Abstract
We used the multilocus sequence typing (MLST) method to evaluate the genetic diversity of 46 Campylobacter jejuni isolates from chickens and to determine the link between quinolone resistance and sequence type (ST). There were a total of 16 ST genotypes, and the majority of them belonged to seven clonal complexes previously identified by using isolates from human disease. The ST-353 complex was the most common complex, whereas the ST-21, ST-42, ST-52, and ST-257 complexes were less well represented. The resistance phenotype varied for each ST, and the Thr-86-Ile substitution in the GyrA protein was the predominant mechanism of resistance to quinolone. Nine of the 14 isolates having the Thr-86-Ile substitution belonged to the ST-353 complex. MLST showed that the emergence of quinolone resistance is not related to the diffusion of a unique clone and that there is no link between ST genotype and quinolone resistance. Based on silent mutations, different variants of the gyrA gene were shown to exist for the same ST. These data provide useful information for understanding the epidemiology of C. jejuni in Senegal.
Campylobacter jejuni is the leading cause of bacterial food-borne diarrheal disease throughout the world (2). In developing countries, the estimated incidence of Campylobacter in diarrhea is between 40,000 and 60,000 per 100,000 for children less than 5 years old and is 90 per 100,000 in the general population (3, 5, 19). Organisms are carried in the intestinal tracts of many animals, including birds, and epidemiological evidence has suggested that this site is a potential reservoir for human infections and an entry point into the food chain (12, 14, 17, 22). Most human infections with C. jejuni are self-limiting and do not require antimicrobial therapy. However, treatment with erythromycin or a fluoroquinolone is needed in individuals with invasive or severe disease, in immunocompromised patients, or in children less than 5 years old. But resistance to fluoroquinolone is increasing throughout the world (10). In Campylobacter, the predominant mechanism of fluoroquinolone resistance is the C-to-T transition at codon 86 in the quinolone resistance-determining region (QRDR) of the gyrA gene, which results in a threonine-to-isoleucine substitution in the functional protein (1, 24).
To study the epidemiology of Campylobacter infections, multiple typing methods have been developed in order to differentiate isolates below the species level (25). The multilocus sequence typing (MLST) method was developed by Dingle et al. (8) to characterize C. jejuni strains and to identify clonal lineages in this species (6, 7, 9, 16). This method uses genetic variations at multiple chromosomal locations and allows generation of sequence data, which may be available via the Internet for comparison with DNA sequences of other strains.
To date, there are little data on the epidemiology of Campylobacter in Senegal; one study was a study of the incidence of quinolone-resistant C. jejuni isolates in commercial chickens, which was 40% (4).
The aims of the present study were (i) to evaluate the genetic diversity of C. jejuni strains isolated from chickens in Senegal and (ii) to determine the link between quinolone resistance and sequence type (ST) genotypes.
MATERIALS AND METHODS
Campylobacter isolates.
The C. jejuni isolates used originated from chicken carcasses and were collected during 2000 to 2002 in the Dakar region (Senegal) and suburbs (4). All of the bacterial isolates had been maintained as stock cultures in glycerol broth at −80°C. A total of 46 isolates that were randomly picked from 99 C. jejuni isolates were included in the study, and these strains were obtained from 14 dispersed collection sites over a 3-year period. Prior to DNA extraction, cultures were removed from storage and allowed to thaw at room temperature. Isolates were cultured on Columbia agar plates containing 5% sheep blood and were incubated under microaerophilic conditions (5% O2, 10% CO2, 85% N2) at 42°C for 48 h.
MLST.
Bacterial DNA was prepared by boiling as reported previously (1). MLST was performed as described by Dingle et al. (8) by using sequences obtained from seven housekeeping genes. PCR products were purified with a QIAquick gel extraction kit (QIAGEN, Germany), and the DNA sequences of both strands were determined with an ABI PRISM 310 DNA sequencer (Applied Biosystems). MLST alleles and sequence types were assigned to the isolates by comparing data to the Campylobacter MLST database (http://campylobacter.mlst.net). The isolates were organized into lineages with the program BURST (based on related STs), which is part of the START (sequence type assignment recombination tests) group of programs. Isolates were grouped together if they shared five, six, or all seven MLST loci. eBURST was used to examine patterns of evolutionary descent (11). Analysis of fixed differences, polymorphic sites, and the ratio of nonsynonymous substitutions to synonymous substitutions was performed with the DnaSP software package (version 4.00), available at http://www.ub.edu/dnasp/ (20). The index of association (IA) was calculated using the program START (15).
