Abstract
We applied nucleic acid-based molecular methods, combined with estimates of biomass (ATP), pigments, and microelectrode measurements of chemical gradients, to map microbial diversity vertically on a millimeter scale in a hypersaline microbial mat from Guerrero Negro, Baja California Sur, Mexico. To identify the constituents of the mat, small-subunit rRNA genes were amplified by PCR from community genomic DNA extracted from layers, cloned, and sequenced. Bacteria dominated the mat and displayed unexpected and unprecedented diversity. The majority (1,336) of the 1,586 bacterial 16S rRNA sequences generated were unique, representing 752 species (≥97% rRNA sequence identity) in 42 of the main bacterial phyla, including 15 novel candidate phyla. The diversity of the mat samples differentiated according to the chemical milieu defined by concentrations of O2 and H2S. Bacteria of the phylum Chloroflexi formed the majority of the biomass by percentage of bulk rRNA and of clones in rRNA gene libraries. This result contradicts the general belief that cyanobacteria dominate these communities. Although cyanobacteria constituted a large fraction of the biomass in the upper few millimeters (>80% of the total rRNA and photosynthetic pigments), Chloroflexi sequences were conspicuous throughout the mat. Filamentous Chloroflexi bacteria were identified by fluorescence in situ hybridization within the polysaccharide sheaths of the prominent cyanobacterium Microcoleus chthonoplastes, in addition to free living in the mat. The biological complexity of the mat far exceeds that observed in other polysaccharide-rich microbial ecosystems, such as the human and mouse distal guts, and suggests that positive feedbacks exist between chemical complexity and biological diversity.
Microbial mats are benthic aquatic ecosystems fueled by light energy and composed of microbial cells attached to extracellular polymeric material and mineralized scaffolds in visible millimeter scale layers (12). Unlike stromatolites, which have a biotic mechanism for calcification (49), microbial mats become layered because of occasional sedimentation and regrowth. Microbial mats and stromatolites are found in the fossil record dating back 3.4 billion years (60) and are thought to have significantly influenced the composition of the atmosphere with production of O2, H2, and CH4 (21). Ancient and modern mats share properties inherent to their structure. For instance, different wavelengths of light penetrate differentially, gas exchange with the atmosphere occurs at the surface, an organic carbon-based matrix provides a scaffold for growth, and sedimentation occasionally buries the surface, which is then overgrown, leading to layering. Comparisons of biosignatures in modern and fossilized mats seek to describe ancient biogeochemical cycles and the microbial activities of ancient communities (34, 57, 58).
The microbial mats within the hypersaline lagoons of the Exportadora de Sal SA saltern in Guerrero Negro, Baja California Sur, Mexico, cover an extensive area of artificial shallow lagoons protected from tidal disturbance by levees. Biogeochemical studies of these mats have shown that oxygen and light, as well as photosynthetic capacity, are rapidly depleted with depth. Degradation of organic matter occurs largely by two processes, fermentation and substrate oxidation through sulfate reduction (12). Both processes take on unusual characteristics in the mats. Fermentation contributes to molecular hydrogen release into the overlying water column, even in the presence of oxygen, such that bubbles of mixed oxygen and hydrogen gases form on the surface of the mat (21). In addition, the highest rates of sulfate reduction occur in the upper, oxygen-rich layers of the mat (5). Known sulfate-reducing bacteria of the delta group of proteobacteria occupy the anoxic zone, however, which suggests that novel groups of sulfate-reducing bacteria reduce sulfate in the mats aerobically (51).
Despite the intriguing biogeochemistry of hypersaline microbial mats and their importance as model systems for studies of the early Earth (21), the composition of the microbiota has not been surveyed comprehensively by culture-independent molecular methods. Classic microbiological studies and limited molecular studies have shown that cyanobacteria dominate the surface layers and revealed five of the other main bacterial phyla (phylogenetic divisions Chloroflexi, Spirochaetes, Proteobacteria, Bacteroidetes, and Firmicutes) and thus indicated a relatively simple community with little deep evolutionary diversity (10, 39-42, 51, 57). The biological simplicity of the mats was the basis for their recommended use as model systems for metagenomic analyses (4). The dominance of cyanobacteria and the biological simplicity of the community have not been verified by culture-independent methods, however.
The aim of this study was a more comprehensive description of the microbial diversity within the mats and how that diversity is distributed in relation to depth and chemical gradients characteristic of the depth profile. We studied a limited area of one mat intensively. In situ gradients of O2 and H2S concentrations and pH, measured on a micrometer scale with microelectrodes, provided a backdrop of vertical chemical gradients onto which we mapped the biological data. ATP concentrations were measured to provide an estimate of living biomass distribution throughout the mat. Pigment concentrations measured by high-pressure liquid chromatography (HPLC) offered a view of the distribution of oxygenic chlorophyll a (Chl a)-containing cyanobacteria in relation to bacteriochlorophyll (BChl)-bearing anoxygenic photosynthetic bacteria. A survey of rRNA genes provided a culture-independent assessment of dominant organisms. For each of 10 layers that divided the mat into a millimeter-centimeter scale depth profile, the composition and diversity of communities were determined by sequence analysis of 16S rRNA genes generated by PCR with universal and bacterium-specific primers from community genomic DNA. We used RNA extraction and quantitative hybridization with group-specific probes as a PCR-independent verification of the abundance of the dominant group identified by sequence analysis and of the cyanobacteria. In addition, we visualized the morphologies and associations of these bacteria by fluorescence in situ hybridization (FISH) with tyramide signal amplification to overcome the intrinsic fluorescence of the mat. These studies collectively revealed unexpected diversity, complexity, and structure within the mat.
MATERIALS AND METHODS
Sample collection.
We studied the microbial mat underlying pond 4 (near 5) of the Exportadora de Sal SA, a solar saltworks located at Guerrero Negro, Baja California Sur, Mexico (see reference 38 for site details). The mat is covered by ∼1 m of brine with a salinity of ∼80‰. Collections were made in June and October of 2001 at 4 a.m. (night) and 1 p.m. (day). We collected replicate cores (1 cm by 6 cm) from a 0.4-m2 area of mat harvested approximately 10 m from the levee. Upon collection, cores were treated randomly in one of three different ways. Cores destined for DNA extraction and ATP and pigment analyses were sliced horizontally in 1-mm increments (from the top to a depth of 6 mm) and 1.5-cm depth increments (remainder of the core, to a depth of approximately 60 mm), and then the corresponding layers from five different cores were pooled, homogenized manually with a polypropylene pellet pestle (Thomas Scientific, Swedesboro, NJ), and frozen in liquid N2. Cores intended for RNA extraction and analysis were sliced lengthwise and frozen in liquid N2. Those intended for microscopy assays were fixed in 4% paraformaldehyde in pond brine for 4 h and preserved in 50:50 10× phosphate-buffered saline-ethanol at 4°C.
In situ microelectrode measurements.
The O2 concentration, H2S concentration, and pH within the microbial mat were measured at 200-μm intervals on the sample collection days in October 2001 with a diver-operated microprofiler (Unisense, Århus, Denmark). A total of 31 profiles were taken over the course of the diel period; the data shown are representative of eight qualitatively similar midafternoon profiles taken at three different mat locations. All profiles were made within the same 0.25-m2 area from which the mat cores were taken. The Clark-type oxygen microelectrode and amperometric sulfide microelectrode were calibrated as previously described (3, 38). A three-point calibration (pHs 4, 7, and 10) was performed for the pH microelectrode.
ATP analysis.
We measured ATP concentrations, a biomarker for biomass (28), in each section of the October and June daytime mat by using the luciferase enzyme assay (Molecular Probes, Carlsbad, CA). ATP was extracted from homogenized mat samples by treatment with 0.5 M H2PO4 for 20 min on ice, followed by centrifugation at 420 × g to remove suspended material and neutralization by addition of 1 M NaOH to a final pH of 7.4. Mat samples were washed twice with a 1.7 M glucose solution by centrifugation at 420 × g, followed by replacement of the supernatant with the glucose solution, prior to extraction, to remove salts that were found to inhibit the luciferase reaction. Light emitted from the ATP-activated luciferase reaction was quantified with a scintillation counter (Beckman Coulter, Inc., Fullerton, CA).
Pigment analysis.
