Abstract
Stable microbial communities associated with health can be disrupted by altered environmental conditions. Periodontal diseases are associated with changes in the resident oral microflora. For example, as gingivitis develops, a key change in the microbial composition of dental plaque is the ascendancy of Actinomyces spp. and gram-negative rods at the expense of Streptococcus spp. We describe the use of an in vitro model to replicate this population shift, first with a dual-species model (Actinomyces naeslundii and Streptococcus sobrinus) and then using a microcosm model of dental plaque. The population shift was induced by environmental changes associated with gingivitis, first by the addition of artificial gingival crevicular fluid and then by a switch to a microaerophilic atmosphere. In addition to the observed population shifts, confocal laser scanning microscopy also revealed structural changes and differences in the distribution of viable and nonviable bacteria associated with the change in environmental conditions. This model provides an appropriate system for the further understanding of microbial population shifts associated with gingivitis and for the testing of, for example, antimicrobial agents.
Bacteria colonize a variety of surfaces of the human body, including the digestive tract, the respiratory tract, the skin, and the oral cavity. These organisms can coexist with the human host in a mutually beneficial way (31) or as commensal organisms which neither benefit nor harm the human host. The normal resident microbiota can prevent colonization of pathogenic organisms by colonization resistance (19), occupying the environmental niche in which these organisms could thrive. This relationship is highly dependent on the balance of host factors, environmental factors, and microbial interactions. Potentially pathogenic bacteria which may be present (in very small proportions) as part of the normal microbiota can become dominant if this equilibrium is disrupted.
Plaque-related diseases, such as gingivitis and periodontitis, are linked to fluctuations in the oral environment which lead to a change in microbial composition. Gingivitis is the result of poor dental hygiene, leading to plaque accumulation and inflammation of the gingiva. It is estimated to affect between 40 and 50% of North American adults (5) and, if left untreated, can lead to the irreversible condition of periodontitis, in which loss of attachment of the tooth to supporting structures occurs, which can ultimately lead to tooth loss. Although not all cases of periodontitis are preceded by gingivitis, the prevention of gingivitis in the general population is thought to be the way forward of reducing the incidence of periodontal diseases on the whole (20).
Oral streptococci tend to dominate the microbiota of most individuals not suffering from any form of periodontal disease. A key change in the microflora of supragingival plaque observed by experimental gingivitis studies is the ascendancy of Actinomyces spp. and gram-negative rods at the expense of Streptococcus spp. (24, 26, 33, 40). Actinomyces spp. are initial colonizers of the tooth surface (19) and are particularly associated with the accumulation phase of plaque development (17). Actinomyces israelii is associated with nonbleeding gingivitis, while Actinomyces viscosus and Actinomyces naeslundii are associated with bleeding gingivitis (33). Gram-negative species indicative of the changes occurring within the plaque environment, such as Fusobacterium nucleatum (25), Prevotella spp. (14), and Capnocytophaga spp. (23), are more frequently isolated from dental plaque associated with gingivitis, due to the development of gradients in oxygen availability and environmental niches that promote their survival.
A key factor associated with gingivitis is increased plaque thickness, allowing the development of gradients in factors such as oxygen potential, pH, and nutrient availability. In addition, protein-based nutrients become available in the form of gingival crevicular fluid (GCF), the flow rate of which increases during the course of gingivitis (7). Sources of nutrients for plaque bacteria include saliva, GCF, and dietary sources (38) as well as nutrients derived from the degradation of the extracellular polysaccharides present in the biofilm matrix by oral bacteria producing enzymes able to break down these complex macromolecules (13). Changes in these nutritional components are likely to influence plaque composition. Inflammation of the marginal gingiva is important in early, supragingival plaque accumulation (6), which suggests that inflammatory factors influence bacteria involved in plaque accumulation. Indeed, Actinomyces spp. are more frequently isolated from subjects with a strong inflammatory response to gingivitis (16).
Due to the variability of the oral environment, there is great heterogeneity of plaque composition between individuals and from different sites within the mouth (1). In vitro models of the oral cavity allow for greater control of the environmental factors which contribute to this variation in plaque composition, such as nutrient source, temperature, pH, oxygen availability, and substrata (37). The constant-depth film fermentor (CDFF) is established as a representative model for dental plaque (21, 27) producing diverse microbial populations (29) that maintain key oral species at levels similar to those observed in vivo. As key parameters can be controlled using this model, changes in plaque composition can be directly linked to specific environmental changes.