Antibiotic susceptibility.
The antibiotic susceptibility of each isolate was determined by the disk diffusion method. The MICs of ciprofloxacin and nalidixic acid were determined by the Etest method and were interpreted according to CLSI (formerly NCCLS) guidelines (18). Isolates were inoculated onto Mueller-Hinton blood agar, and Etest strips (AB Biodisk, Solna, Sweden) were applied to each plate. C. jejuni subsp. jejuni ATCC 33560 (= NCTC 11351) was included as a quality control strain. The plates were incubated at 42°C under microaerophilic conditions (5% O2, 10% CO2, 85% N2) for 24 h. The breakpoint for ciprofloxacin was considered to be an MIC of ≥4 μg/ml.
Determination of mutations in the QRDR of gyrA.
A 220-bp fragment covering the QRDR of gyrA was amplified by PCR by using the primers and conditions described previously, except that the annealing temperature was 50°C (24). After purification (QIAquick gel extraction kit; QIAGEN, Germany), sequencing of the amplified fragment was performed with the same primers using an ABI PRISM 310 DNA sequencer (Applied Biosystems). DNA sequences were compared to the previously published C. jejuni sequence (GenBank accession number L04566) using the CLUSTALW program available at http://bioweb.pasteur.fr/seqanal/interfaces/clustalw.html.
RESULTS
Diversity of MLST alleles.
The number of unique alleles at each locus varied from five for the Asp locus to nine for the Gln locus, and the percentage of variable sites in the data set ranged from 0.6% for the uncA locus to 4.8% for the pgm locus (Table 1). For all seven loci, the ratio of nonsynonymous substitutions to synonymous substitutions was much less than 1.
TABLE 1.
Allelic diversity
| Locus | Fragment size (bp) | No. of alleles | No. of variables sites | % Variable sites | dN/dSa |
|---|---|---|---|---|---|
| aspA | 477 | 5 | 6 | 1.3 | 0.022 |
| glnA | 477 | 9 | 13 | 2.7 | 0.006 |
| gltA | 402 | 6 | 6 | 1.5 | 0.062 |
| glyA | 507 | 6 | 22 | 4.3 | 0.001 |
| pgm | 498 | 8 | 24 | 4.8 | 0.000 |
| tkt | 459 | 7 | 17 | 3.7 | 0.002 |
| uncA | 489 | 4 | 3 | 0.6 | 0.043 |
dN/dS, ratio of nonsynonymous substitutions to synonymous substitutions.
Diversity of STs and lineages.
There were a total of 16 STs among the 46 isolates examined. Several of these STs (ST1035, ST1036, ST1037, ST1038, ST1039, ST1040, ST1041, ST1081, ST1358, ST1359, and ST1370) had not been described previously at the Campylobacter MLST database (http://campylobacter.mlst.net). Using the program BURST, the 46 isolates were grouped into four lineages (Table 2), and seven singleton sequence types were unrelated to any other sequence types based on the fact that the isolates in one group were identical at five, six, or all seven of the MLST loci. The most common ST was ST1036, which comprised 11 isolates (23.91% of the data set), followed by ST1035, which comprised 8 isolates (17.39%), and ST1039 with 5 isolates (10.87%). The seven singleton STs (ST1370, ST1359, ST1358, ST1211, ST1035, ST824, and ST52) included 17 isolates and accounted for 36.95% of the data set. A total of seven clonal complexes were identified (ST-21, ST-22, ST-42, ST-52, ST-257, ST-353, and ST-354 complexes). Within each clonal complex, the number of isolates ranged from 1 (ST-21, ST-42, and ST-257 complexes) to 12 (ST-353 complex). eBURST analysis allowed us to determine the relationships between ST1040 (unassigned) and ST1036 (ST-353 complex) on the one hand and ST1211 (ST-353 complex) on the other hand (Fig. 1). Indeed, the BURST analysis revealed that both ST1040 and ST1036 belong to group 1, whereas ST1211 was a singleton ST. ST1039 and ST1041 were grouped in the same lineage (group 2) but were not assigned to any complex based on the data in the Campylobacter MLST database. The other STs that were not assigned to any complex were ST1035 and ST1358. The IA for the complete data set was 2.028, and the IA was 0.371 when only one representative of each lineage was included.
TABLE 2.
