Abstract
The involvement of coenzyme M in aerobic biodegradation of vinyl chloride and ethene in Pseudomonas putida strain AJ and Ochrobactrum sp. strain TD was demonstrated using PCR, hybridization, and enzyme assays. The results of this study extend the range of eubacteria known to use epoxyalkane:coenzyme M transferase.
Elucidating the pathway for vinyl chloride (VC) biodegradation is likely to improve methods for predicting the fate of this compound in natural and engineered environments. Under aerobic conditions, metabolism of VC and ethene, the nonchlorinated counterpart of VC, begins with epoxide formation (4, 8, 11, 20). The involvement of epoxyalkane:coenzyme M transferase (EaCoMT) in the processing of VC epoxide and ethylene oxide (EtO) has recently been demonstrated with several strains of Mycobacterium and one strain of Nocardioides (5, 6, 15). The EaCoMT gene is located on large linear plasmids in Mycobacterium strains and Nocardioides sp. strain JS614 when they are grown on VC (5, 15). The objective of this study was to determine if coenzyme M (CoM) is involved in the pathway of two other microbes that also grow on VC and ethene, Pseudomonas putida strain AJ and Ochrobactrum sp. strain TD (8).
Chemicals, media, and growth conditions are described elsewhere (7). Ethene, EtO, and VC consumption was quantified by gas chromatography (20). Growth rates were determined based on plots of protein concentration (measured by UV [4, 7, 12]) versus time (4). The EaCoMT assay was performed (6) with minor modifications (7). Cells were harvested at the early to mid-exponential phase (optical density at 600 nm, 0.2 to 0.35), washed, and resuspended in buffer (2, 6, 7).
Agarose plugs were prepared and pulsed-field gel electrophoresis (PFGE), DNA elution, and PCR were performed as described previously (5-8). Southern hybridization and radiolabeling of probe DNA were performed (3, 17) using a probe that was from the 891-bp EaCoMT gene fragment from strain AJ, using an Ambion DECAprime II labeling kit. Partial CoM gene sequences (834 bp) for various Mycobacterium strains were obtained from GenBank (accession no. AY243034 to AY243043).
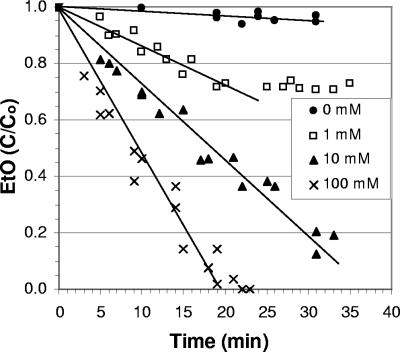
The rates of EtO consumption by extracts of EtO-grown strain AJ increased from 57 ± 26 nmol min−1 mg protein−1 with no CoM added to 670 ± 70, 1,300 ± 80, and 2,600 ± 280 with 1, 10, and 100 mM CoM added, respectively (Fig. 1). For EtO-grown strain TD, the rates of EtO consumption were 86 ± 72, 1,800 ± 230, 3,500 ± 280, and 7,200 ± 710 nmol min−1 mg protein−1 with 0, 1, 10, and 100 mM CoM added, respectively.
FIG. 1.
Effect of CoM concentration (0, 1, 10, and 100 mM) on EaCoMT activity, as indicated by the rate of EtO utilization in cell extracts of VC-grown strain AJ. C/Co is the ratio of remaining EtO to the initial amount (approximately 5 μmol per bottle).
EaCoMT activities were also determined following growth of strains AJ and TD on VC and ethene (Table 1). The activities with 10 mM CoM were 1 to 2 orders of magnitude higher than the activities without CoM. There were statistically significant differences among the EaCoMT activities with CoM for strains AJ and TD grown on VC, ethene, and EtO (α = 0.05, as determined by analysis of variance). Tukey's range test indicated that the EaCoMT activity of strain AJ when it was grown on EtO (1,300 nmol min−1 mg protein−1) was statistically lower than the other activities (range, 3,400 to 4,600 nmol min−1 mg protein−1).
TABLE 1.