Pigments were extracted from June and October daytime samples by sonication in acetone-methanol (7:2, vol/vol) in the dark, followed by centrifugation, filtration (0.45-μm pore size; Whatman), and injection into an HPLC column (25 cm by 4.6 mm; 5-μm Discovery C18; Supelco, Bellefonte, PA) as described in reference 37, with the following modifications. The time at 100% solution B was extended to 16 min to ensure that all long-tailed quinones were completely eluted. The HPLC diode array detection system consisted of an Agilent 1100 series binary pump (model G1312A), a vacuum degasser (model G1379A), a manual injector (model G1328A), and a diode array detector (model G1315B). The data were analyzed with ChemStation for LC 3D software (version A.08.03; Agilent Technologies, Waldbronn, Germany). Pigments were identified by absorption spectra and retention times. The concentrations of undegraded BChls were determined from the areas of the elution peaks with the equation m = FA (emd)−1, where m is the mass of BChl in milligrams, F is the rate of solvent flow through the column (1 ml min−1), A is the area of the elution peak, em is the extinction coefficient in liters per milligram per centimeter, and d is the detection length of the diode array detector (1 cm). The following extinction coefficients (in liters per milligram per centimeter) were used: Chl a, 79.2 (33); BChl a, 60 (45); BChl c, 86 (55); BChl d, 82 (55).
DNA extraction, PCR, and cloning.
DNA was extracted from frozen mat samples by a bead-beating protocol (14). For each sample, six replicate 25-μl PCR operations were performed, with each mixture containing 100 to 200 ng of purified genomic DNA, 100 mM Tris-HCl (pH 8.3), 500 mM KCl, 20 mM MgSO4, 200 μM deoxynucleoside triphosphates, 200 μM each forward and reverse primer, 1 M betaine, 800 μg/ml bovine serum albumin, and 1 U of Taq DNA polymerase (Invitrogen). We used the bacterium-specific forward primer 8F 5′-AGA GTT TGA TCC TGG CTC AG-3′, universal primer 515F 5′-GTG CCA GCM GCC GCG GTA A-3′, and archaeon-specific primer 333Fa 5′-TCC AGG CCC TAC GGG-3′, all coupled with universal reverse primer 1391R 5′-GAC GGG CGG TGW GTR CA-3′ (31). Cycling conditions were 94°C for 2 min, followed by 20 cycles of 94°C for 1 min, 45 s at 55°C, and 2 min at 72°C, with a final extension period of 20 min at 72°C. Replicate PCR products were pooled, and amplicons were gel purified (QIAGEN, Valencia, CA), cloned into TOPO TA pCR4.0, and transformed into Escherichia coli TOP10 cells (Invitrogen, Carlsbad, CA). The majority of the sequences were generated from October daytime samples after a pilot study of June day and night samples did not show statistically significant diurnal differences due to the unexpectedly high diversity (these sequences are included in the final data set). A subset of sequences was generated with forward primer 333Fa, which is usually employed to amplify archaea. Extraction controls (no mat material added) did not produce visible PCR products or colonies. For each sample-primer pair combination, 96 colonies were selected; strands of plasmids were sequenced with vector-specific primers with an ABI 377 DNA sequencer (BigDye Terminator ready reaction mixture; PE Applied Biosystems, Inc.).
Sequence and phylogenetic analysis.
Small-subunit rRNA sequences were edited and assembled into consensus sequences with PHRED and PHRAP aided by XplorSeq (Daniel Frank, unpublished), and bases with a PHRAP quality score of <20 were trimmed. Chimeras were detected with Bellerophon (23). Nonchimeric consensus sequences were named according to the layer they originated from (01 to 10), the time of collection (D for day, N for night), the month of collection (1 = June, 2 = October), the primer pair (X, Z, or ZZ = 8F-1391R, Y or YY = 515F-1391R, B = 333Fa-1391R; different symbols for the same primer pair indicate that different authors generated the sequence), and the clone number (01 to 96). The layers correspond to specific depths (1, 0 to 1 mm; 2, 1 to 2 mm; 3, 2 to 3 mm; 4, 3 to 4 mm; 5, 4 to 5 mm; 6, 5 to 6 mm; 7, 6 to 13 mm; 8, 13 to 26 mm; 9, 26 to 39 mm; 10, 39 to 60 mm). For example, clone 08D2Z44 was collected during the day in October from a 13- to 26-mm depth and generated with 8F-1391R.
Nonchimeric sequences were aligned with the Arb software package (36), based on an initial alignment described in reference 26. The alignment is available at http://Pacelab.colorado.edu/Publications/publications.html. Distance matrices generated in Arb (with Olsen correction) were used to cluster sequences into operational taxonomic units (OTUs) by pairwise identity (% ID) with a furthest-neighbor algorithm and a precision of 0.01 implemented in DOTUR (52). We used DOTUR to determine OTU frequencies in mouse cecal and human colonic 16S rRNA gene sequence data sets with Arb alignments provided by the authors (15, 32). Simpson's diversity index and collector's curves were calculated with EstimateS (8).
Assignment of the majority of sequences to their respective phyla was based on their position after parsimony insertion into the Arb dendrogram (omitting hypervariable portions of the rRNA gene with the lanemaskPH provided with the database; see http://Pacelab.colorado.edu/Publications/publications.html). Sequences that did not fall within described phyla were further characterized. Phylogenetic trees including the novel sequences and reference taxa (20) were constructed by evolutionary distance (test version 4.0b2 of PAUP*, a neighbor-joining algorithm with either Kimura two-parameter correction or maximum-likelihood [ML] correction with an empirically determined gamma distribution model of site-to-site rate variation and empirically determined base frequencies), parsimony (test version 4.0b2 of PAUP*; heuristic search), and ML (fastDNAml) analyses. Bootstrap resampling was used to test the robustness of inferred topologies. Novel candidate phyla, designated GN01 to GN15, were defined by the generally accepted criteria that (i) there must be three or more sequences from independent PCR products, (ii) sequences must be a minimum of 1,000 bp, and (iii) there must be high levels of support in phylogenetic analyses (20, 24, 46). However, four of the novel candidate phyla (GN6, GN12, GN13, and GN14) met only two of these criteria (the sequences were <1,000 bp) and await confirmation with longer sequence reads.
To cluster the communities from each layer, we used the UniFrac computational tool (35). The Arb alignment (excluding hypervariable regions) containing all 1,586 sequences was used to construct an ML tree with RAxML (54). The ML tree was annotated according to the layer from which each sequence was derived, and the fraction of tree branch length unique to any one layer in pairwise comparisons (the UniFrac metric) was calculated. Microbial communities from individual layers were clustered by application of the unweighted-pair group method using average linkages (UPGMA) to the UniFrac metric matrix.
FISH of Chloroflexi.
Mat samples were disrupted gently with a pestle, dehydrated in an ethanol series, and adhered to silane-coated glass slides with Cell-Tak (BD Biosciences, Inc.). Cells were permeabilized with lysozyme (10 mg/ml, 30 min, 37°C), followed by achromopeptidase (60 U/ml, 1 h, 37°C) and mutalysin (10×, 30 min, 37°C) (Sigma Aldrich, Inc.). Hybridizations were performed with a buffer containing 30% formamide, 0.9 M NaCl, 20 mM Tris-HCl (pH 8.0), 0.01% sodium dodecyl sulfate, and 50 ng of biotin-labeled probe for 2 h at 46°C. Signal intensity was boosted with TSA kit no. 22 with horseradish peroxidase-streptavidin and Alexa Fluor 488 tyramide by following the manufacturer's instructions (Molecular Probes, Carlsbad, CA). The probes used in this study were Chloroflexi-specific ChloroflexiB941 (5′-AAA CCA CAC GCT CCG CT-3′) (18) and bacterium-specific EUB338 (5′-GCT GCC TCC CGT AGG AGT-3′) (1). Samples were counterstained with 10 μg of 4′,6′-diamidino-2-phenylindole (DAPI)/ml and mounted with antifadent (CitiFluor Ltd., Leicester, England). Images were generated by laser confocal microscopy (Leica Microsystems, Bannockburn, IL) with a 488-nm excitation laser and a 350-nm excitation laser (DAPI). Probes and protocols were tested against reference strains (Chloroflexi Thermomicrobium roseum ATCC 27502, Chloroflexus aurantiacus ATCC 23779, Roseiflexus castenholzii obtained from S. Hanada [19], and Herpetosiphon aurantiacus ATCC 23781; Chlorobium tepidum ATCC 49652; proteobacteria Pseudomonas aeruginosa ATCC 14205 and E. coli ATCC 29181; Firmicutes Clostridium sporogenes ATCC 13663 and Bacillus subtilis ATCC 12432; and the archaeon Sulfolobus acidocaldarius ATCC 33909).
