kumamoto, Japan, in March 2004. This aquifer has been subjected to monitored natural attenuation from April 2002, after the termination of a 10-year pumping/purge treatment (49). The groundwater sample was obtained from a shallow dug well (well 29) and stored in sterile glass bottles (1 liter) at 4°C during transportation to our laboratory (∼3 days). The headspace in the bottles was minimized to reduce contamination with air. Physical and chemical characteristics of the groundwater (oxidation/reduction potential [ORP], dissolved oxygen concentration [DO], pH, and temperature) were determined as described elsewhere (49) immediately after sampling.
SIP.
Groundwater samples were manipulated in an anaerobic glove box filled with reduced-copper-treated nitrogen gas. An electron acceptor (NaNO3 or Na2SO4) was added to the groundwater at a final concentration of 5 mM from a sterile anaerobic stock solution. Inorganic nutrients (NH4Cl and K2HPO4) were also added from sterile anaerobic stock solutions to final concentrations of 1.0 mg liter−1 and 0.2 mg liter−1, respectively. After the bottles were supplemented with 200 μM [13C6]benzene (Cambridge Isotope Laboratories), they were incubated at 25°C in the dark without shaking. For aerobic incubation of the groundwater, 300 ml of groundwater sample was dispensed into a glass bottle (1-liter capacity), and the headspace was filled with air as the source of oxygen. The inorganic nutrients and [13C6]benzene were added as described above. Sterile controls were prepared under the same conditions by autoclaving the bottles for 20 min at 121°C twice before adding [13C6]benzene.
RNA extraction, ultracentrifugation, and fractionation.
Microbial cells in the groundwater sample were collected on a GV membrane (0.22-μm pore size; Millipore) by vacuum filtration. Total RNA was extracted using a FastRNA Blue kit (Qbiogene) and an RNeasy mini kit (QIAGEN) as described elsewhere (37). RNA purity and quantity were determined by measuring the UV absorption spectrum (46).
13C-labeled RNA and unlabeled RNA were separated by equilibrium density gradient centrifugation and gradient fractionation (37). Total RNA (500 ng) was loaded onto a cesium trifluoroacetate gradient and centrifuged at 64,000 rpm for 36 h at 20°C. Gradients were fractionated from the bottom by displacement with water from the top by using a high-performance liquid chromatography pump at a flow rate of 0.2 ml min−1. The buoyant densities of gradient fractions were determined by weighing specific volumes. RNAs were isolated from gradient fractions by precipitation with 1 volume of isopropyl alcohol and were checked by electrophoresis of agarose gels as described previously (37).
16S rRNA amplification and DGGE analysis.
RNA samples from the equilibrium density gradient fractions were reverse transcribed using the reverse primer 518R (38) and avian myeloblastosis virus reverse transcriptase (Takara). The cDNAs produced were used to amplify the V3 regions of bacterial 16S rRNA genes (corresponding to positions 341 to 534 in the Escherichia coli rRNA sequence) connected to a GC clamp, using PCR primers 357F and 518R (38). PCR was performed as described previously (55), and amplification of PCR products with the expected sizes was confirmed by electrophoresis through 1.5% (wt/vol) agarose gels (LO3; Takara) in Tris-borate-EDTA buffer (46). Denaturing gradient gel electrophoresis (DGGE) was performed with a DCode instrument (Bio-Rad) as described previously (25). Three independent PCR/DGGE analyses were performed for each sample to verify the reproducibility of the method. DGGE profile images were obtained and analyzed using Gel Doc 2000 (Bio-Rad) and Multi-Analyst software (version 1.0.2 for Apple Power PC; Bio-Rad).
Nucleotide sequences of DGGE bands were determined as described previously (55). Sequence homology searches were conducted using the GenBank nucleotide sequence library and the BLAST program (3) through the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST).
Isolation and phylogenetic characterization of bacterial strains.
The groundwater sample incubated with [13C6]benzene under denitrifying conditions was spread onto agar plates containing diluted CGY medium (dCGY medium, containing 0.05% Bacto Casamino Acids, 0.01% Bacto yeast extract, 0.05% glycerol, and 1.5% agar) and incubated aerobically at 25°C. The sample was also spread onto dCGY plates containing 2 mM NaNO3 and incubated anaerobically under an atmosphere of nitrogen at 25°C. Colonies formed on the plates were purified by restreaking on dCGY plates and aerobic or anaerobic incubation.
Purified bacterial strains were grown aerobically in 30 ml of dCGY medium, and genomic DNAs were extracted by using a genomic DNA purification kit (Promega) in accordance with the manufacturer's instructions. The 16S rRNA gene fragments were amplified by PCR, using primers B27f and U1492r (56), and sequenced as described previously (26). PCR amplification and sequencing were repeated several times to check for amplification errors. The determined nucleotide sequences were aligned with the sequences of reference strains in the Rhodocyclaceae family, using Clustal W, version 1.7 (50). A phylogenetic tree was constructed from the evolutionary distance data (28) by the neighbor-joining method (45). The bootstrap resampling method of Felsenstein (18) was used with 1,000 replicates to evaluate the robustness of the branches of the inferred tree.
