Abstract
The detection and prevention of cyanobacterial blooms are important issues in water quality management. As such, the diversity and community dynamics of cyanobacteria during cyanobacterial bloom in the Daechung Reservoir, Korea, were studied by analyzing the intergenic spacer (IGS) region between phycocyanin subunit genes cpcB and cpcA (cpcBA IGS). To amplify the cpcBA IGS from environmental samples, new PCR primers that could cover a wider range of cyanobacteria than previously known primers were designed. In the samples taken around the bloom peak (2 September 2003), seven groups of cpcBA IGS sequences were detected, and none of the amplified cpcBA IGSs was closely related to the cpcBA IGS from chloroplasts. Apart from the Microcystis-, Aphanizomenon (Anabaena)-, Pseudanabaena-, and Planktothrix (Oscillatoria)-like groups, the three other groups of cpcBA IGS sequences were only distantly related to previously reported sequences (<85% similarity to their closest relatives). The most prominent changes during the bloom were the gradual decrease and eventual disappearance of the Aphanizomenon (Anabaena)-like group before the bloom peak and the gradual increase and sudden disappearance of Planktothrix (Oscillatoria)-like groups right after the bloom peak. The community succession profile obtained based on the cpcBA IGS analysis was also supported by a PCR-denaturing gradient gel electrophoresis analysis of the 16S rRNA genes.
Bloom-forming cyanobacteria in freshwater are a serious problem for the management of drinking water, since they produce a wide range of toxic compounds, including neurotoxins and hepatotoxins (5). Thus, proper environmental management of water supplies relies on prior knowledge of cyanobacterial ecology. However, the study of cyanobacterial diversity relies largely on the use of microscopic techniques and is very labor intensive. Moreover, since the isolation of cyanobacteria from samples is not always successful, rapid identification of cyanobacteria without cultivation is important. Molecular technologies based on 16S rRNA gene amplification are already widely employed for the analysis of environmental samples (9, 19). Yet, the 16S rRNA gene is often restricting in regard to resolving bacterial strains due to its slow evolution. Furthermore, when other bacteria are present in samples, selective identification of cyanobacteria can be severely hindered. Indeed, diverse noncyanobacterial prokaryotes are associated with cyanobacterial blooms (9).
Of the functional genes used for the taxonomic study of cyanobacterial strains, including the cpcBA intergenic spacer (IGS) (7, 15, 17, 20, 22), nifH (7), rpoC1 (24), and gyrB (24), the cpcBA IGS is specific to cyanobacteria and has been widely used for the phylogenetic analysis of pure cyanobacterial culture strains. Baker et al. (1, 2) recently employed a PCR amplification method to analyze the cpcBA IGSs from environmental samples, using a primer set previously designed by Neilan et al. (20), and found a limited cyanobacterial diversity. Although the primer set was originally designed with six cpcBA IGSs to study the genetic diversity of several pure culture strains, cpcBA IGS sequence information from various other cyanobacteria has also been deposited in public databases for potential enhanced primer design.
Cyanobacterial blooms capable of producing microcystins are a seasonal problem every summer in the Daechung Reservoir, which is a representative large eutrophic lake in Korea (21). Accordingly, to further elucidate the composition and dynamics of cyanobacteria during bloom, this study investigated the cpcBA IGS diversity in addition to physicochemical and biological factors. To analyze the cpcBA IGS diversity, new degenerate primers were designed based on more than 300 cpcBA IGS sequences that are currently available in public databases. Finally, the cyanobacterial diversity derived from the cpcBA IGS analysis was compared with that determined by 16S rRNA gene PCR-denaturing gradient gel electrophoresis (DGGE).
MATERIALS AND METHODS
Sampling and field survey.
The Daechung Reservoir in Korea is an artificial lake created by the construction of a dam in 1980, and its water is used for drinking, as well as agricultural and industrial uses. The reservoir is a large branch-type lake with a 72-m-high dam and a gross storage capacity of 1,490 Mm3. The water sampling was conducted weekly from a floating wharf about 20 m off shore near the Daechung dam from 15 July to 14 October 2003. Surface water above a depth of 20 cm was collected after some mixing, and then the samples were stored in 20-liter polyethylene bottles at 4°C in the dark and the laboratory analysis performed within 24 h.
Water quality analysis.