Quantitative dot blot hybridization of rRNA from Chloroflexi and cyanobacteria.
To extract bulk RNA from the mat, triplicate mat samples (100 mg) from three depths (0 to 4 mm, 5 to 9 mm, and 20 to 30 mm) were washed with 1.7 M glucose amended with RNase OUT (Sigma Aldrich, St. Louis, MO) to remove excess salts and resuspended in 750 μl of pH 4.7 buffer containing 100 mM EDTA, 20 mM Na-acetate, and 0.5 μl/ml RNase OUT. Aliquots (100 μl) were further homogenized by bead beating (2 min on high with 0.1-mm zirconium beads; Mini-BeadBeater-8; BioSpec Products, Inc., Bartlesville, OK) in Tri-Reagent (Sigma Aldrich, St. Louis, MO). RNA was extracted similarly but without the glucose wash from the pure cultures previously mentioned in the FISH protocol. In addition, RNA was obtained from the following cyanobacteria: Synechococcus sp. strain UTCC477, Lyngbya sp. strain UTCC592, and Oscillatoria sp. strain UTCC487. To obtain small-subunit rRNA from reference organisms without cultured representatives, RNA was synthesized by transcription from rRNA gene clones. We transcribed RNA from two cyanobacterial clones (01D2Z20 and 05D2Z68), and five Chloroflexi clones (10D2Z49, 08D2Z44, 05D2Z83, 05D2Z90, and 05D2Z56) chosen to represent the diversity of those groups. Plasmids with cloned rRNA gene inserts were prepared from E. coli cultures with a QIAGEN mini-prep kit. Plasmids were linearized by restriction enzyme digestion (SpeI, 37°C, 2 h). Transcription reaction mixtures incubated at 37°C overnight contained the following per 100 μl: 6 to 8 μg of linearized plasmid, 52.5 U of yeast pyrophosphatase, 7 μl of RNase OUT T7 RNA polymerase, 28 mM each ribonucleotide, and 0.175 M MgCl2 buffer. Newly synthesized RNA was ethanol precipitated and isolated by electrophoresis in denaturing polyacrylamide gels, followed by nondenaturing ion-exchange chromatography on HiTrap Q HP Sepharose (Amersham Pharmacia) (29).
RNAs from mat samples and reference strains were blotted in triplicate onto nylon membranes (Magna Charge; Micron Separation Inc., Westboro, MA) and bound by baking at 80°C. Probes were labeled with γ-ATP and T4 polynucleotide kinase by following the manufacturer's instructions (Invitrogen, Carlsbad, CA). Washing temperatures for each probe were determined empirically according to reference 11, with modifications. We used 200-μl assay volumes in 96-well plates and controlled the temperature with a thermocycler. Membranes were hybridized as described in reference 11. Signal intensity of the blots was determined with a Phosphorimager (Molecular Dynamics, Carlsbad, CA) and the program ImageQuant (Amersham Biosciences, Piscataway, NJ). Signal intensities were analyzed as described in references 47 and 48.
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to the GenBank database and assigned accession numbers DQ329539 to DQ331020 and DQ397339 to DQ397511.
RESULTS
Depth gradients of H2S, O2, and pH.
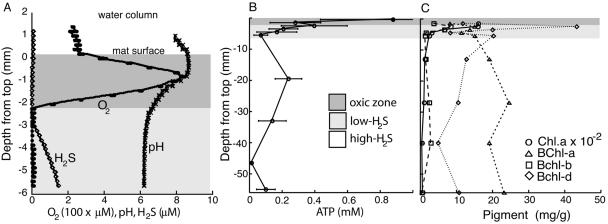
The in situ chemical environment of the mat was profiled by microelectrode measurements of O2 and H2S concentrations and of pH from the surface of the mat to a depth of 6 mm in 200-μm increments. The high spatial resolution of these measurements provides a view of the variability of the local chemistry. Based on our microelectrode measurement profiles, the mat can be divided into three distinct habitats that serve as a backdrop for the spatial organization of the microbial community (Fig. 1A). These zones are the oxic zone, ranging from the top of the mat to a depth of 2 mm and characterized by diurnally fluctuating concentrations of O2; the low-H2S zone, ranging in depth from 2 mm to 6 mm, where H2S levels are drawn down diurnally; and the H2S-rich zone, the largest zone, ranging from 6 mm to the bottom (∼60 mm), where concentrations of H2S are permanently high (Fig. 1A). The coefficient of variation for the O2 measurements was greatest at an average depth of 2.5 mm, where the average O2 concentration was low (28 μM; Fig. 1A). This is an indication that the deepest penetration of oxygen into the mat is also the depth at which oxygen concentrations are the most variable (measured range, 0 to 522 μM) during the day. At night, the mat is completely anoxic.
FIG. 1.
Chemical and biochemical characteristics of the mat as a function of depth. (A) Microelectrode measurements of O2 and H2S concentrations and pH. (B) ATP concentrations. Means of three independent measurements are plotted; the bars show standard errors. (C) Pigment concentrations. For all measurements, October values are plotted; the June values were equivalent (data not shown).
Biomass and pigment distributions by depth.
To determine the depth distribution of biomass in the mat, we measured the concentration of ATP per gram, from the top of the mat to the bottom, in June and October. ATP is the energy currency of all cells, and it therefore serves as a proxy for biomass (28). ATP concentrations were highest in the oxic zone (1,343 ± 400 ng of ATP g−1; Fig. 1B) and tapered off rapidly with depth to 322 ± 50 ng of ATP g−1in the lower, H2S-rich, zone. Integrating by depth, the substantially greater volume of the lower H2S-rich zone results in fivefold greater overall biomass per unit of surface area: 27.8 μmol of ATP cm−2 in the lower zone versus 5.0 μmol of ATP cm−2 for the combined oxic and low-H2S zones. The ATP concentrations and depth profiles of the October and June sampling dates were similar.
We determined the concentrations and distributions of photosynthetic pigments by HPLC. Pigment concentrations were highest in the oxic zone, as expected, yet we detected pigments at all depths (Fig. 1C). The cyanobacterial pigment Chl a was an order of magnitude more abundant than BChls (BChls a, d, and c) in the oxic zone.
Estimated richness and coverage.
Two rRNA gene clone libraries were constructed from each of the 10 layers, one with bacterium-specific primers (8F and 1391R) and one with universal primers (515F and 1391R). Restriction fragment length polymorphism screening revealed unexpectedly high levels of diversity for each 96-clone library; therefore, all clones were sequenced bidirectionally. When the universal libraries from all layers were combined, the bacteria/archaea/eucarya ratio was 57:7:1. In this report, we focus on the bacterial sequences. Archaeal and eucaryal sequence data will be described elsewhere (J. Spear, R. Ley, and N. R. Pace, unpublished). Bacterial sequences encountered in the libraries constructed with archaeal primers were included in the final data set discussed here. The two forward primers (515F and 8F) used to generate the majority of the sequences yielded an equivalent proportion of bacterial phyla (χ2, P < 0.05), an indication that our coverage was not significantly biased by the primer pairs that we used. For subsequent analyses, all sequences were combined on the basis of the layer from which they originated.
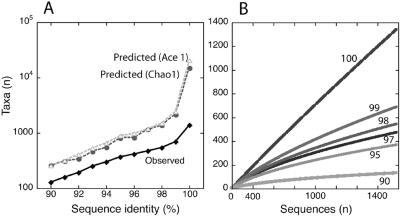
To assess the coverage and the richness of clone libraries combined by layer, we employed collector's curves for coverage, the nonparametric estimators Chao1 and Ace1 for richness, and the computed Simpson index to estimate the evenness of community composition (see reference 52). The total bacterial 16S rRNA gene sequences (n = 1,586) consisted of 1,336 unique sequences and 752 phylotypes defined by a minimum threshold of 99% ID. In all subsequent analyses, we report masked sequence pairwise identities (% IDs). The hypervariable regions of sequences were masked for alignment purposes because these regions cannot be aligned with certainty, particularly across the large phylogenetic distances encountered in this data set. Generally, 99% ID is equivalent to ∼97% ID when the entire sequence length is used (e.g., see the data set of 11,831 bacterial 16S rRNA genes in reference 15), often taken to indicate species level variation. Figure 2A shows the frequency of observed taxa with % ID thresholds ranging from 90% to 100%. The Chao1 and the Ace1 richness estimates for phylotypes ranging from 90% ID to 100% ID yielded equivalent richness curves (Fig. 2A). Both richness estimators indicated >10,000 unique sequences based on the distribution of observed sequences. Collector's curves for taxa with >90% ID indicate that our coverage of the diversity was not comprehensive, since the curves did not begin to become asymptotic (Fig. 2B). Together, the richness estimators and the collector's curves indicate a high degree of diversity, most of which likely remains undescribed. Bacterial diversity was uneven in the upper layers sampled, which had a subset of dominant bacteria, and became more even with depth (Simpson's index versus depth, R2 = 0.63, P < 0.005).