MLST lineages, ST, gyrA mutations, and resistance phenotypes
| BURST lineage | Clonal complex | ST | Isolate | Year | gyrA mutationsa | Phenotypeb
|
|
|---|---|---|---|---|---|---|---|
| Ciprofloxacin | Nalidixic acid | ||||||
| Group 1 | 353 | 1036 | 83B | 2001 | His-81, Ser-119 | S | S |
| 104C | 2002 | His-81, Ser-119 | S | S | |||
| 116B | 2002 | His-81, Ser-119, Thr-86-Ile, Gly-110 | R | R | |||
| 65B | 2001 | His-81, Ser-119, Thr-86-Ile | R | R | |||
| 48A | 2000 | His-81, Ser-119, Thr-86-Ile | R | R | |||
| 48B | 2000 | His-81, Ser-119, Thr-86-Ile | R | R | |||
| 65A | 2001 | His-81, Ser-119, Thr-86-Ile | R | R | |||
| 65C | 2001 | His-81, Ser-119, Thr-86-Ile | R | R | |||
| F19 | 2001 | His-81, Ser-119, Thr-86-Ile | R | R | |||
| 118C | 2002 | His-81, Ser-119, Thr-86-Ile, Gly-110 | R | R | |||
| 82A | 2001 | His-81, Ser-119, Thr-86-Ile | R | R | |||
| Unassigned | 1040 | 113B | 2002 | His-81, Ser-119, Thr-86-Ile | R | R | |
| 93A | 2001 | His-81, Ser-119 | S | S | |||
| 112A | 2002 | His-81, Ser-119, Thr-86-Ile | R | R | |||
| Group 2 | Unassigned | 1039 | 50C | 2000 | His-81, Ser-119 | S | S |
| 75A | 2001 | His-81, Ser-119 | S | S | |||
| 95B | 2001 | His-81, Ser-119 | S | S | |||
| 95A | 2001 | His-81, Ser-119 | R | R | |||
| 57A | 2001 | His-81, Ser-119 | S | S | |||
| 1041 | 62A | 2001 | His-81, Ser-119 | S | S | ||
| 64C | 2001 | His-81, Ser-119 | S | S | |||
| F18 | 2001 | His-81, Ser-119, Thr-86-Ala, Gly-110 | R | R | |||
| Group 3 | 22 | 1081 | 118B | 2002 | None | S | S |
| 660 | 118A | 2002 | None | S | S | ||
| 22 | 117C | 2002 | None | S | S | ||
| Group 4 | 354 | 1037 | 106B | 2002 | His-81, Ser-119, Thr-86-Ile | R | R |
| 1038 | 113A | 2002 | His-81, Ser-119 | S | S | ||
| 115B | 2002 | His-81, Ser-119 | S | S | |||
| V34A | 2002 | His-81, Ser-119 | S | S | |||
| Singleton STs | 52 | 52 | 44B | 2000 | His-81, Ser-119 | S | S |
| 47C | 2000 | His-81, Ser-119 | S | S | |||
| 72B | 2001 | His-81, Ser-119 | S | S | |||
| 55A | 2001 | His-81, Ser-119 | S | S | |||
| 257 | 824 | 103C | 2002 | His-81, Ser-119 | S | S | |
| 21 | 1359 | 90B | 2001 | His-81, Ser-119 | S | S | |
| 353 | 1211 | 115A | 2002 | His-81, Ser-119 | S | S | |
| 42 | 1370 | 41B | 2000 | His-81, Ser-119 | S | S | |
| Unassigned | 1358 | 84C | 2001 | His-81, Ser-119 | R | R | |
| 1035 | 130A | 2000 | His-81, Ser-119, Gly-110 | S | S | ||
| 76C | 2001 | His-81, Ser-119, Thr-86-Ala, Gly-110 | S | I | |||
| 110B | 2002 | His-81, Ser-119, Thr-86-Ile, Gly-110 | R | R | |||
| 76A | 2001 | His-81, Ser-119, Thr-86-Ala, Gly-110 | S | I | |||
| 76B | 2001 | His-81, Ser-119, Thr-86-Ala, Gly-110 | S | I | |||
| 78C | 2001 | His-81, Ser-119, Thr-86-Ala, Gly-110 | R | R | |||
| 78A | 2001 | His-81, Ser-119, Thr-86-Ala, Gly-110 | R | R | |||
| 110A | 2002 | His-81, Ser-119, Thr-86-Ile | R | R | |||
Silent mutations are His-81 (CAC→CAT), Ser-119 (AGT→AGC), and Gly-110 (GGC→GGT).