The water temperature, pH, and conductivity were all measured in situ, using a YSI meter (63/100 FT; YSI Inc., Yellow Springs, OH), while the dissolved oxygen (DO) and turbidity were measured with a DO meter (95/100 FT; YSI Inc., Yellow Springs, OH) and a turbidimeter (DRT-15CE; HF Scientific Inc., Fort Meyers, FL), respectively. The Secchi depth was measured using a Secchi disk. The total N (TN) and P (TP) were determined after persulfate oxidation to nitrate (6) and orthophosphate (18), respectively. The resulting nitrate was then determined by a second-derivative method (4), while the orthophosphate was determined using an ascorbic acid method (8). The total dissolved nitrogen (TDN) and total dissolved phosphorus (TDP) were determined after filtering the water sample through a GF/C filter (Whatman Ltd., Maidstone, United Kingdom) and persulfate oxidation. The total particulate nitrogen (TPN) and total particulate phosphorus (TPP) were obtained by subtracting the TDN from the TN and the TDP from the TP, respectively.
The samples used for plankton identification and enumeration were preserved in Lugol's solution and enumerated with a hemocytometer (Fuchs-Rosenthal; Paul Marienfeld GmbH & Co., Lauda-Königshofen, Germany) under phase-contrast microscopy (Microphot-FXA; Nikon Corp., Tokyo, Japan). The chlorophyll a was extracted using a chloroform-methanol mixture (2:1 [vol/vol]) and measured with a fluorometer (Turner 450; Barnstead/Thermolyne, Dubuque, IA) (29). To determine the picocyanobacterial chlorophyll a, the large cyanobacteria were removed using a filter with a 3-μm pore size, and only the fraction that remained on a 0.2-μm filter was used (10).
Primer design for PCR amplification of cpcBA IGS.
To monitor the diversity and community composition of the cyanobacteria based on the cpcBA IGS, a PCR primer set was specifically designed for cyanobacterial cpcBA IGS amplification. Among all known cpcBA IGS sequences (more than 300 sequences), 24 representative cpcBA IGSs from diverse cyanobacteria (Cylindrospermopsis raciborskii, GenBank accession number AF426795; Cylindrospermopsis raciborskii, AF426792; Arthrospira sp. strain Maxima, AJ401168; Microcystis aeruginosa EAWAG175, AJ003181; Fischerella sp., M75599; Nostoc sp. strain PCC 7120, X05239; Anabaena lemmermannii BC Ana 0018, AY886908; Nodularia spumigena nsb105, AF101444; Synechococcus elongatus, D13173; Synechocystis sp. strain PCC 9413, AF068771; Pseudanabaena sp., M99426; Calothrix sp., M36276; Fremyella diplosiphon, X07012; Synechocystis sp. strain PCC 6803, U34930; Chroococcus dispersus, AJ003184; Agmenellum quadruplicatum, K02660; Synechococcus sp. strain WH7803, X59809; Synechococcus sp. strain PCC 7942, AB008546; Synechococcus elongatus PCC 6301, AP008231; Anacystis nidulans, M94218; Oscillatoria sp. strain PCC 6304, AJ401186; Planktothrix rubescens BC-Pla 9307, AJ131820; Planktothrix sp. strain FP1, AF212923; Gloeobacter violaceus PCC 7421, BA000045; Microcystis sp. strain KLL MG-K, AY524850; Microcystis sp. strain KLL MB-J, AY524849; Microcystis sp. strain KLL MB-K, AY524848; Aphanizomenon sp. Norman Lake isolate, AJ243969; and Aphanizomenon flos-aquae, AJ243971) were aligned by using Clustal X (27). In addition, two conserved regions of cpcB and cpcA were chosen to design the primer set (named CPC1F and CPC1R, respectively) used in this study.
The genomic DNAs from these bacteria were extracted as described previously (13), and the PCR was performed with a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems, Foster City, CA) using a 20-μl (total volume) reaction mixture containing 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl, 0.1% Triton X-100, pH 9.0), four deoxyribonucleoside triphosphates at concentrations of 1 mM, 1.5 mM MgCl2, each primer at a concentration of 1 μM, 4 μg of bovine serum albumin (Roche Diagnostics Corp., Indianapolis, IN), and 2.5 U of Taq DNA polymerase (Perkin-Elmer, Norwalk, CT). The amplification conditions were 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final extension step of 72°C for 7 min.
cpcBA IGS and 16S rRNA gene analysis of bloom samples.
For genomic DNA extraction from the bloom samples, about 500 ml of the water samples was filtered using 0.22-μm membrane filter (Millipore System, Bedford, MA) and the dozens of resulting filter papers were stored at −65°C until required. The bulk community DNA was directly extracted from the filter by grinding the frozen samples and treating them with sodium dodecyl sulfate for cell lysis (13). The conditions used for cpcBA IGS amplification from the environmental samples were the same as those used in the PCR primer evaluation described above. The amplified PCR products were directly ligated into a pCR II vector obtained from Invitrogen (San Diego, CA). The ligation and transformation were carried out as described previously (28). A total of 100 clones per sample were screened, and 50 positive clones per sample were randomly selected and sequenced using the primers CPC1F and CPC1R. The diversity coverage by the clone libraries was analyzed using a rarefaction method, as previously described (25).