EaCoMT activity in cell extracts of strains AJ and TD
| Strain | Substrate | EaCoMT activitya (nmol min−1 mg protein−1)
|
|
|---|---|---|---|
| 10 mM CoM added | No CoM added | ||
| AJ | VC | 4,400 ± 580 | 46 ± 77 |
| Ethene | 3,400 ± 690 | 100 ± 31 | |
| EtO | 1,300 ± 80 | 57 ± 26 | |
| LBB, following growth on VC | 250 ± 46 | 140 ± 76 | |
| LBB, following growth on ethene | 190 ± 37 | 180 ± 20 | |
| LBB, following growth on EtO | 120 ± 79 | 28 ± 26 | |
| TD | VC | 3,700 ± 700 | 95 ± 74 |
| Ethene | 4,600 ± 700 | 130 ± 83 | |
| EtO | 3,500 ± 280 | 86 ± 72 | |
| LBB, following growth on VC | 270 ± 89 | 100 ± 67 | |
| LBB, following growth on ethene | 300 ± 40 | 87 ± 43 | |
| LBB, following growth on EtO | 160 ± 16 | 150 ± 20 | |
| No cells | None | 25 ± 5 | 6 ± 4 |
The values are means ± standard deviations of triplicate measurements.
Table 1 also shows the EaCoMT activities with 10 mM CoM for Luria-Bertani broth (LBB)-grown strains AJ and TD following growth on VC, ethene, and EtO. Growth in LBB caused the EaCoMT activities to decrease by 1 order of magnitude, although the activities of LBB-grown cells were greater than the activities of controls with no cells. The EaCoMT activities of LBB-grown cells with CoM were higher than the activities of LBB-grown cells with no CoM, although the value for only one of the LBB-grown treatments with CoM was statistically greater (α = 0.05, as determined by the t test) than the value for the corresponding treatment with no CoM (i.e., strain TD grown in LBB following growth on ethene).
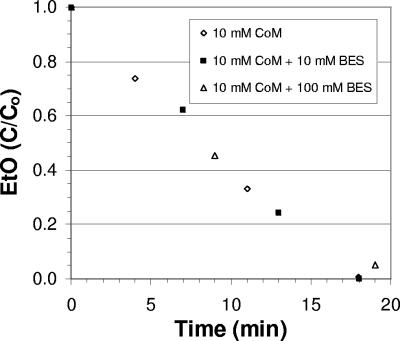
CoM is a methyl carrier in the final step of methanogenesis (18, 21), which is inhibited by bromoethanesulfonate due to its reactivity with methyl-CoM reductase (10). Since bromoethanesulfonate inhibits reductase activity, it should not inhibit aerobic metabolism of VC, ethene, and EtO, which involves only transferase activity. Adding bromoethanesulfonate did not alter the EaCoMT activities with extracts of ethene-grown strain AJ containing 10 mM CoM (Fig. 2).
FIG. 2.
Effect of 0, 10, and 100 mM bromoethanesulfonate (BES) on EaCoMT activity, as indicated by the rate of EtO utilization in cell extracts of ethene-grown strain AJ. C/Co is the ratio of remaining EtO to the initial amount (approximately 5 μmol per bottle).
The EaCoMT assay was performed with glutathione in place of CoM. There was no difference in the EaCoMT activities between extracts with and without glutathione, indicating that glutathione is not involved in catabolism of EtO in strains AJ and TD (data not shown). Previous studies have shown that glutathione is a cofactor in epoxide degradation (19).
The maximum specific substrate utilization rates for strain AJ were 18, 22, and 62 nmol min−1 mg protein−1 when VC, ethene, and EtO were used as the substrates, respectively. For strain TD, the maximum specific substrate utilization rates were 20, 15, and 84 nmol min−1 mg protein−1 with VC, ethene, and EtO, respectively. Since the EaCoMT activities of strains AJ and TD (Table 1) were at least 10 times higher than the maximum substrate utilization rates measured for whole cells, it was evident that EaCoMT is not a limiting factor during growth. The maximum substrate utilization rate determined in this study for strain AJ growing on VC was higher than the maximum rate reported previously (1.41 μmol VC mg biomass−1 day−1 or 1.8 nmol mg protein−1 min−1) (8). Differences in the incubation temperature (22°C in the previous study and 30°C in this study) and in the assay procedures (steady-state reactor biomass in the previous study and exponentially growing cells in this study) likely contributed to the different rates. Variations in kinetic parameters are often a consequence of differences in the physiological state of the cells used (9).
DNA was eluted from plugs prepared from strains AJ and TD following growth on VC, ethene, and EtO. The DNA was subjected to PCR using primers based on conserved regions of the EaCoMT gene. Cells grown on VC, ethene, and EtO yielded a single PCR product of the expected size (891 bp), as did ethene-grown Mycobacterium sp. strain JS60. No product was obtained with a control containing no DNA (data not shown).
Growth of strains AJ and TD in LBB cures the large linear plasmid(s) required for use of VC and ethene as growth substrates (7), as well as for use of EtO by strains AJ (8) and TD (data not shown). Following growth of both strains in LBB, the cultures were also subjected to PCR, and a band of the expected size was recovered (data not shown), confirming the presence of the EaCoMT gene even after the large linear plasmids were cured. Sequencing of the 891-bp fragment confirmed the presence of the EaCoMT gene. However, there were differences in the sequence of the EaCoMT gene following growth in LBB compared to growth on VC (differences in four amino acids for strain AJ and in five amino acids for strain TD, based on the 891-bp fragment).