R, resistant; S, susceptible; I, intermediate.
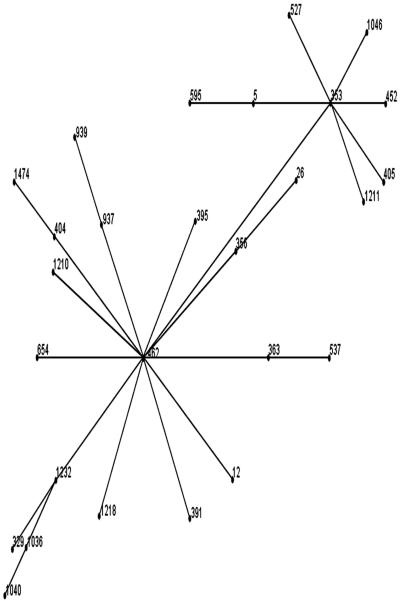
FIG. 1.
Relationship between isolates of the ST-353 complex from the Campylobacter MLST database and ST1040 as determined by eBURST analysis. ST1036 is the ancestor of ST1040.
Resistance phenotypes and mutations of the gyrA QRDR.
Twenty-four isolates were susceptible to both ciprofloxacin and nalidixic acid, 3 isolates were susceptible to ciprofloxacin but had intermediate susceptibility to nalidixic acid, and 19 isolates were resistant to both antibiotics (Table 2). The great majority of quinolone-resistant isolates had a change from C to T in codon 86 of the gyrA gene, resulting in a threonine-to-isoleucine substitution in the functional protein. Another substitution in the same codon was the Thr-86-Ala substitution observed in nalidixic acid-resistant isolates and isolates with intermediate susceptibility. Forty-three isolates had silent mutations at His-81 (CAC→CAT) and Ser-119 (AGT→AGC), and 10 isolates had an additional silent mutation at Gly-110 (GGC→GGT).
Relationships between ST, resistance phenotypes, and gyrA mutations.
In all groups except group 3, the resistance phenotype varied within each ST (Table 2). However, certain phenotypes were predominant. In group 1, the most common phenotype was double resistance to ciprofloxacin and nalidixic acid, whereas groups 2 and 4 contained a majority of susceptible isolates. In singleton STs, the resistance phenotypes were diverse. Three phenotypes were identified among isolates assigned to ST1035, while ST52 contained only susceptible isolates. Eleven of the 14 isolates having the Thr-86-Ile substitution belonged to group 1. Based on silent mutations, different variants of the gyrA gene were shown to exist for the same ST genotype; the first variant had a silent mutation at His-81 and Ser-119, and the second variant had an additional silent mutation at Gly-110.
DISCUSSION
The isolates analyzed in this study originated only from chickens, in contrast to other studies in which isolates having diverse origins were examined (8, 16). This study is the first study in which the MLST approach was used to examine the clonal relationships of C. jejuni isolates in Senegal. Although the number of isolates was relatively small, the diversity of the isolates was established by identification of various genotypes and clonal complexes.
The allelic diversity observed here was similar to the diversity observed previously in farm animals by Colles et al. (6) with regard to the number of alleles and nucleotide variability; the only exception was the uncA locus, for which Colles et al. found a higher degree of diversity.
In this study, we identified seven clonal complexes, and all of these complexes have been associated with human disease (9, 21). Consistent with our results, Dingle et al. (8) identified six clonal complexes in 34 isolates obtained from chickens. In contrast to previous studies in which the ST-21 complex was the largest complex (6, 7, 8), this complex appears to be uncommon in Senegalese chickens since its ST1359 genotype occurred only once in the data. The other complexes that were less well represented were ST-42, ST-52, and ST-257. In Europe, these complexes were identified for various isolates from farm animals (6, 16, 21) and also for human isolates (6, 21), suggesting that they have a wide distribution. The ST-353 complex, the most common complex in this study, and the ST-22 and ST-354 complexes have all been found previously in poultry (16). Other studies have shown that these complexes contain isolates associated with human gastroenteritis (7, 9, 16). Indeed, a search of the Campylobacter MLST database revealed that these complexes contained a majority of the isolates obtained from human disease. Despite the lack of MLST data for humans in Senegal, one could envisage that these complexes represent pathogen strains.