The cpcBA IGS sequences obtained in this study were aligned with all those available in current databases by using the Clustal X program (27) and then edited using BioEdit (12). The initial phylogenetic trees were based on all available sequences and constructed using the neighbor-joining DNA distance program (23) in MEGA 2 (16), with bootstrap values based on 1,000 replications (11). Based on the initial phylogenetic results, appropriate subsets of cpcBA IGS sequences were selected and subjected to a final phylogenetic analysis.
Bacterial diversity analysis based on 16S rRNA gene PCR-DGGE was performed as described by Ishii and Fukui (14) and Muyzer et al. (19). A 560-bp (bp 341 to 907 in Escherichia coli numbering) fragment of the 16S rRNA gene from four samples used in the cpcBA IGS analysis was amplified and analyzed using DGGE. The bands were excised from the DGGE gel and incubated in distilled water for 24 h at 4°C. The eluent was then reamplified for sequencing with the primers without a GC clamp.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the clones are as follows: P1-5R, AY942919; P1-30, AY942910; P2-20, AY942916; P2-31, AY942914; P2-48, AY942917; P2-52, AY942907; P3-26, AY942908; P3-36, AY942918; P3-51, AY942909; P3-59, AY942915; P4-9, AY942906; P4-21, AY942905; P4-25, AY942912; P4-29, AY942913; P4-30, AY942911; P4-56, AY942920; 16S-1, AY942899; 16S-2, AY942896; 16S-3, AY942902; 16S-4, AY942903; 16S-5, AY942894; 16S-6, AY942895; 16S-7, AY942897; 16S-8, AY942900; 16S-9, AY942904; 16S-10, AY942901; and 16S-11, AY942898.
RESULTS AND DISCUSSION
Physicochemical and biological characteristics of blooming water body.
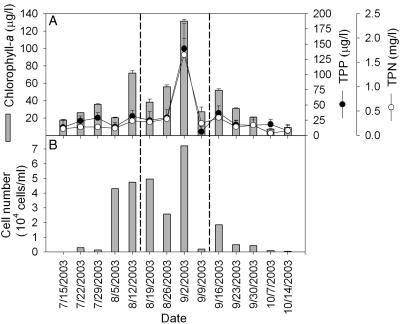
The chlorophyll a concentration, an indicator of a trophic state, revealed that the Daechung Reservoir was in a eutrophic state that became more severe in summer and early autumn (Fig. 1A). The cell count profile, water turbidity, Secchi depth, and chlorophyll a (Fig. 1B and Table 1) collectively indicated that the most significant bloom occurred around 2 September 2003. Four samples out of a total of 14 were collected before and after this bloom peak to analyze the cyanobacterial community. In the bloom sample taken on 2 September 2003, several characteristics of the water body were significantly different from those taken at different times. Most notably, the peak of the bloom was associated with the highest concentrations of TP and TN. It has already been reported that increased concentrations of N and P after rainfall influence cyanobacterial growth in Daechung Reservoir (21). As such, the apparent peak of cyanobacterial bloom that occurred on 2 September 2003 is an annual phenomenon, which is generally observed at the transition point between summer and autumn.
FIG. 1.
Changes in chlorophyll a, TPP, and TPN concentrations (A) and number of total phytoplankton (B) in Daechung Reservoir. The sampling was carried out at 1-week intervals. The samples between the dashed lines were used for further molecular analysis. The error bars indicate standard deviations (n = 3).
TABLE 1.