BTX degradation test (cold test).
All manipulations were performed in an anaerobic glove box. A sterile basal salt medium (26) (30 ml) was prepared in a 70-ml glass bottle and supplemented with NaNO3 (2 mM), titanium chloride (pH 7.0, 2 mM), and resazurin (2 mg liter−1). The medium was inoculated with approximately 106 cells of a bacterial strain pregrown in dCGY medium, and the bottle was sealed with a Teflon-coated butyl rubber septum (approximately 3 mm in thickness) secured with a crimped aluminum cap. Benzene was added to the bottle at a final concentration of 15 μM from an anaerobic stock solution (3 mM in sterilized water) by use of a syringe. Alternatively, toluene or m-, p-, or o-xylene was added to the bottle at a final concentration of 100 μM, using a microsyringe. Uninoculated controls were prepared in the same manner. The bottle was incubated at 25°C in the dark without shaking. The stopper-sealed orifice of the bottle was always kept below the surface of the medium to avoid adsorption of benzene to the rubber stopper (1, 41, 42, 43). All incubations were prepared in triplicate. Samples were taken from the bottle by use of a syringe at appropriate intervals and analyzed as described below.
Radiorespirometry.
Radiorespirometry was conducted to investigate the mineralization of benzene by bacterial isolates. Isolated strains were grown in dCGY medium, and the cells were collected by centrifugation. They were inoculated anaerobically at 106 cells ml−1 in 30 ml of basal salt medium containing 2 mM NaNO3 as described above. The culture was supplemented with 37 kBq [14C]benzene from an anaerobic labeled stock solution prepared by diluting universally labeled [14C]benzene (2,220 MBq mmol−1; Sigma) with anoxic sterile water to give a final radioactivity of 148 kBq ml−1. The bottle was incubated at 25°C in the dark without shaking, as described above. The culture was prepared in triplicate. To quantify the 14CO2 produced from [14C]benzene, the medium was flushed with reduced-copper-treated nitrogen gas (>99.999% pure) through a series of traps comprised of a first trap containing 10 ml of cold benzene (for trapping the labeled benzene) and second and third traps containing 10 ml of 0.4 N NaOH (for trapping 14CO2). Before being flushed, the medium was acidified with 7 ml of 2 N H2SO4 to convert carbonate to CO2. For measurements of the radioactivity in the 14CO2 traps, 3 ml of solution was sampled from each trap, mixed with 10 ml of scintillation cocktail (ScintiVers; Perkin-Elmer), and subjected to analysis in a model 1900CA Tri-Carb liquid scintillation analyzer (Perkin-Elmer). The efficiencies of the CO2 traps were checked by using NaH14CO3 (24). A mineralization ratio (MR) was estimated by using the equation MR (%) = (R − Rc)/Rb × 100, where R is the radioactivity in the CO2 trap for a bacterial culture, Rc is the radioactivity in the CO2 trap for the uninoculated control, and Rb is the radioactivity of [14C]benzene initially added to the culture.
Analytical procedures.
Concentrations of sulfate and nitrate were determined by ion chromatography with an ICA-2000 ion analyzer (DKK Toa). A total direct count of microbial cells in a liquid sample was determined by fluorescence microscopy after staining with 4′,6′-diamidino-2-phenylindole (DAPI) (55). In order to determine BTX concentrations, a headspace sample (100 μl) was analyzed on a gas chromatograph (Shimadzu GC-17A) equipped with a 30-m DB-624 capillary column (0.53 mm in diameter, 3 μm in film thickness; J&W Scientific) and a flame ionization detector. The injector and detector temperatures were 120°C and 160°C, respectively, while the column temperature was 90°C. The carrier gas was nitrogen at a flow rate of 30 ml min−1.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported here have been submitted to the DDBJ, EMBL, and GenBank databases under accession no. AB241391 to AB241407.
RESULTS AND DISCUSSION
RNA-SIP of benzene-degrading bacteria.
BTX have constantly been detected in groundwater sampled from well 29 during natural attenuation, with xylene concentrations consistently being higher than benzene and toluene concentrations (49). During this period, nitrate and sulfate concentrations in groundwater were below the detection limits (<0.1 mg liter−1), and the DO concentration was always below 0.5 mg liter−1 (49). The characteristics of the groundwater obtained for the present study are summarized in Table 1.
TABLE 1.