FIG. 2.
Bacterial diversity within the hypersaline microbial mat. (A) Observed and predicted (Chao1, Ace1) numbers of taxa with minimum thresholds ranging from 90 to 100% ID for masked sequences. (B) Collector's curves for taxa (OTUs) with minimum thresholds of 90, 95, 97, 98, 99, and 100% ID.
Bacterial diversity and distribution.
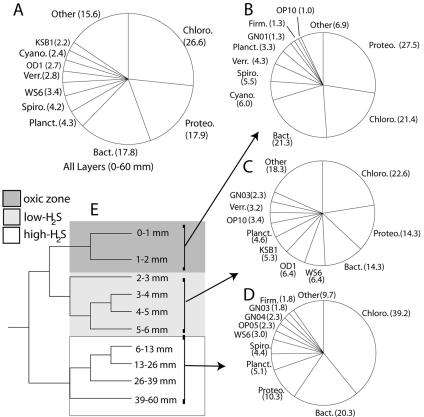
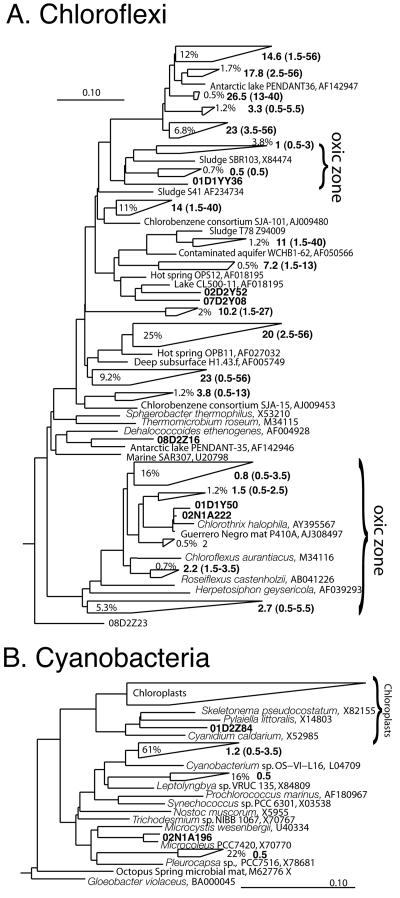
The phylum Chloroflexi dominated clone libraries numerically and included the highest proportion of sequences, on average (Fig. 3A), from each chemically defined zone (Fig. 3B, C, and D). Proteobacteria and Bacteroidetes were the second most represented phyla in each zone. Cyanobacterial sequences comprised, at most, 10% of the total sequences in a chemical zone (Fig. 3B) and were obtained from the oxic zone only. Figure 4 shows the phylogenetic relationships of the Chloroflexi and cyanobacterial sequences obtained from the mat in relation to cultured representatives and clones obtained by culture-independent methods from other environments. Twenty-four percent of the Chloroflexi sequences were obtained from the surface layers and were close relatives of known photosynthetic organisms, such as Chloroflexus and Chlorothrix spp., previously described for these mats (30, 41) (Fig. 4A). Another group of Chloroflexi sequences (4.5% of Chloroflexi sequences) with no cultured close relative was obtained only from the surface layers (Fig. 4A). This result indicates that the Chloroflexi may include additional unrecognized photosynthetic members. The other Chloroflexi groups recovered from the mat had average depth distributions extending below the oxic zone. These groups include oxygen-tolerant members, however, since members of all groups were obtained from the oxic zone. The majority of sequences have no closely related cultured representatives from which properties can be inferred. Sixty-one percent of the cyanobacterial sequences obtained from the mat were members of a novel group of cyanobacteria whose closest relative was obtained from a hot spring mat (Fig. 4B). Sixteen percent formed a group affiliated with Leptolyngya spp.
FIG. 3.
Bacterial diversity in the mat. Proportions of bacterial phyla in the total data set (A), in the oxic zone (0 to 2 mm) (B), in the low-H2S zone (2 to 6 mm) (C), and in the H2S-rich zone (6 to 60 mm) (D) are shown. Others: cyanobacteria, KSB1, OP10, GN03, OP5, GN1, Firmicutes, OP11, GN04, GN05, GN09, GN10, WS1, WS2, GN2, Deinococcus-Thermus, GN07, Haloanaerobiales, GN06, GN11, BRC1, OP8, OS-K, GN12, GN13, GN14, actinobacteria, GN15, WS3, GN8, OP9, TM6, and VadinBE97. Abbreviations: Chloro., Chloroflexi; Cyano., Cyanobacteria; Verr., Verrucomicrobia; Planct., Planctomycetales; Spiro., Spirochaetales; Firm., Firmicutes, Bact., Bacteroidetes; Proteo., Proteobacteria. (E) Bacterial community clustering by layer studied (UPGMA tree of UniFrac metric based on 1,585 16S rRNA gene sequences). Shaded areas refer to the different chemical milieus identified by the microelectrode measurements in Fig. 1A.
FIG. 4.
Diagrammatic phylogenetic trees of microbial mat sequences and their cultured and uncultured relatives with associated GenBank accession numbers. Reference sequences of cultured representatives are shown in italics. Wedges represent groups of microbial mat sequences, and single sequences are indicated by their clone names. The length of the top and bottom edges represents the range of sequence divergence. The average depth from which sequences were obtained is indicated next to the wedge, with the total depth range in parentheses. (A) Chloroflexi sequences. Percentages indicate the fraction of Chloroflexi sequences within a given sequence cluster. “Oxic zone” indicates clusters of sequences obtained from surface layers exclusively. (B) Cyanobacterial sequences. Percentages indicate the fraction of cyanobacterial sequences within a given sequence cluster.
In addition to Chloroflexi and Cyanobacteria, the 16S rRNA gene sequence analysis revealed 28 other previously described phyla (Fig. 3). Half of these were candidate phyla, so termed because they are known only by 16S rRNA gene sequences and do not contain representatives that have been cultured in the laboratory and from which physiological and metabolic properties can be inferred. This study expanded the known habitat space and diversity of several candidate phyla. For example, candidate phylum KSB1, previously represented by <10 sequences encountered in surveys of brackish water sediments (59), cave sediments (22), and a bioreactor treating 4-methylbenzoate (62), was expanded significantly with the addition of 35 sequences from the mat.
The majority of sequences could be assigned unambiguously to known phyla; however, 119 sequences remained (7.5% of the total) that were not affiliated with any known phylum. Phylogenetic analysis of these unaffiliated sequences revealed 15 novel candidate phyla termed GN01 to GN15. Several of the candidate phyla include sequences that were previously deposited in GenBank and for which there is no described affiliation (Table 1; a dendrogram is available in the Arb database at http://Pacelab.colorado.edu/Publications/publications.html). Most of the novel GN candidate phyla include sequences derived from several different layers and therefore several separate PCR products. Novel GN candidate phyla were detected in all layers of the mat, and more than half were detected in the oxic zone (Table 1).
TABLE 1.