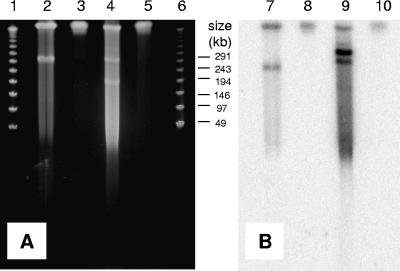
Southern hybridization was performed to determine if the EaCoMT gene was located on the plasmid. Agarose plugs of strains AJ and TD grown on VC and in LBB were subjected to contour-clamped homogeneous electric field PFGE (Fig. 3A) and then exposed to the EaCoMT probe (Fig. 3B). A hybridization signal for VC-grown strains AJ and TD corresponded to an approximately 260-kb plasmid (Fig. 3B, lanes 7 and 9). The strongest hybridization signal for VC-grown strain TD did not correspond to a plasmid from PFGE (lane 9). In addition, a hybridization signal was observed for both strains (lanes 7 and 9) in the compression zone (i.e., large, intact, high-molecular-weight DNA). The compression zone for strains AJ and TD following growth in LBB also exhibited hybridization (lanes 8 and 10). Compression zone binding was also observed in VC-grown Mycobacterium strains and propylene-grown Rhodococcus sp. strain B276 (5, 16), possibly due to nonspecific binding to the EaCoMT probe and unresolved plasmid DNA (5). Alternatively, plasmid DNA may have integrated into chromosomal DNA via insertion elements. Strain TD also showed the strongest hybridization signal that did not correspond to a plasmid (lane 9). Similar results were observed with VC-grown Mycobacterium sp. strain JS616 (5).
FIG. 3.
Contour-clamped homogeneous electric field PFGE of strains AJ and TD grown on VC and in LBB (A) and subsequent Southern hybridization of the EaCoMT gene of VC- and LBB-grown strains AJ and TD (B). Lane 1, marker; lane 2, strain AJ grown on VC; lane 3, strain AJ grown in LBB following growth on VC; lane 4, strain TD grown on VC; lane 5, strain TD grown in LBB following growth on VC; lane 6, marker; lane 7, strain AJ grown on VC; lane 8, strain AJ grown in LBB following growth on VC; lane 9, strain TD grown on VC; lane 10, strain TD grown in LBB following growth on VC.
Retention of the EaCoMT gene even after curing of the plasmids is consistent with the higher levels of EaCoMT activity in cells grown in LBB and exposed to 10 mM CoM than in cells not exposed to CoM, even after the abiotic activity with no cells present was subtracted (Table 1). Differences in the EaCoMT gene sequence of LBB-grown cells may have contributed to the decrease in EaCoMT activity compared to the activity of cells grown on VC, ethene, or EtO. When Nocardioides sp. strain JS614 was grown in Trypticase soy broth or on acetate, it also retained low levels of EaCoMT activity (15). This was attributed to the presence of two active EaCoMT genes in strain JS614, one of which contains a small deletion (15). However, multiple PCR bands were not observed for strains AJ and TD.
The sequences of the EaCoMT gene of VC-grown strains AJ and TD were compared to the sequences of various strains of Mycobacterium, Rhodococcus, Xanthobacter, and Nocardioides. Strains AJ and TD clustered with Mycobacterium strains JS60 and JS621 (see Fig. S1 in the supplemental material). The high degree of homology among EaCoMT sequences is interesting when how geographically diverse the microbes are is considered (4, 8), although there is considerable variability among EaCoMT rates (5, 6).
This study demonstrated that EaCoMT is used by P. putida strain AJ and Ochrobactrum sp. strain TD during metabolism of VC, ethene, and EtO, thereby extending the range of eubacteria known to use CoM during catabolism of alkenes (1, 4, 5, 13-15).
Nucleotide sequence accession numbers.
Sequences of the EaCoMT genes in strain AJ grown on VC (accession no. AY858984) and in LBB (accession no. DQ370435) and in strain TD grown on VC (accession no. AY858985) and in LBB (accession no. DQ370436) have been deposited in the GenBank database.