Characteristics of contaminated groundwater used in this study
| Parameter | Valuea |
|---|---|
| Temp (°C) | 16 |
| pH | 6.8 |
| ORP (mV) | −62 |
| DO (mg liter−1) | 4.3 |
| Nitrate (mg liter−1) | ND |
| Sulfate (mg liter−1) | ND |
| Benzene (mg liter−1) | 0.02 |
| Toluene (mg liter−1) | 0.27 |
| Xylene (mg liter−1) | 8.3 |
| Total cell count (cells ml−1) | 5.6 × 106 |
ND, not detected (the lower detection limit was 0.1 mg liter−1).
Groundwater samples were supplemented with [13C6]benzene (200 μM, equivalent to approximately 16 mg liter−1) and an electron acceptor (nitrate, sulfate, or oxygen). Changes in concentrations of benzene and xylene are presented in Fig. 1. Toluene concentrations are not presented in this figure, since they were very low (Table 1) and were below the detection limit from day 7 on under all conditions. Figure 1 shows that benzene was degraded under all conditions, albeit relatively slowly under denitrifying and sulfidogenic conditions. Although xylene was rapidly degraded under aerobic conditions, no significant degradation was observed under sulfidogenic conditions. In the sterile controls, benzene and xylene were not significantly decreased (data not shown).
FIG. 1.
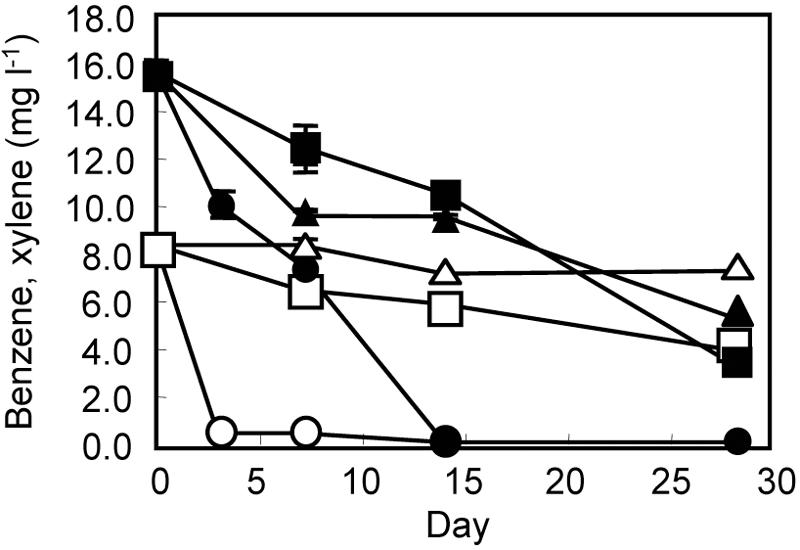
Degradation of benzene and xylene during incubation of groundwater for SIP. Closed squares, benzene under denitrifying conditions; open squares, xylene under denitrifying conditions; closed triangles, benzene under sulfidogenic conditions; open triangles, xylene under sulfidogenic conditions; closed circles, benzene under aerobic conditions; open circles, xylene under aerobic conditions. Data points are means of triplicate experiments, and error bars represent standard deviations.
During the 28-day incubation under denitrifying conditions, 1.8 mM of nitrate and 0.13 mM of oxygen (equivalent to 4.3 mg liter−1) were consumed in association with losses of 132 μM of benzene, 2.9 μM of toluene, and 78.3 μM of xylene. Assuming that all oxygen molecules were used for benzene degradation, this could result in the loss of 17.3 μM of benzene, as estimated according to equation 1:
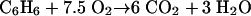 |
(1) |
The remaining benzene plus toluene and xylene were likely degraded in association with denitrification. According to equations 2 to 4 (48), the degradation of these hydrocarbons could consume 1.4 mM of nitrate, which is 77.8% of the actual consumption of nitrate.
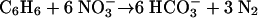 |
(2) |
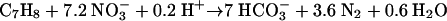 |
(3) |
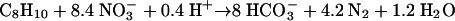 |
(4) |
The remaining nitrate may have been utilized for the oxidation of other undefined organic matters in the groundwater. This estimation indicates that a significant portion of benzene was degraded in association with denitrification.