Mat layers from which sequences forming candidate phyla GN01 to GN15 were obtaineda
| Phylum | Depth(s) in mm (no. of sequences) | Accession no. of clones from other studies |
|---|---|---|
| GN14 | 0-1 (2) | |
| GN07 | 0-1 (2), 3-4 (1), 4-5 (1) | |
| GN03 | 0-1 (2), 2-3 (1), 4-5 (7), 5-6 (9), 6-13 (2), 13-26 (5) | |
| GN10 | 1-2 (3), 2-3 (1), 3-4 (1), 4-5 (1) | |
| GN01 | 1-2 (8), 2-3 (1), 3-4 (6), 4-5 (1), 5-6 (6) | |
| GN05 | 1-2 (1), 2-3 (1), 3-4 (3), 4-5 (4), 5-6 (2), 26-39 (1), 39-60 (1) | DQ154857 |
| GN09 | 2-3 (1), 3-4 (1), 4-5 (1), 5-6 (1), 26-39 (6) | |
| GN08 | 3-4 (1) | AJ441248 |
| GN11 | 3-4 (1), 4-5 (1), 5-6 (1) | AY255001 |
| GN02 | 3-4 (2), 4-5 (2), 13-26 (1) | |
| GN12 | 4-5 (1), 5-6 (1) | |
| GN13 | 5-6 (1), 6-13 (1) | |
| GN06 | 5-6 (2), 26-39 (1) | AB089123, DQ154831, AB218870 |
| GN04 | 5-6 (3), 6-13 (1), 13-26 (3), 26-39 (4), 39-60 (2) | AF323768 |
| GN15 | 26-39 (2) | AJ567570 |
Values indicate the depths in the mat from which candidate phylum clones were obtained; the number of sequences from each depth is in parentheses. The GenBank accession numbers are those of clones from other studies without prior phylum affiliations that are included in GN candidate phyla. See http://Pacelab.colorado.edu/Publications/publications.html for the Arb dendrogram showing these phyla in the context of previously described phyla.
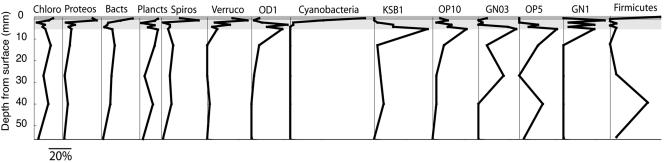
Figure 5 shows the depth distribution of members of the 14 most commonly observed bacterial phyla. The abundance of each is shown as the percentage of sequences at each depth, calculated as a fraction of the total number of sequences obtained for any given phylum. The Chloroflexi bacteria are distributed fairly evenly with depth, except for a relative reduction in abundance in the transitional low-H2S zone. The phyla that were most abundant in the oxic zone included Cyanobacteria, Proteobacteria, Bacteroidetes, Spirochaetes, Verrucomicrobia, and candidate phylum GN01. Several known candidate phyla (KSB1, OP10, OP5, and OD1) had their highest abundances in the low-H2S zone, just below the oxic layer. Firmicutes exhibited a marked bimodal distribution, abundant in the oxic zone and in the lower portion of the mat.
FIG. 5.
Depth distributions of the 10 most abundant phyla in the mat. Points indicate the percentage of sequences within each phylum (not the percentage of total sequences) obtained at each depth. The bar indicates 20% of sequences within each group. Shaded areas: see legend to Fig. 2E.
Community similarities.
In order to compare the communities within the chemical zones characterized by the microelectrode measurements of O2 and H2S, we used the recently developed UniFrac metric analysis (35). UniFrac measures the phylogenetic distance between pairs of communities represented by sequences in a phylogenetic tree as the fraction of branch length of the tree that leads to descendants from either one community or the other but not both (35). A phylogenetic tree containing all of the microbial mat sequences (n = 1,585) was generated, each sequence was annotated according to the analytical layer from which it was derived, and the UniFrac for all combinations of pairs of communities (10 × 10) was computed. The layers were then clustered according to their pairwise UniFrac metrics by UPGMA. This analysis revealed three main clusters of related communities that match the zones delimited by the chemistry of the mat. The communities in layers 0 to 1 mm and 1 to 2 mm clustered together, as did the four communities obtained from layers 2 to 6 mm, and the four communities from the lower layers clustered together (6 to 60 mm; Fig. 3E). The robustness of the inferred UniFrac tree topology to the presence of specific communities represented was confirmed by jackknife analysis (P < 0.001).
Quantitative RNA hybridizations with probes specific for Chloroflexi bacteria and cyanobacteria.
In order to test the abundances of Chloroflexi bacteria and cyanobacteria indicated by the clone library results that may be subject to PCR and other biases (24), we conducted quantitative rRNA dot blot hybridizations with oligonucleotide probes based on the sequences. We used pure cultures of members of the phyla Chloroflexi and Cyanobacteria to generate reference rRNAs. To represent novel Chloroflexi clades detected by rRNA gene analysis for which no cultures are available, we synthesized rRNA by in vitro transcription from selected clones. Overall, the majority of the community rRNA extracted from the mat hybridized with the Chloroflexi-specific probe, confirming the dominance of Chloroflexi observed in the rRNA clone libraries. However, the majority of the total rRNA in the oxic zone was cyanobacterial (87% ± 2.7%), while Chloroflexi likely comprised the majority of the remainder (22% ± 1.7%). Together, these percentages exceed 100%, which indicates that some nonspecific binding of the probes may have occurred. In the low-H2S zone, the proportion of cyanobacterial rRNA dropped to 28% ± 3.6% while the proportion of Chloroflexi rRNA increased to 41% ± 0.8%. Deep in the H2S-rich zone of the mat, cyanobacterial abundance declined further to 10% ± 2.2%, while Chloroflexi remained a dominant proportion of the overall biomass (32% ± 4.5%).
Visualization of Chloroflexi bacteria by FISH.
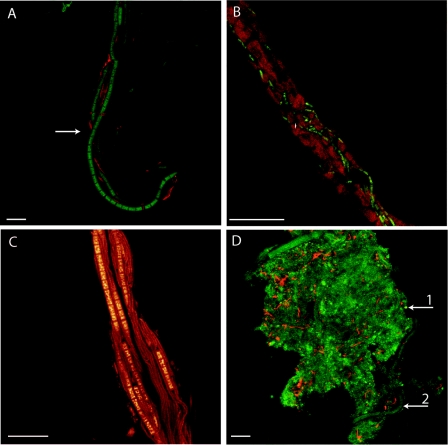
Because of their abundance, the Chloroflexi bacteria must have a major influence on the nature of this mat. In order to gain insight into the ecology and spatial organization of these organisms, we visualized the Chloroflexi bacteria in the upper and lower layers of the mat with Chloroflexi-specific fluorescent oligonucleotide probes by in situ hybridization and confocal microscopy. Tyramide signal amplification combined with the use of Alexa Fluors boosted the signal above the high levels of background autofluorescence. Chloroflexi bacteria were seen within the exopolysaccharide sheaths of the conspicuous filamentous cyanobacterium Microcoleus chthonoplastes in the upper layers of the mat (Fig. 6A, B, and C). The invasion of the trichomes seemed specific for Chloroflexi bacteria. No other bacteria were seen within the sheaths, although many were visualized by DAPI staining on the outside of the sheaths (not shown). The filaments of cyanobacterial cells were often observed disrupted in the presence of the Chloroflexi bacteria (Fig. 6B). In the lower, anoxic zones of the mat, the Chloroflexi bacteria were visualized as filamentous bacteria pervading the polymeric matrix of the mat, but they were not observed inside sheaths (Fig. 6D).
FIG. 6.
Chloroflexi bacteria and the cyanobacterium M. chthonoplastes in the mat visualized by laser confocal microscopy. (A) Chloroflexi bacteria (red, FISH Chloroflexi probe) entwined with M. chthonoplastes (green, DAPI) at a 1-mm depth. The arrow indicates the edge of the polysaccharide sheath. (B) Chloroflexi bacteria (green, Chloroflexi probe) and M. chthonoplastes (green, autofluorescence of Chl a). (C) Chloroflexi bacteria (thin filaments) and M. chthonoplastes (thick filaments), DAPI stained. (D) Chloroflexi filaments (red, Chloroflexi probe) and polysaccharide material (dull green) at a 50-mm depth. Non-Chloroflexi bacteria are visible as bright green spots (arrow 1). Arrow 2 indicates a buried M. chthonoplastes filament. Scale bars, 10 μm.