Biogeochemical characteristics of bloom water
| Property (unit) | Value on sampling date (mo/day):
|
|||
|---|---|---|---|---|
| 8/19 | 8/26 | 9/2 | 9/9 | |
| Turbidity (NTUa) | 8.33 | 14.33 | 33.33 | 12.00 |
| Secchi depth (m) | 1.33 | 1.10 | 1.00 | 1.20 |
| Chlorophyll a (μg/liter) | 38.4 | 56.1 | 131.5 | 27.3 |
| pH | 9.06 | 9.73 | 9.26 | 8.79 |
| Water temp (°C) | 25.2 | 26.5 | 24.8 | 25.5 |
| Conductivity (μS/cm) | 88.3 | 97.0 | 88.3 | 91.0 |
| DO (mg/liter) | 8.50 | 11.63 | 9.60 | 12.43 |
| Nutrient concnb | ||||
| TN (mg/liter) | 1.21 (0.06) | 1.10 (0.03) | 2.59 (0.34) | 0.91 (0.02) |
| TDN (mg/liter) | 0.93 (0.02) | 0.76 (0.03) | 0.93 (0.04) | 0.66 (0.03) |
| TPN (mg/liter) | 0.28 (0.06) | 0.34 (0.04) | 1.66 (0.34) | 0.25 (0.03) |
| TP (μg/liter) | 49.4 (7.0) | 42.0 (9.6) | 154.3 (41.0) | 17.0 (8.0) |
| TDP (μg/liter) | 24.2 (12.2) | 12.6 (8.6) | 11.6 (2.3) | 10.9 (2.7) |
| TPP (μg/liter) | 24.9 (14.1) | 29.4 (12.9) | 142.7 (41.1) | 6.2 (8.4) |
| Cell count (% or no./ml)c | ||||
| Cyanobacteria | 98.6 | 95.2 | 95.8 | 74.2 |
| Phormidium | 0 | 0 | 0 | 375 |
| Planktothrix | 40,250 | 18,313 | 44,125 | 750 |
| Microcystis | 9,375 | 7,500 | 28,125 | 406 |
| Anabaena | 0 | 0 | 83 | 113 |
| Chroococcus | 0 | 0 | 0 | 306 |
| Green algae | 1.2 | 3.8 | 4.1 | 19.9 |
| Chlamydomonas | 0 | 400 | 0 | 0 |
| Scenedesmus | 63 | 125 | 0 | 25 |
| Chlorella | 344 | 463 | 3,125 | 500 |
| Staurastrum | 219 | 50 | 0 | 0 |
| Diatoms | 0.1 | 0.9 | 0.0 | 5.7 |
| Synedra | 63 | 38 | 0 | 13 |
| Aulacoseira | 0 | 0 | 0 | 25 |
| Fragilaria | 0 | 225 | 0 | 113 |
| Melosira | 0 | 0 | 0 | 31 |
| Surirella | 0 | 13 | 0 | 0 |
| Asterionella | 0 | 0 | 0 | 56 |
| Nitzschia | 0 | 0 | 0 | 6 |
NTU, nephelometric turbidity units.
The nutrient concentrations were measured three times, and the averages and standard deviations (in parentheses) are shown.
The relative values of the cell counts for cyanobacteria, green algae, and diatoms are indicated as percentages; counts for individual groups are indicated as number of cells per milliliter.
By microscopic counting, the cyanobacterial counts were found to remain high (accounting for 74 to 99%) during the bloom season (Table 1), thereby directly contributing to the bloom peak on 2 September 2003 and the higher chlorophyll a concentration observed at this time. However, the most significant change at the bloom peak, as determined by the microscopic analysis, was a prominent increase in the mass of Planktothrix and Microcystis. After the bloom peak, there was a decrease in both the relative and absolute numbers of cyanobacteria in the total algae (Table 1). Nonetheless, the eukaryotic algae (green algae and diatoms) did not significantly affect the bloom, and there was no significant difference in any other physicochemical properties, such as the pH, water temperature, DO, TDN, and TDP, during the bloom.
Design of primers for PCR amplification of the cpcBA IGS.
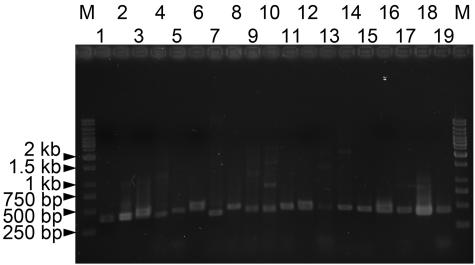
Although the reported cpcB sequences are quite diverse (nucleotide similarity of 266 bp at the 3′ end of the cpcB gene, 72.3 ± 7.4% [mean ± standard deviation]; range of pairwise similarity, 56 to 98%), several highly conserved regions were useful for the primer design. The priming site of PCbF (20), positioned at nucleotide 250 according to the numbering for Microcystis sp. strain KLL MB-J (accession no. AY524849) cpcB, was selected for the design of the new forward primer CPC1F (5′-GGCKGCYTGYYTRCGYGACATGGA-3′). Since several mismatches were found in the PCbF primer, degeneracy was applied to increase the coverage of CPC1F. In addition, a new priming site near the 5′ end of cpcA (nucleotide 43 according to the numbering for Microcystis sp. strain KLL MB-J [accession no. AY524849] cpcA) was explored for the reverse primer PC1R (5′-GCHGATWCYCAAGGNCGYTT-3′). Among the primer sets previously reported (3, 20) and those designed in this study, the CPC1F-CPC1R pair produced the most successful amplification of representative test strains from the cyanobacterial collections of PCC, UTEX, NIES, SAG, and the Korean Collection for Type Cultures (KCTC) (data not shown). As shown in Fig. 2, cpcBA IGSs were amplified from diverse cyanobacteria. As expected, due to size variations in the IGS region, the amplicon size ranged between 400 and 500 bp, indicating that the newly designed primer set was able to cover cpcBA IGSs from diverse cyanobacteria.