This work is the first to describe an in vitro model for evaluating the bacterial population shifts associated with the onset of gingivitis, first in a dual-species system and then with a more complex microcosm model, by changing environmental parameters. As such shifts in microbial composition may also influence changes in biofilm structure, confocal laser scanning microscopy (CLSM) in combination with viability staining was used to visualize any structural changes and the spatial distribution of live and dead bacteria within these biofilms.
MATERIALS AND METHODS
Bacterial strains.
Actinomyces naeslundii NCTC 10301 and Streptococcus sobrinus NCTC 12279 were grown anaerobically in brain heart infusion broth (Oxoid, Hampshire, United Kingdom) at 37°C to a concentration of approximately 1 × 108 CFU ml−1.
Inoculation of dual-species and microcosm biofilms.
The CDFF was inoculated with 500 ml of artificial saliva (27) containing 10 ml each of A. naeslundii and Streptococcus sobrinus cultures in brain heart infusion broth for the dual-species experiments. This mixture was pumped in at a continuous rate of 1 ml min−1 for 8 h using a peristaltic pump.
In order to produce microcosm biofilms, whole saliva was obtained from 15 individuals and used to create a pooled stock to be used as the inoculum for each experiment. Briefly, equal amounts of saliva from each individual were added to the pool along with glycerol (final concentration, 10% [vol/vol]). This pool was then split into 1-ml aliquots which were stored at −80°C. These 1-ml aliquots were added to 500 ml of artificial saliva and then pumped into the CDFF at a constant rate under microaerophilic conditions (2% O2, 3% CO2, and 95% N at 200 × 105 Pa) for the 8-h inoculation period before being switched back to an aerobic atmosphere once inoculation was ceased.
In vitro model parameters.
The biofilms were grown as previously described (29) using the CDFF. The system was maintained at a constant temperature of 36°C (representative of the oral cavity) by being housed in an incubator. The artificial saliva growth medium was pumped in at a flow rate of 0.72 liter day−1, which is representative of the daily salivary flow rate (4, 10, 15). An aerobic atmosphere was maintained by exposure to the environment via a filtered air inlet in the top plate. After initial comparisons of bovine enamel and hydroxyapatite, as substrata showed no difference in total counts or species proportions, hydroxyapatite disks (5-mm diameter) were used and recessed to a depth of 600 μm. This increased recess depth was used to allow thicker biofilms to develop, as would be seen in interdental regions of supragingival plaque associated with poor dental hygiene. Biofilms were removed aseptically from the CDFF during the course of each run via the sampling port and subjected to various analyses.
Gingivitis conditions.
To provide nutrients associated with the onset of gingivitis, an artificial GCF formulation (37) comprised of 60% RPMI tissue culture medium (used here to provide nutrients present in tissue exudates, which are a major component of GCF) and 40% horse serum, with 0.5 μg ml−1 menadione and 5.0 μg ml−1 haemin (all Sigma, Dorset, United Kingdom), was pumped into the artificial saliva formulation as it flowed into the CDFF at a rate of 50 μl min−1, an approximation of the flow rate of GCF into saliva during gingivitis (7). This was accompanied by a switch to a more microaerophilic environment by pumping in a microaerophilic gas mixture (2% O2, 3% CO2, and 95% N at 200 × 105 Pa), with an oxygen content associated with periodontal disease (18), via a filtered air inlet in the top plate at a rate of 200 cm3 min−1.
Cultural analysis.
After aseptic removal from the CDFF, biofilms were placed in 1 ml of sterile phosphate-buffered saline containing five glass beads and vortexed for 1 min to create a homogeneous suspension. This suspension was then serially diluted and plated onto appropriate media to give viable counts and species proportions. Serial dilutions were plated onto fastidious anaerobe agar (Bioconnections, Leeds, United Kingdom) to give total anaerobic counts and on Columbia blood agar (Oxoid, Hampshire, United Kingdom) to give total aerobic counts. Actinomyces spp. were selected for on cadmium fluoride-acriflavine-tellurite plates (41), and Streptococcus spp. were selected for on mitis salivarius agar (BD Biosciences, Oxford, United Kingdom). For microcosm plaques, gram-negative species were selected for on Columbia blood agar with a gram-negative supplement (Oxoid), Veillonella spp. were selected for on Veillonella agar (BD Biosciences), and Lactobacillus spp. on Rogosa agar (Oxoid). All plates were incubated anaerobically at 37°C for 4 days, except fastidious anaerobe agar, cadmium fluoride-acriflavine-tellurite plates, and gram-negative selective agar, which were incubated for up to 2 weeks to allow slower-growing colonies to become distinct. Columbia blood agar plates were incubated aerobically at 37°C overnight. On all selective media, species were confirmed by colony morphology and Gram reaction.
Biofilm visualization.