DISCUSSION
Microbial mats occur worldwide in shallow aquatic environments where high salinity or temperature precludes the establishment of algae or aquatic plants that may overgrow the mats or of grazers that may otherwise consume them (17). Prior to the evolution of such organisms, microbial mat-like structures were widespread around shallow seas and lakes and are thought to have contributed significantly to the evolution of Earth's atmosphere (21). The biogeochemical cycling of carbon, oxygen, and hydrogen has been studied extensively in the hypersaline mats of Guerrero Negro, Baja California Sur, Mexico (5, 12, 13, 61), yet the identities of the organisms that drive these cycles are known primarily from culture and microscopy studies (9) and only limited molecular analyses have been performed (39-41, 51). This study is the most extensive rRNA-based survey of a microbial mat so far conducted. Furthermore, we integrated a variety of molecular and chemical analytical approaches to characterize the depth profile of the mat. In contrast to the long-held view that these mats are dominated by cyanobacteria and are biologically simple, our results indicate an overall unexpected dominance of Chloroflexi bacteria and a remarkably high level of diversity. We found that cyanobacteria dominate the biomass in the upper 2 mm; however, the dominance of Chloroflexi bacteria below 2 mm, where the majority of the biomass resides, makes them the most abundant type of bacterium overall. In the upper 2 mm, the Chloroflexi bacteria can be seen intertwined within the exopolysaccharide sheaths of the dominant cyanobacterium. Together, our results indicate that the photosynthetic activity of the cyanobacteria sustains a highly diverse and structured community.
Chemically defined niches and biomass distribution.
Our microelectrode measurements of H2, O2, and pH provided a detailed view of the chemical environment in the upper 6 mm of the mat. These chemical profiles defined three distinct chemical niche spaces onto which to map the biological diversity of the mat. These three zones, the oxic zone, the low-H2S zone, and the H2S-rich zone, have been described previously with coarser spatial resolution (5).
In previous studies, the density of the cells was observed qualitatively to be highest in the upper few millimeters and to taper off with depth (10). Bulk RNA concentrations have also been shown to be higher by an order of magnitude in the upper 1 mm than in the underlying layers (51). We measured ATP concentrations in the mat as a proxy for biomass. ATP concentrations approached those of pure cell paste in the uppermost millimeters of the mat. The upper few millimeters are considered to be the highly active zone of the mat, where carbon flux driven by photosynthesis is highest. However, even though ATP concentrations were lowest below the oxic zone, the total amount of ATP present per unit of area in the lower portion of the mat is highest below the oxic zone. Therefore, more overall biomass resides in the dark sulfidic part of the mat than in the cell-dense upper few millimeters, where most photosynthesis occurs.
Pigment depth profiles showed that the cyanobacterial pigment Chl a was by far the most abundant photosynthetic pigment. It is therefore likely that cyanobacteria fix more inorganic C than the nonoxygenic phototrophic bacteria (e.g., Chloroflexi bacteria and proteobacteria) and thus the cyanobacteria presumably provide the main sustenance of the mat, as previously thought. We measured peak concentrations of the BChls directly under the peak concentration of Chl a. This segregation with depth is consistent with the specialization of BChls for longer, more deeply penetrating wavelengths and the previously reported vertical stratification of different phototrophic organisms (10, 56). The photosynthetic Chloroflexi bacteria (e.g., Chlorothrix halophila and relatives), which are similar in properties to green sulfur bacteria by having chlorosomes, may also contribute in a substantial way to the primary productivity of the mat. Taken together, the biomass and pigment depth distributions indicate that photosynthetic activity within the oxic zone fuels the underlying, larger biomass. Such an inverted pyramid model of trophic levels, where consumer biomass apparently outweighs producer biomass, has recently been described for the stromatolites (calcified mats) of Sharks's Bay, Australia (44) and may be common in microbial ecosystems where primary producers exude polysaccharides.
Diversity mapped onto chemical gradients.
We generated 1,586 bacterial 16S rRNA gene sequences from the entire depth profile of the mat, although this number appears to undersample woefully the diversity present in the mat. Together, the diversity estimates based on our sequence coverage and the collector's curves all indicate a highly diverse community, most of which still awaits description. Despite the low sampling coverage, the high proportion of known and new bacterial phyla in this hypersaline mat makes it the most biologically diverse environment yet characterized. This study has encountered the highest number of confirmed and potential candidate phyla so far observed in a single environment (25). This is remarkable, particularly at a time when the discovery rate of new phyla was thought to be tapering off (46), and suggests that diversity and complexity are a feature of the microbial mat ecosystem.
Previous microscopy-based studies have indicated a vertical stratification of the microbial community with depth, down to about 6 mm, the maximum depth studied (10). Cyanobacteria, particularly the filamentous, exopolysaccharide sheath-forming species M. chthonoplastes, were observed by microscopy in the upper 2 mm. Other, anoxygenic photosynthetic bacteria such as Chloroflexus-like Chloroflexi bacteria were observed below the cyanobacteria along with other undefined microbiotas with various morphologies (10). We found cyanobacterial rRNA genes in the uppermost layers only, and the majority of the RNA in the upper few millimeters was cyanobacterial. Furthermore, cyanobacterial Chl a was far more abundant in the upper few millimeters than BChls c and d, ascribed to the Chloroflexi bacteria. The discrepancy between Chloroflexi bacterial and cyanobacterial representation in 16S rRNA gene libraries versus bulk rRNA and pigment profiles in the upper millimeters could be due to the comparatively large size of cyanobacteria. For instance, M. chthonoplastes cells are two to three times wider than Chloroflexi bacterial cells, which may result in a higher ratio of expressed rRNA to rRNA gene copy number. Nonetheless, overall, the Chloroflexi bacteria dominated the mat biomass, with the exception the top 2 mm, where cyanobacteria were a larger proportion of the biomass. Popular reference to the mats as “cyanobacterial” reflects the focus of most previous work on the top few millimeters.
The majority of the novel candidate phyla that we describe from the mat were detected at a variety of depths. Candidate phyla are known from their constituent 16S rRNA gene sequences alone, and therefore the physiological attributes of the bacteria within them can be gleaned only from the context in which they were discovered. The depth distributions of the novel GN candidate phyla provide testable hypotheses about the physiologies of the organisms. More importantly, the spatial distribution of the candidate phyla can help direct efforts to bring representatives into culture for physiologic studies. The middle, low-H2S zone harbored 9 of the 15 GN candidate phyla, indicating that such organisms are anaerobic and H2S tolerant yet may rely on the diurnal variation in H2S levels and the H2S-O2 interface. In contrast, candidate phyla GN4 and GN12 were detected only well below the oxic layer and are likely to be composed of strict anaerobes. GN14 was observed only in the top layer (day and night libraries), which suggests that it could be a novel photosynthetic group.
The three distinct zones delineated by concentration gradients of O2 and H2S harbored distinct bacterial communities. The UniFrac analysis of the phylogenetic tree representing the entire data set resulted in clusters of the 10 separate samples according to the depth distributions of the three chemical habitats delineated by our in situ microelectrode measurements. This observation implies that related bacteria occupy similar chemical niches. Therefore, despite the potential for horizontal gene transfer to confer many physiological traits on distantly related bacteria, phylogenetic groups appear to share physiological properties that are manifested as chemical niche preferences at particular depths in the mat. For many of the novel phylotypes observed, the depth at which they were found is consistent with what is known about the physiologies of their close relatives. For instance, close relatives of known photosynthetic organisms, such as Chloroflexi relatives of the photosynthetic Chloroflexus and Chlorothrix spp. previously described for these mats (30, 41), were abundant in the upper zone, where light penetrates. Similarly, close relatives of known sulfate-reducing members of the delta group of proteobacteria were most abundant in the oxic zone, which also is where rates of sulfate reduction are highest (39). Members of the phylum Bacteroidetes, known to degrade polysaccharide anaerobically (63), were abundant throughout the dark anoxic zone.
Chloroflexi bacteria and cyanobacteria: symbiosis or antagonism?
A previously described feature of hypersaline microbial mats is the close physical interaction between the cyanobacterium M. chthonoplastes and a thinner filamentous bacterium also inside the polysaccharide sheath. The thinner partner was thought to be a member of either Proteobacteria or Chloroflexi based on physiological properties and transmission electron microscopy (9). We used FISH with Chloroflexi bacterium-specific probes and laser confocal microscopy to confirm that the filamentous bacteria inside the M. chthonoplastes sheaths were Chloroflexi bacteria (Fig. 6A to C). The association is most often observed at 0.3 to 1.2 mm (5, 9), corresponding to the zone below the region of maximal oxygenic photosynthesis. Indeed, lower in the mat, Chloroflexi bacteria are free living (Fig. 6D). The depth location of the association and physiological experiments (9) have suggested a cometabolism of sulfur: the Chloroflexi bacteria may draw down levels of H2S stressful for the cyanobacterium (38), which excretes organic carbon used by the Chloroflexi bacteria. However, we observed that M. chthonoplastes filaments were often disrupted when Chloroflexi bacteria were present (Fig. 6B), suggesting the alternative view that Chloroflexi bacteria may parasitize the cyanobacterium under H2S stress. The tight physical association of these bacteria from deeply divergent lineages is an example of the physical and chemical microbial interactions that build complexity in the mat.