FIG. 2.
Photographs of ethidium bromide-stained gels showing amplification products of cpcBA IGSs from reference cyanobacteria. The strains used were as follows: 1, Aphanothece nidulans KCTC AG10041; 2, Aphanothece naegelii KCTC AG10042; 3, Microcystis aeruginosa UTEX 2385; 4, Microcystis sp. strain PCC 7806; 5, Synechococcus sp. strain PCC 7002; 6, Synechocystis sp. strain PCC 6803; 7, Merismopedia tenuissima NIES 230; 8, Arthrospira maxima SAG 49.88; 9, Arthrospira platensis NIES 39; 10, Oscillatoria tenuis UTEX 1566; 11, Oscillatoria tenuis NIES 33; 12, Planktothrix agardhii NIES 204; 13, Anabaena flos-aquae UTEX 2517; 14, Anabaena affinis KCTC AG10008; 15, Anabaena sp. strain KCTC AG10059; 16, Aphanizomenon flos-aquae NIES 81; 17, Nodularia spumigena UTEX 2092; 18, Nostoc sp. strain PCC 7120; 19, Chlorogloeopsis sp. strain PCC 9212. Lanes M, markers (1-kb ladder). The numbers on the left indicate the sizes of the markers after electrophoresis.
cpcBA IGS diversity among cyanobacterial bloom samples.
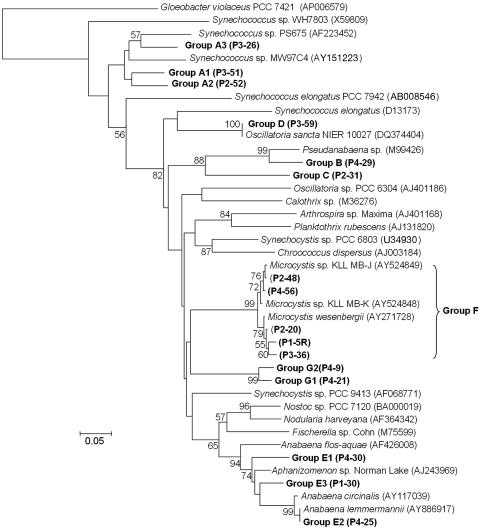
cpcBA IGSs were successfully amplified within 25 cycles from all four bloom samples. For the screened clones, all the sequences were related to the cpcBA IGS, and the rarefaction analysis showed that the number of clones reached a hypothetical saturation value (data not shown), where 50 clones covered most of the cpcBA IGS diversity within each sample. Based on the phylogenetic tree, seven groups of cpcBA IGS sequences were found from the libraries. The cpcB gene part of the PCR product was also selected for further phylogenetic analysis, and although the seven distinct groups of cpcB genes in the phylogenetic tree were widely distributed (Fig. 3), there was little variation in the sequences within each group. Furthermore, since the IGS part did not vary among subgroups, it is possible that each group of sequences was from a genetically identical ecotype. Only the Microcystis-like group (group F) exhibited some variations in the cpcBA IGS sequence, with a >95% intragroup sequence similarity.
FIG. 3.
Phylogenetic relationships of cpcB genes cloned from Daechung Reservoir. The reference sequences were from GenBank. The nucleotide sequences (267 bp from the 3′ end) from cpcB were used for the phylogenetic analysis. cpcB genes with a similarity of more than 95% are grouped. A representative clone from each group is indicated in parentheses. The bar indicates 0.05 substitution per nucleotide position. The local bootstrap probabilities are indicated at the nodes if they are larger than 50%.
With the exception of the Microcystis (group F, >95% similarity to closest relatives)-, Aphanizomenon (Anabaena) (group E, 90% similarity)-, Pseudanabaena (group B, 89% similarity)-, and Planktothrix (Oscillatoria) (group D, 100% similarity)-like groups, all the remaining groups of cpcBA IGS sequences were only distantly related to previously reported sequences (about <85% similarity to closest relatives). None of the amplified cpcBA IGSs was closely related to cpcBA IGSs from chloroplasts. However, the amplification of chloroplast cpcBA IGSs should be excluded, since the primer sites had several mismatches in the chloroplasts (1- to 5-bp mismatches in the forward and reverse primers) (data not shown). Consequently, this suggests that the present protocol could be useful for the specific analysis of cyanobacteria in the presence of other bacteria and eukaryotic algae in environmental samples.
Dynamics of the cyanobacterial community during bloom.