A number of new STs were identified for the first time in this study (ST1035, ST1036, ST1037, ST1038, ST1039, ST1040, ST1041, ST1081, ST1358, ST1359, and ST1370), and some of them were not assigned to any known complex (ST1035, ST1039, ST1040, ST1041, and ST1358). In group 1, for instance, MLST showed that isolates with the ST1040 genotype are single-locus variants of ST1036, which is a member of the ST-353 complex. eBURST analysis of all ST-353 complex isolates in the MLST database with ST1040 indicated that ST1040 was derived from ST1036 (Fig. 1); the only difference was a single nucleotide polymorphism in the gltA gene sequence. Therefore, ST1040 probably diverged from ST1036 very recently. Interestingly, based on eBURST and BURST analyses of all ST-353 complex isolates in the MLST database it also appeared that ST462 rather than ST353 was the founder of the complex. As more MLST data are added to the database, more new links should be seen, and it is also likely that conclusions concerning the clonal complex founder will have to be modified. Also, eBURST analysis showed the relationship between ST1211 and ST1036. ST1211 was grouped among the singleton STs by BURST analysis, even though both ST1211 and ST1036 belonged to the ST-353 complex, and we explained this classification by the absence of intermediate STs (ST1232, ST462, and ST353, not present among our isolates) between ST1211 and ST1036 and also by the fact that only three of the seven MLST loci are shared by the two STs. Singleton STs were not negligible in our study (36.95% of the data set), and they represent distant genotypes not related to any other genotypes. Thus, we believe that with more MLST data, new linkages (not seen in this study) will be discovered. Another finding was that group 2 contained only STs (ST1039 and ST1041) that were not assigned to any complex. Perhaps these genotypes were the result of recombination events (23). Based on the complete data set, linkage analysis revealed that the IA was 2.028, which is comparable to the value found by Dingle et al. (8) (2.016). In our analysis, the IA was 0.371 when only one representative of each lineage was included, whereas Dingle et al. (8) obtained an IA of 0.56 and concluded that this indicated that there was a weakly clonal population. In addition, Suerbaum et al. (23) obtained an IA of 0.256, which was comparable to our value, and suggested that there was a limited amount of recombination. Although the number of isolates analyzed in this study was too small to draw any definite conclusions, certain STs (ST1035, ST1036, and ST1039) and clonal complexes (ST-353, ST-354, and ST-52 complexes) seem to be predominant, even though other genotypes and clonal complexes may still be present in Senegalese chickens.
As previously reported (1, 24), the Thr-86-Ile substitution in the GyrA protein was the major mechanism of quinolone resistance, although a small number of isolates had the Thr-86-Ala substitution. In Senegal, fluoroquinolones (enrofloxacin and norfloxacin) were introduced into poultry production in 1996 (4), and their use became an important selective factor in the spread of quinolone-resistant clones. The fact that quinolone resistance was observed in STs of different MLST lineages (except the ST-22 complex) suggests that the emergence of quinolone resistance is not related to the diffusion of a unique clone but rather is the result of quinolone selection pressure on strains from different clones. Nevertheless, there might be a relationship between isolates carrying the Thr-86-Ile substitution and the ST-353 complex. It also appeared that there were different variants of the gyrA gene for the same ST genotype. This illustrates the variability of the C. jejuni genome outside the MLST loci. Previous studies have argued that gyrA polymorphism could be useful in epidemiological studies (13, 26). We believe that this polymorphism may provide information that supplements the data obtained by the MLST approach for understanding the epidemiology of this pathogen. Generally speaking, isolates that had the same ST genotype had variable resistance phenotypes, suggesting that there was no link between ST genotype and quinolone resistance.
To date, there is little information concerning the epidemiology of Campylobacter in Senegal, and in particular, there are no data on human isolates due to the absence of systematic investigations of Campylobacter in medical laboratories. This report provides more insight into the molecular biology of this bacterium in poultry production. Further studies with a larger number of isolates, including isolates from humans and several time frames, should provide more data on the epidemiology of Campylobacter in Senegal.
Acknowledgments
We thank Rokhaya Mbaye and Fatou Kiné Loum for their technical help. We also thank Amy Gassama, Pierre Nabeth, Bianca Paglietti, and Elisa Deriu for useful discussions.
This work was supported by the Pasteur Institute in Dakar, Senegal.