Comparisons with other systems: chemical diversity drives microbial diversity.
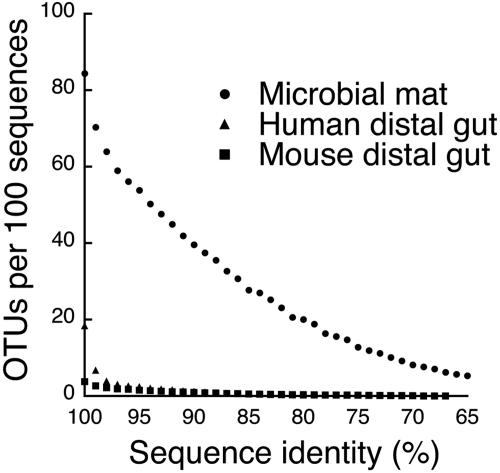
Comparisons of diversity levels between environments with contrasting characteristics can yield valuable insight and testable hypotheses regarding the rules that govern diversity distributions. For instance, as shown in Fig. 7, the very high level of phylum diversity that we found in this study is a sharp contrast to the relatively low level encountered in the human and mouse distal guts. Furthermore, whereas the diversity in the distal gut is characterized by a few deep lineages that diversified at phylogenetically “shallow” levels (more closely related, e.g., at the species-strain level) (2), the microbiota of the microbial mat is far more diverse at all phylogenetic levels (Fig. 7). The difference in phylogenetic structure between these two microbial systems is most likely due to the far greater diversity of chemical niches in the mat, which allow more opportunities for specialization within the microbiota. The mammalian distal gut, like microbial mats, is an energy-rich microbial ecosystem where polysaccharides form scaffolds that provide attachment sites and nutrients for bacteria (53). Fermentation dominates organic matter diagenesis in both systems, although sulfate reduction and methanogenesis also occur in both systems (12, 43). In microbial mats where sulfide production levels are high, however, a larger proportion of electrons produced by diagenesis is likely shunted to sulfate. The prominence of the sulfur cycle in mats allows the exploitation by the microbiota of many intermediate chemical moieties (61). Another niche available in mats but not in the distal gut is phototrophy: the differential depth penetration of light wavelengths creates a stratification of phototrophs adapted to the use of particular wavelengths of light (6, 7, 27, 50, 56). Additionally, the frequent washout inherent to the gut undoubtedly is a powerful selection force against slow-growing microbes and the accretion of community structures important in mats. Thus, the relative complexity of the microbiota in microbial mats probably correlates with broad niche space. Furthermore, biological diversity itself can drive diversity through niche creation (16); therefore, chemical and biological diversity is expected to form positive feedback loops.
FIG. 7.
Phylogenetic structure of microbial mat (n = 1,585; this study), human colonic (n = 11,831) (15), and mouse cecal (n = 5,088) (32) 16S rRNA gene sequence data sets. Sequences were clustered into phylotypes based on percent sequence identity (OTUs with similarity thresholds ranging from 65% ID to 100% ID). The ratio of phylotypes at each threshold to the total sequence in each data set is plotted.
Microbial mats have been called simple systems based on microscopy and culture studies. However, our molecular analysis revealed that the hypersaline microbial mats of Guerrero Negro harbor the most complex bacterial assemblage documented to date in any environment, with 42 phyla, including 15 novel candidate phyla. Microbial mats are hot spots of bacterial diversity and constitute a rich reservoir of gene diversity for future studies of bacterial evolution and genomic diversity.
Acknowledgments
We thank D. DesMarais, A. Dalby, L. Angenent, S. Hanada, A. St. Amand, and J. Gordon for valuable assistance and the Exportadora de Sal of Guerrero Negro, Baja California Sur, Mexico, for access to sites.
R. Ley was supported in part by an NRC-NASA Astrobiology Institutes postdoctoral associateship, and J. Spear was supported by an Agouron Institute postdoctoral fellowship. This work was supported by the NASA Cooperative Agreement with the University of Colorado Center for Astrobiology to N. R. Pace.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisolm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 3.Bebout, B. M., S. P. Carpenter, D. J. DesMarais, M. Discipulo, T. Embaye, F. Garcia-Pichel, T. M. Hoehler, M. Hogan, L. L. Jahnke, R. M. Keller, S. R. Miller, L. E. Prufert-Bebout, C. Raleigh, M. Rothrock, and K. Turk. 2002. Long-term manipulations of intact microbial mat communities in a greenhouse collaboratory: simulating Earth's present and past field environments. Astrobiology 2:383-402. [DOI] [PubMed] [Google Scholar]
- 4.Buckley, M. R. 2004. The global genome question: microbes as the key to understanding evolution and ecology. American Academy of Microbiology, Washington, D.C. [PubMed]
- 5.Canfield, D. E., and D. J. DesMarais. 1993. Biogeochemical cycles of carbon, sulfur, and free oxygen in a microbial mat. Geochim. Cosmochim. Acta 57:3971-3984. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, Y. 1984. The Solar Lake cyanobacterial mats: strategies of photosynthetic life under sulfide, p. 133-148. In Y. Cohen, R. W. Castenholz, and H. O. Halvorson (ed.), Microbial mats: stromatolites. Alan R. Liss, Inc., New York, N.Y.
- 7.Cohen, Y., and E. Rosenberg. 1989. Microbial mats: physiological ecology of benthic microbial communities. American Society for Microbiology, Washington, D.C.
- 8.Colwell, R. K. 2005. EstimateS: statistical estimation of species richness and shared species from samples. Version 7.5. User's guide and application published at: http://purl.oclc.org/estimates.
- 9.D'Amelio, E. D., Y. Cohen, and D. J. DesMarais. 1987. Association of a new type of gliding, filamentous, purple phototrophic bacterium inside bundles of Microcoleus chthonoplastes in hypersaline cyanobacterial mats. Arch. Microbiol. 147:213-220. [DOI] [PubMed] [Google Scholar]
- 10.D'Amelio D'Antoni, E., Y. Cohen, and D. J. DesMarais. 1989. Comparative functional ultrastructure of two hypersaline submerged cyanobacterial mats: Guerrero Negro, Baja California Sur, Mexico, and Solar Lake, Sinai, Egypt, p. 97-113. In Y. Cohen and E. Rosenberg (ed.), Microbial mats: physiological ecology of benthic microbial communities. American Society for Microbiology, Washington, D.C.
- 11.de los Reyes, F. L., W. Ritter, and L. Raskin. 1997. Group-specific small-subunit rRNA hybridization probes to characterize filamentous foaming in activated sludge systems. Appl. Environ. Microbiol. 63:1107-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DesMarais, D. J. 1995. The biogeochemistry of hypersaline microbial mats. Adv. Microb. Ecol. 14:251-274. [DOI] [PubMed] [Google Scholar]
- 13.DesMarais, D. J. 2003. Biogeochemistry of hypersaline microbial mats illustrates the dynamics of modern microbial ecosystems and the early evolution of the biosphere. Biol. Bull. 204:160-167. [DOI] [PubMed] [Google Scholar]
- 14.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emerson, B. C., and N. Kolm. 2005. Species diversity can drive speciation. Nature 434:1015-1017. [DOI] [PubMed] [Google Scholar]
- 17.Garrett, P. 1970. Phanerozoic stromatolites: noncompetitive ecological restriction by grazing and burrowing animals. Science 169:171-173. [DOI] [PubMed] [Google Scholar]
- 18.Gich, F., J. Garcia-Gil, and J. Overmann. 2001. Previously unknown and phylogenetically diverse members of the green nonsulfur bacteria are indigenous to freshwater lakes. Arch. Microbiol. 177:1-10. [DOI] [PubMed] [Google Scholar]
- 19.Hanada, S., S. Takaichi, K. Matsuura, and K. Nakamura. 2002. Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int. J. Syst. Evol. Microbiol. 52:187-193. [DOI] [PubMed] [Google Scholar]
- 20.Harris, J. K., S. T. Kelley, and N. R. Pace. 2004. New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 70:845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoehler, T. M., B. M. Bebout, and D. J. DesMarais. 2001. The role of microbial mats in the production of reduced gases on the early Earth. Nature 412:324-327. [DOI] [PubMed] [Google Scholar]
- 22.Holmes, A. J., N. A. Tujula, M. Holley, A. Contos, J. M. James, P. Rogers, and M. R. Gillings. 2001. Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ. Microbiol. 3:256-264. [DOI] [PubMed] [Google Scholar]
- 23.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 24.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hugenholtz, P., and T. Huber. 2003. Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int. J. Syst. Evol. Microbiol. 53:289-293. [DOI] [PubMed] [Google Scholar]
- 27.Jοrgensen, B. B. 1989. Light penetration, absorption, and action spectra in cyanobacterial mats, p. 123-137. In Y. Cohen and E. Rosenberg (ed.), Microbial mats: physiological ecology of benthic microbial communities. American Society for Microbiology, Washington, D.C.