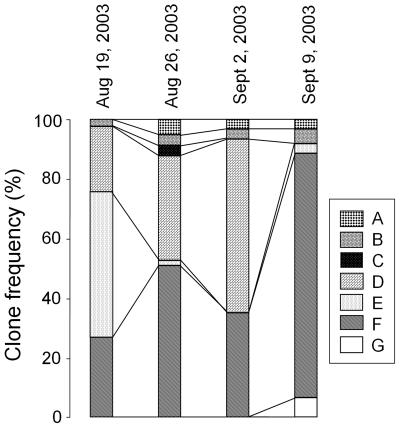
To monitor the changes in the cyanobacterial community during cyanobacterial bloom, the relative composition of the cpcBA IGS sequences in each sample was analyzed. Groups D, E, and F were the major components of the cyanobacterial community on 19 August 2003, as shown in Fig. 4. The two most significant changes around the bloom peak on 2 September 2003 were the gradual decrease to extinction of group E and gradual increase and sudden disappearance of group D on 9 September. Meanwhile, group F steadily increased and finally predominated on 9 September. Interestingly, after the bloom peak, the cyanobacterial community profile did not return to that observed before the peak, as group F accounted for up to 85% of all the clones. However, the total cyanobacterial count, including Microcystis, decreased (down to 30% of that at the bloom peak), as shown in Table 1.
FIG. 4.
Distribution dynamics of the cpcBA IGS sequence group during cyanobacterial bloom in Daechung Reservoir. The frequency distribution was calculated based on 50 clones analyzed from each sample. The dates of sampling are indicated.
This relative increase of Microcystis-like cyanobacteria correlates well with the increased microcystin concentration measured after the bloom peak. Oh et al. (21) also observed that the particulate microcystin concentration in the Daechung Reservoir started to increase from early September after the bloom period. This phenomenon could be explained by an increase of Microcystis in the water body, as observed in this study. Thus, the Aphanizomenon (Anabaena)-like cyanobacteria were not involved in toxin production. The complete disappearance of group D also seemed to be an important marker for the transition of the cyanobacterial community from summer to autumn. The other groups (A, B, C, and G) had only a minor presence during the bloom period. Thus, the present cpcBA IGS-based approach revealed that the changes in the cyanobacterial community in response to environmental stimuli were more dynamic than those observed by microscopy.
Comparison of cpcBA IGS and 16S rRNA gene analyses for cyanobacterial bloom study.
The cpcBA IGS-based method was compared with a 16S rRNA gene-based method using 16S rRNA gene PCR-DGGE. The best matches with the 16S rRNA gene sequences from the DGGE bands were identified using BLAST (Table 2). As expected, 16S rRNA genes for bacteria and chloroplasts were amplified together with those for cyanobacteria. Dynamic changes in the bacterial community were observed during the bloom period in the DGGE profile (Fig. 5), and based on a comparison of the phylogenies of the 16S rRNA and cpcBA IGS and their succession patterns, tentative correlations were found between the cpcBA IGS sequence groups and cyanobacterial DGGE bands. For example, the sequences of DGGE band 10 and cpcBA IGS group F, which closely matched those of Microcystis (>98% and 100% similarities, respectively), were present throughout the bloom period, while the sequences of DGGE band 8 and cpcBA IGS group E, which closely matched those of Aphanizomenon and Anabaena, occupied a major proportion on 19 August 2003. Group E and band 8 were present as a minor component in the sample taken from 26 August to 9 September 2003. However, no Aphanizomenon- or Anabaena-like microorganisms were observed by microscopy on 19 August 2003 (Table 1). Considering that Aphanizomenon and Anabaena were observed in over 10% of cyanobacteria on 12 August 2003, Aphanizomenon and Anabaena were presumed to have existed on 19 August, even at low densities, but were not observed by microscopic enumeration. These results indicate that molecular techniques are more sensitive than microscopic observation for community study. Another reason could be the different sample volumes for each analysis, because about a 10-times-greater volume was used for molecular methods than for microscopy. Small flock formation by rarely occurring Aphanizomenon and Anabaena and their heterogeneous distribution could have amplified such a discrepancy.
TABLE 2.
Identities of bands obtained from DGGE analysis of bloom samplesa
| Band no. | Closest relative | GenBank accession no. | Similarity (%) |
|---|---|---|---|
| 1 | Oscillatoria sanctab | AY074801 | 99 |
| 2 | Aulacoseira ambigua chloroplast | AJ536463 | 97 |
| 3 | CFB group bacteriumc | AF236016 | 95 |
| 4 | Uncultured Bacteroidetes | AJ583816 | 97 |
| 5 | Uncultured phytoplankton ESR 3 | AF268287 | 96 |
| 6 | Uncultured phytoplankton ESR 3 | AF268287 | 97 |
| 7 | Chlamydomonas reinhardtii chloroplast | BK000554 | 85 |
| 8 | Aphanizomenon flos-aquae | AJ630443 | 100 |
| 9 | Uncultured Fibrobacteres bacterium | AY509521 | 99 |
| 10 | Microcystis flos-aquae | AF139327 | 100 |
| 11 | Oscillatoria sanctab | AY074801 | 100 |
See Fig. 5.