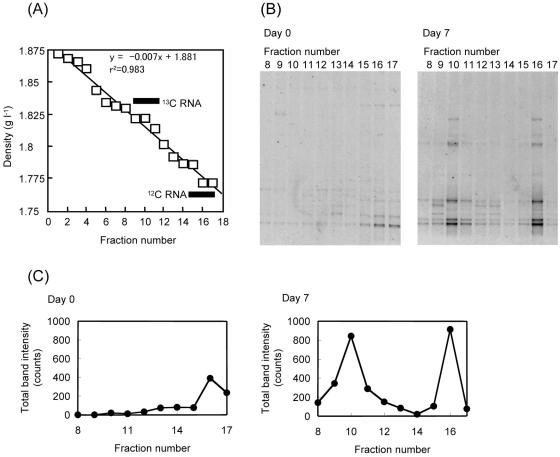
RNAs were extracted from the incubated groundwater at 7-day intervals (from day 0 to day 21), fractionated, and subjected to reverse transcription-PCR (RT-PCR) of 16S rRNA fragments (Fig. 2). Figure 2A confirms that the formation and fractionation of density gradients were successful. Based on known density values (37), we estimated that the 13C-RNA and 12C-RNA fractions were positioned around fractions 10 and 16, respectively. Figure 2B presents DGGE patterns of RT-PCR products from different fractions for denitrifying incubation on days 0 and 7. As shown in this figure, bacterial species that utilized [13C6]benzene were also detected in the 12C-RNA fraction, because the groundwater was originally contaminated with benzene. Total DGGE band intensities of these DGGE profiles were plotted in Fig. 2C. It is shown that only one peak was seen around the 12C-RNA fractions on day 0, while two peaks appeared around the 12C-RNA and 13C-RNA fractions on day 7. This observation indicates that 13C in benzene was incorporated into rRNA during the 7-day incubation under denitrifying conditions. Similarly, 13C incorporation within 7 days was also observed under aerobic and sulfidogenic conditions (data not shown).
FIG. 2.
(A) Separation of 13C-RNA and 12C-RNA by density gradient centrifugation and fractionation. Black bars indicate estimated positions of the 13C-RNA and 12C-RNA fractions. (B) DGGE profiles of RT-PCR-amplified community rRNAs present in fractions 8 to 17 from panel A for day 0 and day 7 samples incubated under denitrifying conditions. (C) Densitometry plots with total band intensities of DGGE profile lanes in panel B.
Figure 3 shows changes in DGGE patterns of the 13C-RNA fractions (corresponding to fraction 10 in Fig. 2) during 21-day incubations. DGGE analysis was not conducted for aerobic incubation on day 21, since all benzene disappeared by day 14. It was found that several bands, such as bands 2 and 3, commonly appeared under different conditions, while some bands were specific, e.g., band 4 was seen under denitrifying conditions and bands 11 and 12 were seen under sulfidogenic conditions. In addition, many unique bands appeared under aerobic conditions. It is likely that the common bands (bands 2 and 3) represented organisms that utilized oxygen molecules (presumably contaminated during sampling) to assimilate benzene. In contrast, comparisons of bands occurring under the different electron-accepting conditions allowed us to identify phylotypes that specifically assimilated benzene under certain electron-accepting conditions, such as band 4, representing organisms assimilating benzene under denitrifying conditions.
FIG. 3.
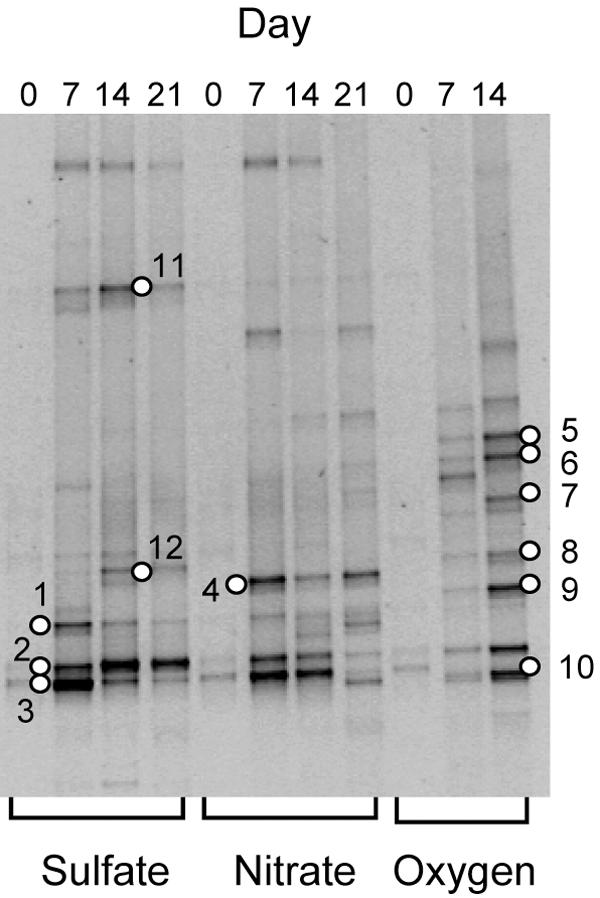
Time courses of DGGE profiles for 13C-labeled community RNAs obtained from groundwater incubated under sulfidogenic (sulfate), denitrifying (nitrate), and aerobic (oxygen) conditions. The numbered bands were excised for DNA sequencing analysis.