- 28.Karl, D. M. 1980. Cellular nucleotide measurements and applications in microbial ecology. Microbiol. Rev. 44:739-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazantsev, A. V., A. A. Krivenko, D. J. Harrington, S. R. Holbrook, P. D. Adams, and N. R. Pace. 2005. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl. Acad. Sci. USA 102:13392-13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klappenbach, J. A., and B. K. Pierson. 2004. Phylogenetic and physiological characterization of a filamentous anoxygenic photoautotrophic bacterium ‘Candidatus Chlorothrix halophila’ gen. nov., sp. nov., recovered from hypersaline microbial mats. Arch. Microbiol. 181:17-25. [DOI] [PubMed] [Google Scholar]
- 31.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 32.Ley, R. E., F. Backhed, P. Turnbaugh, C. Lozupone, R. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102:11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lichtenthaler, H. K., C. Buschmann, U. Rinderle, and G. Schmuck. 1986. Application of chlorophyll fluorescence in ecophysiology. Radiat. Environ. Biophys. 25:297-308. [DOI] [PubMed] [Google Scholar]
- 34.Logan, G. A., C. R. Calver, P. Gorjan, R. E. Summons, J. M. Hayes, and M. R. Walter. 1999. Terminal Proterozoic mid-shelf benthic microbial mats in the Centralian Superbasin and their environmental significance. Geochim. Cosmochim. Acta 63:1345-1358. [DOI] [PubMed] [Google Scholar]
- 35.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maresca, J. A., A. Gomez Maqueo Chew, M. R. Ponsati, N. U. Frigaard, J. G. Ormerod, and D. A. Bryant. 2004. The bchU gene of Chlorobium tepidum encodes the C-20 methyltransferase in bacteriochlorophyll c biosynthesis. J. Bacteriol. 186:2558-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, S. R., and B. M. Bebout. 2004. Variation in sulfide tolerance of photosystem II in phylogenetically diverse cyanobacteria from sulfidic habitats. Appl. Environ. Microbiol. 70:736-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minz, D., S. Fishbain, S. J. Green, M. Gerard, Y. Cohen, B. E. Rittmann, and D. A. Stahl. 1999. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl. Environ. Microbiol. 65:4659-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minz, D., J. L. Flax, S. J. Green, M. Gerard, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfate reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nubel, U., M. M. Bateson, M. T. Madigan, M. Kuhl, and D. M. Ward. 2001. Diversity and distribution in hypersaline microbial mats of bacteria related to Chloroflexus spp. Appl. Environ. Microbiol. 67:4365-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nubel, U., F. Garcia-Pichel, M. Kuhl, and G. Muyzer. 1999. Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats. Appl. Environ. Microbiol. 65:422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oremland, R. S., and G. M. King. 1989. Methanogenesis in hypersaline environments, p. 180-190. In Y. Cohen and E. Rosenberg (ed.), Microbial mats: physiological ecology of benthic microbial communities. American Society for Microbiology, Washington, D.C.
- 44.Papineau, D., J. J. Walker, S. J. Mojzsis, and N. R. Pace. 2005. Composition and structure of microbial communities from stromatolites of Hamelin Pool in Shark Bay, Western Australia. Appl. Environ. Microbiol. 71:4822-4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Permentier, H. P., S. Neerken, K. A. Schmidt, J. Overmann, and J. Amesz. 2000. Energy transfer and charge separation in the purple non-sulfur bacterium Roseospirillum parvum. Biochim. Biophys. Acta 1460:338-345. [DOI] [PubMed] [Google Scholar]
- 46.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 47.Raskin, L., L. K. Poulsen, D. R. Noguera, B. E. Rittmann, and D. A. Stahl. 1994. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 60:1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reid, R. P., P. T. Visscher, A. W. Decho, J. F. Stolz, B. M. Bebout, C. Dupraz, L. G. Macintyre, H. W. Paerl, J. L. Pinckney, L. Prufert-Bebout, T. F. Steppe, and D. J. DesMarais. 2000. The role of microbes in accretion, lamination and early lithification of modern marine stromatolites. Nature 406:989-992. [DOI] [PubMed] [Google Scholar]
- 50.Revsbech, N. P., P. B. Christensen, and L. P. Nielsen. 1989. Microelectrode analysis of photosynthetic and respiratory processes in microbial mats, p. 153-162. In Y. Cohen and E. Rosenberg (ed.), Microbial mats: physiological ecology of benthic microbial communities. American Society of Microbiology, Washington, D.C.
- 51.Risatti, J. B., W. C. Capman, and D. A. Stahl. 1994. Community structure of a microbial mat: the phylogenetic dimension. Proc. Natl. Acad. Sci. USA 9117:10173-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonnenburg, J. L., J. Xu, D. D. Leip, C. H. Chen, B. P. Westover, J. Weatherford, J. D. Buhler, and J. I. Gordon. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955-1959. [DOI] [PubMed] [Google Scholar]
- 54.Stamatakis, A., T. Ludwig, and H. Meier. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456-463. [DOI] [PubMed] [Google Scholar]
- 55.Stanier, R. Y., and J. H. C. Smith. 1960. The chlorophylls of green bacteria. Biochim. Biophys. Acta 41:478-484. [DOI] [PubMed] [Google Scholar]
- 56.Stolz, J. F. 1990. Distribution of phototrophic microbes in the flat laminated microbial mat at Laguna Figueroa, Baja California, Mexico. BioSystems 23:345-357. [DOI] [PubMed] [Google Scholar]
- 57.Summons, R. E., L. L. Jahnke, J. M. Hope, and G. A. Logan. 1999. 2-Methylhopanoids as biomarkers for cyanobacterial oxygenic photosynthesis. Nature 400:554-557. [DOI] [PubMed] [Google Scholar]
- 58.Summons, R. E., L. L. Jahnke, and B. R. Simoneit. 1996. Lipid biomarkers for bacterial ecosystems: studies of cultured organisms, hydrothermal environments and ancient sediments. Ciba Found. Symp. 202:174-193. [DOI] [PubMed] [Google Scholar]
- 59.Tanner, M. A., C. L. Everett, W. J. Coleman, M. M. Yang, and D. C. Youvan. 2000. Complex microbial consortia inhabiting hydrogen sulfide-rich black mud from marine coastal environments. Biotechnology et alia 8:1-16. [Online.] http://www.et-al.com/searchable/abstracts/ComplexMicrobial.htm. [Google Scholar]
- 60.Tice, M. M., and D. R. Lowe. 2004. Photosynthetic microbial mats in the 3,416-Myr-old ocean. Nature 431:549-552. [DOI] [PubMed] [Google Scholar]
- 61.Visscher, P. T., L. K. Baumgartner, D. H. Buckley, D. R. Rogers, M. E. Hogan, C. D. Raleigh, K. A. Turk, and D. J. DesMarais. 2003. Dimethyl sulphide and methanethiol formation in microbial mats: potential pathways for biogenic signatures. Environ. Microbiol. 5:296-308. [DOI] [PubMed] [Google Scholar]
- 62.Wu, J., W. Liu, I. Tseng, and S. Cheng. 2001. Characterization of a 4-methylbenzoate-degrading methanogenic consortium as determined by small-subunit rDNA sequence analysis. J. Biosci. Bioeng. 91:449-455. [DOI] [PubMed] [Google Scholar]
- 63.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]