Supplementary Material
Acknowledgments
Jim C. Spain (Department of Civil & Environmental Engineering, Georgia Institute of Technology) generously provided Mycobacterium sp. strain JS60. William C. Bridges, Jr. (Department of Applied Economics and Statistics, Clemson University) provided assistance with the statistical analyses.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allen, J. R., D. D. Clark, J. G. Krum, and S. A. Ensign. 1999. A role for coenzyme M (2-mercaptoethanesulfonic acid) in a bacterial pathway of aliphatic epoxide carboxylation. Proc. Natl. Acad. Sci. USA 96:8432-8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, J. R., and S. A. Ensign. 1997. Purification to homogeneity and reconstitution of the individual components of the epoxide carboxylase multiprotein enzyme complex from Xanthobacter strain Py2. J. Biol. Chem. 272:32121-32128. [DOI] [PubMed] [Google Scholar]
- 3.Chomczynski, P. 1992. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal. Biochem. 201:134-139. [DOI] [PubMed] [Google Scholar]
- 4.Coleman, N. V., T. E. Mattes, J. M. Gossett, and J. C. Spain. 2002. Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl. Environ. Microbiol. 68:6162-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman, N. V., and J. C. Spain. 2003. Distribution of the coenzyme M pathway of epoxide metabolism among ethene- and vinyl chloride-degrading Mycobacterium strains. Appl. Environ. Microbiol. 69:6041-6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, N. V., and J. C. Spain. 2003. Epoxyalkane:coenzyme M transferase in the ethene and vinyl chloride biodegradation pathways of Mycobacterium strain JS60. J. Bacteriol. 185:5536-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danko, A. S. 2005. Use of vinyl chloride as a growth substrate by aerobic bacteria: pathway analysis and involvement of linear plasmids. Ph.D. thesis. Clemson University, Clemson, S.C.
- 8.Danko, A. S., M. Luo, C. E. Bagwell, R. L. Brigmon, and D. L. Freedman. 2004. Involvement of linear plasmids in aerobic biodegradation of vinyl chloride. Appl. Environ. Microbiol. 70:6092-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grady, C. P. L. J., B. F. Smets, and D. S. Barbeau. 1996. Variability in kinetic parameter estimates: possible causes and a proposed terminology. Water Res. 30:742-748. [Google Scholar]
- 10.Gunsalus, R. P., J. A. Romesser, and R. S. Wolfe. 1978. Preparation of coenzyme M analogues and their activity in the methyl coenzyme M reductase system of Methanobacterium thermoautotrophicum. Biochemistry 17:2374-2377. [DOI] [PubMed] [Google Scholar]
- 11.Hartmans, S., and J. A. M. de Bont. 1992. Aerobic vinyl chloride metabolism in Mycobacterium aurum L1. Appl. Environ. Microbiol. 58:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalb, V. F., and R. W. Bernlohr. 1977. A new spectrophotometric assay for protein in cell extracts. Anal. Biochem. 82:362-371. [DOI] [PubMed] [Google Scholar]
- 13.Krum, J. G., and S. A. Ensign. 2001. Evidence that a linear megaplasmid encodes enzymes of aliphatic alkene and epoxide metabolism and coenzyme M (2-mercaptoethanesulfonate) biosynthesis in Xanthobacter strain Py2. J. Bacteriol. 183:2172-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krum, J. G., and S. A. Ensign. 2000. Heterologous expression of bacterial epoxyalkane:coenzyme M transferase and inducible coenzyme M biosynthesis in Xanthobacter strain Py2 and Rhodococcus rhodochrous B276. J. Bacteriol. 182:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattes, T. E., N. V. Coleman, J. C. Spain, and J. M. Gossett. 2005. Physiological and molecular genetic analyses of vinyl chloride and ethene biodegradation in Nocardioides sp. strain JS614. Arch. Microbiol. 183:95-106. [DOI] [PubMed] [Google Scholar]
- 16.Saeki, H., M. Akira, B. Keizo, B. Averhoff, and G. Gottschalk. 1999. Degradation of trichloroethene by a linear-plasmid-encoded alkene monooxygenase in Rhodococcus corallinus (Nocardia corallina) B-276. Microbiology 145:1721-1730. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Thauer, R. K. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144:2377-2406. [DOI] [PubMed] [Google Scholar]
- 19.van Hylckama Vlieg, J. E. T., J. Kingma, A. J. van den Wijngaard, and D. B. Janssen. 1998. A glutathione S-transferase with activity towards cis-1,2-dichloroepoxyethane is involved in isoprene utilization by Rhodococcus sp. strain AD45. Appl. Environ. Microbiol. 64:2800-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verce, M. F., R. L. Ulrich, and D. L. Freedman. 2000. Characterization of an isolate that uses vinyl chloride as a growth substrate under aerobic conditions. Appl. Environ. Microbiol. 66:3535-3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolfe, R. S. 1991. My kind of biology. Annu. Rev. Microbiol. 45:1-35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.