The nucleotide sequences of major bands were determined (Table 2), and band 4 was assigned to Azoarcus, a genus known to include bacteria capable of metabolizing aromatic hydrocarbons, such as toluene, xylene, and ethylbenzene, under denitrifying conditions (8, 19, 22, 42, 47, 48, 58). Recently, Ulrich and Edwards demonstrated that the dominant microbial population in a benzene-degrading denitrifying enrichment culture was closely related to Azoarcus species (52). In our previous study (49), an Azoarcus phylotype was widely detected by the PCR-DGGE analysis of groundwater taken from a gasoline-contaminated aquifer. In this study, we found that the nucleotide sequence of band 4 (Fig. 3) was 100% identical to that of the Azoarcus phylotype retrieved from the gasoline-contaminated aquifer, suggesting that the Azoarcus population represented by band 4 was at least partly responsible for mineralization of benzene in the gasoline-contaminated groundwater.
TABLE 2.
Sequence analysis of major DGGE bands appearing in 13C fractions (Fig. 3)
| DGGE band | Organism with closest sequence (accession no.) | Homology (%) | Phylogenetic group |
|---|---|---|---|
| 1 | Dechloromonas sp. strain JJ (AY032611) | 96 | Betaproteobacteria |
| 2 | Aquaspirillum sp. strain 412 (AY780907) | 95 | Betaproteobacteria |
| 3 | Comamonas aquatica (AJ430346) | 95 | Betaproteobacteria |
| 4 | Azoarcus evansii (X77679) | 100 | Betaproteobacteria |
| 5 | Zoogloea resiniphila (AJ505854) | 99 | Betaproteobacteria |
| 6 | Hylemonella sp. strain WQH1 (AJ565430) | 97 | Betaproteobacteria |
| 7 | Novosphingobium hassiacum (AJ416411) | 100 | Alphaproteobacteria |
| 8 | Aquaspirillum delicatum (AF078756) | 98 | Betaproteobacteria |
| 9 | Zoogloea resiniphila (AJ011506) | 98 | Betaproteobacteria |
| 10 | Xanthobacter autotrophicus (U62888) | 100 | Alphaproteobacteria |
| 11 | Aquaspirillum sp. strain 412 (AY780907) | 95 | Betaproteobacteria |
| 12 | Uncultured bacterium vadinHB76 (AJ583208) | 92 | Bacteria |
Isolation of Azoarcus strains and phylogenetic analysis.
In order to examine if bacteria represented by the Azoarcus phylotype were actually capable of degrading benzene under denitrifying conditions, we attempted to isolate them for axenic benzene degradation tests. The groundwater incubated for SIP under denitrifying conditions for 28 days was spread onto dCGY plates and incubated under aerobic and denitrifying conditions. We used dCGY medium because a previous study demonstrated that this medium facilitated the isolation of bacteria affiliated with the Betaproteobacteria from activated sludge (54). A total of 60 colonies (40 from aerobic and 20 from denitrifying plates) were picked and restreaked on the same medium for purification, and cells on the new plates were subjected to DGGE analysis of PCR-amplified 16S rRNA gene fragments. In the DGGE analysis, band positions were compared with that of band 4 in Fig. 3 on the same gels, and nucleotide sequences of bands migrating to the same position as band 4 were determined to confirm their identity. As a result, we obtained five colonies whose 16S rRNA gene fragments were 100% identical in nucleotide sequence to that in band 4; these were strains DN11, DN15, and DN47, isolated under denitrifying conditions, and strains AN9 and AN21, isolated under aerobic conditions. All of them were capable of aerobic growth in dCGY medium and were therefore maintained under aerobic conditions.
Sequence analysis of the 16S rRNA gene fragments (>1,450 bp) found that their sequences included several mismatches (i.e., they had >99% sequence identity to each other), except for AN9 and AN21, whose 16S rRNA gene sequences were completely identical. Phylogenetic comparison of the 16S rRNA gene sequences with those of reference Azoarcus strains showed that these isolates were affiliated with the genus Azoarcus and were particularly related to Azoarcus evansii (99% sequence identity over 1,480 bp), which metabolizes such aromatic compounds as benzoate, toluene, and phenol under denitrifying conditions (4), and Azoarcus sp. strain ToN1 (99% sequence identity over 1,470 bp), which degrades toluene (42).
Anaerobic benzene degradation in axenic cultures.
The five isolated strains were examined for the ability to degrade BTX under denitrifying conditions (Table 3). We found that four strains, but not strain AN21, degraded toluene and m-xylene, while only two strains (DN11 and AN9) were capable of anaerobic benzene degradation. This observation revealed the variation in degradative capacity among these closely related Azoarcus strains. In particular, although the 16S rRNA gene sequences of strains AN9 and AN21 were 100% identical, their degradative capacities were very different. From these data and the 16S rRNA gene sequences, we tentatively identified that the five isolates are five different strains.
TABLE 3.