REFERENCES
- 1.Bachoual, R., S. Ouabdesselam, F. Mory, C. Lascols, C. J. Soussy, and J. Tankovic. 2001. Single or double mutational alterations of gyrA associated with fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Microb. Drug Resist. 7:257-261. [DOI] [PubMed] [Google Scholar]
- 2.Blaser, M. J. 1997. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176(Suppl. 2):S103-S105. [DOI] [PubMed] [Google Scholar]
- 3.Butzler, J. P. 2004. Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 10:868-876. [DOI] [PubMed] [Google Scholar]
- 4.Cardinale, E., J. A. Dromigny, F. Tall, M. Ndiaye, M. Konte, and J. D. Perrier-Gros-Claude. 2003. Fluoroquinolone susceptibility of Campylobacter strains, Senegal. Emerg. Infect. Dis. 9:1479-1481. [DOI] [PubMed] [Google Scholar]
- 5.Coker, A. O., R. D. Isophi, B. N. Thomas, K. O. Amisu, and L. Obi. 2002. Human campylobacteriosis in developing countries. Emerg. Infect. Dis. 8:237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colles, F. M., K. Jones, R. M. Harding, and M. C. J. Maiden. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. A. Wareing, and M. C. J. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duim, B., P. C. R. Godschalk, N. V. D. Braak, K. E. Dingle, J. R. Dijkstra, E. Leyde, J. V. D. Plas, F. M. Colles, H. P. Endtz, J. A. Wagenaar, M. C. J. Maiden, and A. V. Belkum. 2003. Molecular evidence for dissemination of unique Campylobacter jejuni clones in Curaçao, Netherlands Antilles. J. Clin. Microbiol. 41:5593-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engberg, J., F. M. Aarestrup, D. E. Taylor, P. Gerner-Smidt, and I. Nachamkin. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillepsie, I. A., S. O'Brien, J. A. Frost, G. K. Adak, P. Horby, and A. V. Swan. 2002. A case-case comparison of Campylobacter coli and C. jejuni infections: a tool for generating hypothesis. Emerg. Infect. Dis. 8:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hakanen, A., J. Jalava, P. Kotilainen, H. Jousimies-Somer, A. Siitonen, and P. Huovinen. 2002. gyrA polymorphism in Campylobacter jejuni: detection of gyrA mutations in 162 C. jejuni isolates by single-strand conformation polymorphism and DNA sequencing. Antimicrob. Agents Chemother. 46:2644-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanninen, M. L., P. Perko-Makela, A. Pitkala, and H. Rautelin. 2000. A three-year study of Campylobacter jejuni genotypes in humans with domestically acquired infections and in chicken samples from the Helsinki area. J. Clin. Microbiol. 38:1998-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 16.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore, J. E., L. O' Riordan, D. R. Wareing, R. Doyle, J. Lanser, T. Stanley, M. Matsuda, T. Matsui, and P. G. Murphy. 2003. Phenotypic and genotypic relationship between Campylobacter spp. isolated from humans and chickens in Northern Ireland—a comparison of three phenotyping and two genotyping schemes. Int. J. Hyg. Environ. Health 206:211-216. [DOI] [PubMed] [Google Scholar]
- 18.NCCLS. 2004. Performance standards for antimicrobial susceptibility testing: 14th informational supplement. M100-S14, vol. 24, no. 1. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.Rao, M. R., A. B. Naficy, S. J. Savarino, R. Abu-Elyazeed, T. F. Wierzba, L. F. Peruski, et al. 2001. Pathogenicity and convalescent excretion of Campylobacter in rural Egyptian children. Am. J. Epidemiol. 154:166-173. [DOI] [PubMed] [Google Scholar]
- 20.Rosa, J., J. C. Sanchez-Delbarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 21.Schouls, L. M., S. Reulen, B. Duim, J. A. Wagenaar, R. J. L. Willems, K. E. Dingle, F. M. Colles, and J. D. A. Van Embden. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanley, K., and K. Jones. 2003. Cattle and sheep farms as reservoirs of Campylobacter. J. Appl. Microbiol. 94:104S-113S. [DOI] [PubMed] [Google Scholar]
- 23.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, Y., W. M. Huang, and D. E. Taylor. 1993. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 37:457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter species. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zirnstein, G., Y. Li, B. Swaminathan, and F. Angulo. 1999. Ciprofloxacin resistance in Campylobacter isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J. Clin. Microbiol. 37:3276-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]