This isolate was deposited under the name Oscillatoria sancta (unpublished), which was verified as a Planktothrix-like cyanobacterium (see the text for details).
CFB, Cytophaga-Flavobacterium-Bacteroides.
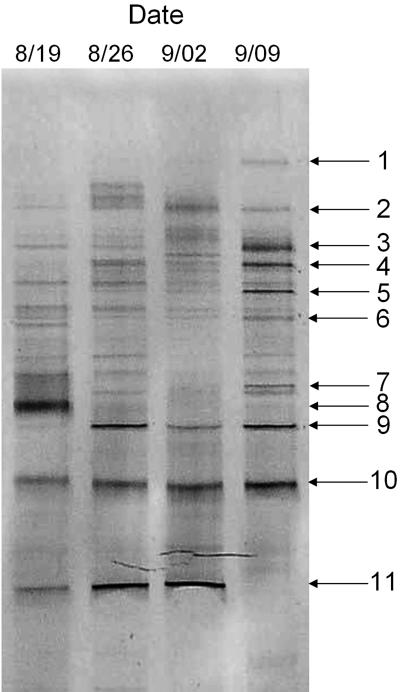
FIG. 5.
16S rRNA gene-DGGE profile during cyanobacterial bloom in Daechung Reservoir. The reproducibility of the DGGE profile was confirmed by three replications of the PCR-DGGE of the genomic DNA. The dates of sampling are indicated.
The pattern of DGGE band 11 and profile of cpcBA IGS group D were highly related to those of Planktothrix (Oscillatoria)-like cyanobacteria identified in the microscopic analysis (Table 1). The sequences of DGGE band 11 and cpcBA IGS group D (P3-59) perfectly matched those of Oscillatoria sancta NIER 10027, which was isolated in Daechung Reservoir (Table 2 and Fig. 3). Because most members of Oscillatoria were taxonomically revised as members of Planktothrix and O. sancta NIER 10027 showed the characteristics of Planktothrix mougeotii (26), O. sancta NIER 10027 could be regarded as Planktothrix. Group D and band 11 were absent in the sample taken on 9 September 2003. The DGGE band 3, 4, and 9 sequences were from uncultured bacteria, whereas the DGGE band 2 and 7 sequences were from eukaryotic algal chloroplasts. Therefore, these comparison data indicate that the cpcBA IGS analysis correlated well with the 16S rRNA gene analysis. The small discrepancies observed between the molecular and microscopic analyses may have reflected the presence of picocyanobacteria that were missed during the microscopic counting. The presence of picocyanobacteria (fewer than 5% of the clones) was also indirectly supported based on the chlorophyll a fraction of picocyanobacteria, which was at 4.90% ± 0.72% and 5.09% ± 0.44% on 24 August and 31 August 2005, respectively.
Improvement of water quality management depends on the development of proper methods to analyze cyanobacterial diversity and determine the cause of toxin production from blooms. In the present study, a molecular analysis of the cpcBA IGS was shown to be efficient for analyzing cyanobacterial diversity, not least because it can exclude other bacteria. Thus, the present protocol could complement conventional tools, such as morphology-based microscopic analyses, the direct determination of toxins, and 16S rRNA gene analyses. Furthermore, since the IGS length of each group of cyanobacteria is different, the proposed cpcBA IGS amplification protocol could also be directly coupled to length polymorphism analysis techniques, such as terminal restriction fragment length polymorphism, to facilitate rapid monitoring of the cyanobacterial community.
Acknowledgments
This study was supported by grants from the Carbon Dioxide Reduction and Sequestration Research Center, a 21st Century Frontier Program funded by the Korean Ministry of Science and Technology (MOST), the KRIBB Research Initiative Program, and the MOST/KOSEF to the Environmental Biotechnology National Core Research Center (R15-2003-012-02001-0).