Degradative capacities of Azoarcus strains isolated from groundwater
| Strain | Cold test resulta
|
MRb (%) | ||||
|---|---|---|---|---|---|---|
| Benzene | Toluene | o-Xylene | m-Xylene | p-Xylene | ||
| DN11 | + | ++ | − | ++ | − | 102 ± 19 |
| DN15 | − | ++ | − | ++ | − | NA |
| DN47 | − | ++ | − | ++ | − | NA |
| AN9 | + | ++ | − | ++ | − | 72 ± 14 |
| AN21 | − | − | − | − | − | 3.7 ± 4.4 |
−, no substantial degradation was observed after 30 days; +, 20% to 90% of the amended substrate was degraded; ++, >90% of the substrate was degraded.
MR of benzene, determined by a radiorespirometry experiment as described in the text. Data are means ± standard deviations (n = 3). NA, not analyzed.
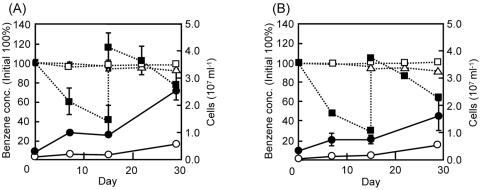
Figure 4 presents growth curves for strains DN11 and AN9 grown in basal salt medium supplemented with 15 μM benzene as the sole carbon source. It is shown that benzene was degraded, concomitant with growth, when the culture was supplemented with nitrate. Although small amounts of growth were observed in the absence of nitrate, they were not associated with benzene degradation and were probably due to nutrient carryover from the preculture. After the degradation test, the purity of the culture was confirmed by PCR-DGGE analysis (data not shown).
FIG. 4.
Growth and benzene degradation by strains DN11 (A) and AN9 (B). The cultures were refed benzene at the time point indicated with vertical lines. Residual benzene was measured in the nitrate-amended (closed squares), unamended (open squares), and uninoculated (open triangles) cultures. Cell numbers are given for the nitrate-amended (closed circles) and unamended (open circles) cultures. Data points are means of triplicate experiments, and error bars represent standard deviations.
In order to investigate if benzene was anaerobically mineralized (converted to CO2) by strains DN11 and AN9, radiorespirometry experiments were conducted, using [14C]benzene under denitrifying conditions. Table 3 shows that large fractions of radioactivity initially added as [14C]benzene were recovered in the CO2 trap. In this experiment, significant amounts of radioactivity were not recovered in the CO2 trap in the absence of nitrate (data not shown), showing that benzene mineralization was coupled to denitrification.
In conclusion, the present study successfully used the RNA-SIP technique to demonstrate that the Azoarcus population is involved in benzene degradation in the aquifer. However, among the five isolated strains affiliated with the Azoarcus phylotype, only two strains were capable of anaerobic benzene degradation. These data indicate that functional heterogeneity exists among strains with the Azoarcus phylotype, and more specific molecular markers than the 16S rRNA gene will be necessary for discriminating benzene-degrading Azoarcus populations from other closely related Azoarcus populations in the gasoline-contaminated aquifer.
Although several studies have obtained enrichment cultures degrading benzene under anaerobic conditions (10, 20, 22, 35, 42, 49, 51, 52), no study has succeeded in isolating benzene-degrading strains from them. We have also attempted to isolate anaerobic benzene-degrading bacteria by using agar plates containing minimal medium after enrichment in liquid cultures from gasoline-contaminated groundwater; however, all these trials were in vain. In contrast, the present study combined RNA-SIP with DGGE screening of heterotrophic bacteria, resulting in the isolation of two denitrifying benzene-degrading bacteria. Accordingly, we suggest that RNA-SIP identification coupled to phylogenetic screening of nonselective isolates facilitates the isolation of enrichment/isolation-resistant microorganisms with a specific function.
Acknowledgments
We are grateful to Hiromi Awabuchi for technical assistance.
This work was supported by the New Energy and Industrial Technology Development Organization (NEDO).
REFERENCES
- 1.Aeckersberg, F., F. Bak, and F. Widdel. 1991. Anaerobic oxidation of saturated hydrocarbons to CO2 by a new type of sulfate-reducing bacterium. Arch. Microbiol. 156:5-14. [Google Scholar]
- 2.Agency for Toxic Substances and Disease Registry (ATSDR). 1997. Toxicological profile for benzene. U.S. Public Health Service, U.S. Department of Health and Human Services, Atlanta, Ga.
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Anders, H.-J., A. Kaetzke, P. Kampfer, W. Ludwig, and G. Fuchs. 1995. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K172 and KB740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of proteobacteria. J. Int. Syst. Bacteriol. 45:327-333. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, R. T., and D. R. Lovley. 1997. Ecology and biogeochemistry of in situ groundwater bioremediation. Adv. Microb. Ecol. 15:289-350. [Google Scholar]
- 6.Anderson, R. T., and D. R. Lovley. 1999. Naphthalene and benzene degradation under Fe(III)-reducing conditions in petroleum-contaminated aquifers. Bioremed. J. 3:121-135. [Google Scholar]
- 7.Anderson, R. T., J. N. Rooney-Varge, C. V. Gaw, and D. R. Lovley. 1998. Anaerobic benzene oxidation in the Fe(III) reduction zone of petroleum-contaminated aquifers. Environ. Sci. Technol. 32:1222-1229. [Google Scholar]
- 8.Ball, H. A., H. A. Johnson, M. Reinhard, and A. M. Spormann. 1996. Initial reactions in anaerobic ethylbenzene oxidation by a denitrifying bacterium, strain EB1. J. Bacteriol. 178:5755-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. de Graaf, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801-804. [Google Scholar]
- 10.Burland, S. I., and E. A. Edwards. 1999. Anaerobic benzene biodegradation linked to nitrate reduction. Appl. Environ. Microbiol. 65:529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakraborty, R., and J. D. Coates. 2005. Hydroxylation and carboxylation: two crucial steps of anaerobic benzene degradation by Dechloromonas strain RCB. Appl. Environ. Microbiol. 71:5427-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraborty, R., S. M. O'Connor, E. Chan, and J. D. Coates. 2005. Anaerobic degradation of benzene, toluene, ethylbenzene, and xylene compounds by Dechloromonas strain RCB. Appl. Environ. Microbiol. 71:8649-8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen, T., P. Kjeldsen, H. Albrechtsen, and G. Heron. 1994. Attenuation of pollutants in landfill leachate polluted aquifers. Crit. Rev. Environ. Sci. Technol. 24:119-202. [Google Scholar]
- 14.Coates, J. D., R. Chakraborty, J. G. Lack, S. M. O'Connor, K. A. Cole, K. S. Bender, and L. A. Achenbach. 2001. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature 411:1039-1043. [DOI] [PubMed] [Google Scholar]
- 15.Dean, B. J. 1985. Recent findings on the genetic toxicology of benzene, toluene, xylenes and phenols. Mutat. Res. 154:336-341. [DOI] [PubMed] [Google Scholar]
- 16.Dunn, W. J., III, J. H. Block, and R. S. Pearlman. 1986. Partition coefficient, determination and estimation. Pergamon Press, New York, N.Y.
- 17.Edwards, E. A., and D. Grbic-Galic. 1992. Complete mineralization of benzene by aquifer microorganisms under strictly anaerobic conditions. Appl. Environ. Microbiol. 58:2663-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 19.Fries, M. R., J. Chee-Sandford, and J. M. Tiedje. 1994. Isolation, characterization, and distribution of denitrifying toluene degraders from a variety of habitats. Appl. Environ. Microbiol. 60:2802-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallagher, E., L. McGuinness, C. Phelps, L. Y. Young, and L. J. Kerkhof. 2005. 13C-carrier DNA shortens the incubation time needed to detect benzoate-utilizing denitrifying bacteria by stable-isotope probing. Appl. Environ. Microbiol. 71:5192-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grbic-Galic, D., and T. Vogel. 1987. Transformation of toluene and benzene by mixed methanogenic cultures. Appl. Environ. Microbiol. 53:254-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hess, A., B. Zarda, D. Hahn, A. Haner, D. Stax, P. Hohener, and J. Zeyer. 1997. In situ analysis of denitrifying toluene- and m-xylene-degrading bacteria in a diesel fuel-contaminated laboratory aquifer column. Appl. Environ. Microbiol. 63:2136-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahn, M. K., S. B. Haderlein, and R. U. Meckenstock. 2005. Anaerobic degradation of benzene, toluene, ethylbenzene, and o-xylene in sediment-free iron-reducing enrichment cultures. Appl. Environ. Microbiol. 71:3355-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanaly, R. A., R. Bartha, K. Watanabe, and S. Harayama. 2000. Rapid mineralization of benzo[a]pyrene by a microbial consortium growing on diesel fuel. Appl. Environ. Microbiol. 66:4205-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasai, Y., H. Kishira, K. Syutsubo, and S. Harayama. 2001. Molecular detection of marine bacterial populations on beaches contaminated by Nakhodka tanker oil-spill accident. Environ. Microbiol. 3:1-10. [DOI] [PubMed] [Google Scholar]
- 26.Kasai, Y., Y. Takahata, T. Hoaki, and K. Watanabe. 2005. Physiological and molecular characterization of a microbial community established in unsaturated, petroleum-contaminated soil. Environ. Microbiol. 7:806-818. [DOI] [PubMed] [Google Scholar]
- 27.Kazumi, J., M. E. Caldwell, J. M. Suflita, D. R. Lovley, and L. Y. Young. 1997. Anaerobic degradation of benzene in diverse anoxic environments. Environ. Sci. Technol. 31:813-818. [Google Scholar]