REFERENCES
- 1.Baker, J. A., B. Entsch, B. A. Neilan, and D. B. McKay. 2002. Monitoring changing toxigenicity of a cyanobacterial bloom by molecular methods. Appl. Environ. Microbiol. 68:6070-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, J. A., B. A. Neilan, B. Entsch, and D. B. McKay. 2001. Identification of cyanobacteria and their toxigenicity in environmental samples by rapid molecular analysis. Environ. Toxicol. 16:472-482. [PubMed] [Google Scholar]
- 3.Crosbie, N. D., M. Pockl, and T. Weisse. 2003. Dispersal and phylogenetic diversity of nonmarine picocyanobacteria, inferred from 16S rRNA gene and cpcBA-intergenic spacer sequence analysis. Appl. Environ. Microbiol. 69:5716-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crumpton, W. G., T. M. Isenhart, and P. D. Mitchell. 1992. Nitrate and organic N analyses with second derivative spectroscopy. Limnol. Oceanogr. 37:907-913. [Google Scholar]
- 5.de Figueiredo, D. R., U. M. Azeiteiro, S. M. Esteves, J. M. G. Fernando, and M. J. Pereira. 2004. Microcystin-producing blooms—a serious global public health issue. Ecotoxicol. Environ. Saf. 59:151-163. [DOI] [PubMed] [Google Scholar]
- 6.D'Elia, C. F., P. A. Steudler, and N. Corwin. 1977. Determination of total nitrogen in aqueous samples using persulfate digestion. Limnol. Oceanogr. 22:760-764. [Google Scholar]
- 7.Dyble, J., H. W. Paerl, and B. A. Neilan. 2002. Genetic characterization of Cylindrospermopsis raciborskii (cyanobacteria) isolates from diverse geographic origins based on nifH and cpcBA-IGS nucleotide sequence analysis. Appl. Environ. Microbiol. 68:2567-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaton, A. D., L. S. Clesceri, and A. E. Greenberg (ed.). 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, D.C.
- 9.Eiler, A., and S. Bertilsson. 2004. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ. Microbiol. 6:1228-1243. [DOI] [PubMed] [Google Scholar]
- 10.Ernst, A., S. Becker, U. I. A. Wollenzien, and C. Postius. 2003. Ecosystem-dependent adaptive radiations of picocyanobacteria inferred from 16S rRNA and ITS-1 sequence analysis. Microbiology 149:217-228. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 12.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 13.Hurt, R. A., X. Qiu, L. Wu, Y. Roh, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishii, K., and M. Fukui. 2001. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl. Environ. Microbiol. 67:3753-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janson, S., and E. Graneli. 2002. Phylogenetic analyses of nitrogen fixing cyanobacteria from the Baltic Sea reveal sequence anomalies in the phycocyanin operon. Int. J. Syst. Evol. Microbiol. 52:1397-1404. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 17.Kurmayer, R., G. Christiansen, J. Fastner, and T. Börner. 2004. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ. Microbiol. 6:831-841. [DOI] [PubMed] [Google Scholar]
- 18.Menzel, D. W., and N. Corwin. 1965. The measurement of total phosphorus in seawater based on the liberation of organically bound fractions by persulfate oxidation. Limnol. Oceanogr. 10:280-282. [Google Scholar]
- 19.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neilan, B. A., D. Jabobs, and A. E. Goodman. 1995. Genetic diversity and phylogeny of toxic cyanobacteria determined by DNA polymorphism within the phycocyanin locus. Appl. Environ. Microbiol. 61:3875-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh, H.-M., S. J. Lee, J.-H. Kim, H.-S. Kim, and B.-D. Yoon. 2001. Seasonal variation and indirect monitoring of microcystin concentration in Daechung Reservoir, Korea. Appl. Environ. Microbiol. 67:1484-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson, B. R., N. Tezuka, and M. M. Watanabe. 2001. Phylogenetic analyses of Synechococcus strains (cyanobacteria) using sequences of 16S rDNA and part of the phycocyanin operon reveal multiple evolutionary lines and reflect phycobilin content. Int. J. Syst. Evol. Microbiol. 51:861-871. [DOI] [PubMed] [Google Scholar]
- 23.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 24.Seo, P.-S., and A. Yokota. 2003. The phylogenetic relationships of cyanobacteria inferred from 16S rRNA, gyrB, and rpoC1 gene sequences. J. Gen. Appl. Microbiol. 49:191-203. [DOI] [PubMed] [Google Scholar]
- 25.Simberloff, D. 1978. Use of rarefaction and related methods, p. 150-165. In K. L. Dickson, J. Cairns, and R. J. Livingston (ed.), Biological data in water pollution assessment: quantitative and statistical analyses. American Society for Testing and Materials, Philadelphia, Pa.
- 26.Suda, S., M. M. Watanabe, S. Otsuka, A. Mahakahant, W. Yongmanitchai, N. Nopartnaraporn, Y. Liu, and J. G. Day. 2002. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int. J. Syst. Evol. Microbiol. 52:1577-1595. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waser, M., D. Hess-Bienz, K. Davies, and M. Solioz. 1992. Cloning and disruption of a putative NaH-antiporter gene of Enterococcus hirae. J. Biol. Chem. 267:5396-5400. [PubMed] [Google Scholar]
- 29.Wood, L. W. 1985. Chloroform-methanol extraction of chlorophyll-a. Can. J. Fish. Aquat. Sci. 42:38-43. [Google Scholar]