- 28.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 29.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielson, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography: a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovely, D. R. 1997. Potential for anaerobic bioremediation of BTEX in petroleum-contaminated aquifers. J. Ind. Microbiol. Biotechnol. 18:75-81. [Google Scholar]
- 31.Lovley, D. R., J. D. Coates, J. C. Woodward, and E. J. P. Phillips. 1995. Benzene oxidation coupled to sulfate reduction. Appl. Environ. Microbiol. 61:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovley, D. R., J. C. Woodward, and F. H. Chapelle. 1994. Stimulated anoxic biodegradation of aromatic hydrocarbons using Fe(III) ligands. Nature 370:128-131. [DOI] [PubMed] [Google Scholar]
- 33.Lovley, D. R., J. C. Woodward, and F. H. Chapelle. 1996. Rapid anaerobic benzene degradation with a variety of chelated Fe(III) forms. Appl. Environ. Microbiol. 62:288-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lueders, T., B. Wagner, P. Claus, and M. W. Friedrich. 2004. Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ. Microbiol. 6:60-72. [DOI] [PubMed] [Google Scholar]
- 35.Lueders, T., M. Manefield, and M. W. Friedrich. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73-78. [DOI] [PubMed] [Google Scholar]
- 36.Mancini, S. A., A. C. Ulrich, G. Lacrampe-Couloume, B. Sleep, E. A. Edwards, and B. S. Lollar. 2003. Carbon and hydrogen isotopic fractionation during anaerobic biodegradation of benzene. Appl. Environ. Microbiol. 69:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orphan, V. J., C. H. House, K. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-486. [DOI] [PubMed] [Google Scholar]
- 40.Phelps, C. D., L. J. Kerkhof, and L. Y. Young. 1998. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol. Ecol. 27:269-279. [Google Scholar]
- 41.Rabus, R., R. Nordhaus, W. Ludwig, and F. Widdel. 1993. Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl. Environ. Microbiol. 59:1444-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rabus, R., and F. Widdel. 1995. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch. Microbiol. 163:96-103. [DOI] [PubMed] [Google Scholar]
- 43.Rabus, R., H. Wilkes, A. Schramm, G. Harms, A. Behrends, R. Amann, and F. Widdel. 1999. Anaerobic utilization of alkylbenzenes and n-alkanes from crude oil in an enrichment culture of denitrifying bacteria affiliating with the β-subclass of proteobacteria. Environ. Microbiol. 1:145-157. [DOI] [PubMed] [Google Scholar]
- 44.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Song, B., M. M. Haggbom, J. Zhou, J. M. Tiedje, and N. J. Palleroni. 1999. Taxonomic characterization of denitrifying bacteria that degrade aromatic compounds and description of Azoarcus toluvorans sp. nov. and Azoarcus toluclasticus sp. nov. Int. J. Syst. Bacteriol. 49:1129-1140. [DOI] [PubMed] [Google Scholar]
- 48.Spormann, A. M., and F. Widdel. 2001. Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria. Biodegradation 11:85-105. [DOI] [PubMed] [Google Scholar]
- 49.Takahata, Y., Y. Kasai, and K. Watanabe. 2004. Assessment of chemical and microbiological signatures during natural attenuation of gasoline-contaminated groundwater, p. 827-831. In Proceedings of the European Symposium on Environmental Biotechnology. A. A. Balkema Publishers, London, United Kingdom.
- 50.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulrich, A. C., H. R. Beller, and E. A. Edwards. 2005. Metabolites detected during biodegradation of 13C6-benzene in nitrate-reducing and methanogenic enrichment cultures. Environ. Sci. Technol. 39:6681-6691. [DOI] [PubMed] [Google Scholar]
- 52.Ulrich, A. C., and E. A. Edwards. 2003. Physiological and molecular characterization of anaerobic benzene-degrading mixed cultures. Environ. Microbiol. 5:92-102. [DOI] [PubMed] [Google Scholar]
- 53.U.S. Environmental Protection Agency. 2002. Integrated risk information system (IRIS) on benzene. National Center for Environmental Assessment, Washington, D.C.
- 54.Watanabe, K., M. Teramoto, and S. Harayama. 1999. An outbreak of nonflocculating catabolic populations caused the breakdown of a phenol-digesting activated-sludge process. Appl. Environ. Microbiol. 65:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe, K., S. Yamamoto, S. Hino, and S. Harayama. 1998. Population dynamics of phenol-degrading bacteria in activated sludge determined by gyrB-targeted quantitative PCR. Appl. Environ. Microbiol. 64:1203-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe, K., Y. Kodama, and N. Kaku. 2002. Diversity and abundance of bacterial populations in groundwater accumulating in an underground crude-oil storage cavity. BMC Microbiol. 2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiner, J., and D. R. Lovley. 1998. Rapid benzene degradation in methanogenic sediments from a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 64:1937-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou, J., M. R. Fries, J. C. Chee-Sanford, and J. M. Tiedje. 1995. Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth on toluene and description of Azoarcus tolulyticus sp. nov. Int. J. Syst. Bacteriol. 45:500-506. [DOI] [PubMed] [Google Scholar]