Biofilms were analyzed by CLSM using methods described previously (12). Briefly, the hydroxyapatite disks supporting the biofilms were placed onto a 5-cm (diameter) petri dish and held in place by vacuum grease, biofilm side up. The biofilms were carefully submerged in 8 ml of a 1:4,000 dilution of BacLight LIVE/DEAD viability stain (Molecular Probes, Oregon) in water and then incubated in the dark for 10 min. Replicates were examined with a Radiance 3000 confocal laser scan head (Bio-Rad GmbH, Jena, Germany) in conjunction with a BX51 stereomicroscope (Olympus UK Ltd., Southall, United Kingdom), equipped with a 40× HCX water immersion dipping lens with a numerical aperture of 0.8 μm. The lasers used were a helium neon (543 nm) laser and an argon (488 nm) laser. The thickness of each confocal optical section was 3 μm. The resulting collections of confocal optical sections were collected by Bio-Rad LaserSharp software as stacks of images (*.PIC files) and archived onto optical disks. At each time point, three biofilms were examined by CLSM, and for each disk, at least three different points within the biofilm were observed.
Image analysis.
All images were analyzed using Image J 1.22d software (NIH). Three-dimensional images were created from the live (green) and dead (red) color channels using the three-dimensional project tool of Image J and then combined to create a single RGB (red-green-blue) stack using RGB merge, allowing the spatial visualization of live and dead bacteria within the biofilm structure. To examine the relative intensity of fluorescence for the live and dead channels through each optical section of the biofilms, intensity profiles were generated using the plot z-axis tool of Image J. The intensity values obtained were normalized against the maximum image intensity value for each channel as described previously (11).
RESULTS
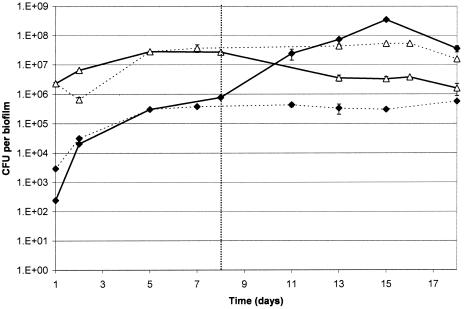
In dual-species biofilms grown aerobically and fed with artificial saliva, S. sobrinus was the dominant organism throughout, achieving a stable population after 7 days, with viable counts of 5.4 × 107 CFU per biofilm. In contrast, Actinomyces spp. reached a maximum viable count of 5.7 × 105 CFU per biofilm, although the counts were also stable by day 7 (Fig. 1). With the addition of artificial GCF and microaerophilic gas into the system, A. naeslundii became the dominant organism, with counts of 3.5 × 108 CFU per biofilm, representing more than a 2-log10 increase in numbers. S. sobrinus counts reached a minimum of 1.6 × 106 CFU per biofilm, representing a log10 reduction. In repeat experiments where the times of addition of artificial GCF and microaerophilic gas were varied, the increase in A. naeslundii counts always corresponded to the time environmental conditions were changed (data not shown).
FIG. 1.
Species counts in dual-species biofilms. ⧫, A. naeslundii; ▵, S. sobrinus. Solid lines represent runs in which artificial GCF addition and a switch to microaerophilic conditions were commenced on day 8. Dashed lines represent runs in which no change in conditions was implemented. Error bars represent the standard deviations (n = 4).
Table 1 shows the selected species proportions from a typical microcosm dental plaque experiment before and after the addition of artificial GCF and microaerophilic gas. Before the change in environmental conditions, Streptococcus spp. dominated the biofilms and accounted for almost 60% of the total cultivable organisms present on the anaerobic agar plates. After the addition, Actinomyces spp. were the dominant genera, increasing from 4 to 70% of the total anaerobes. Veillonella and Lactobacillus proportions increased as did gram-negative species, which showed a 10-fold increase and accounted for a greater proportion of cultivable species than Streptococcus spp.
TABLE 1.
Proportions of selected species before and after the addition of artificial GCF and microaerophilic gas
| Bacteria | Biofilm formation (%)a
|
|
|---|---|---|
| Before addition | After addition | |
| Streptococcus spp. | 58.4 (7.1) | 5.3 (0.9) |
| Actinomyces spp. | 4.5 (0.3) | 70.1 (15.8) |
| Lactobacillus spp. | 0.9 (0.2) | 2.1 (0.29) |
| Veillonella spp. | 0.8 (0.1) | 1.5 (0.53) |
| Gram-negative species | 0.5 (0.04) | 5.7 (0.60) |
The percentage of total cultivable anaerobes at day 7 (before addition) and day 14 (after addition). Data are means and standard deviations (n = 4).
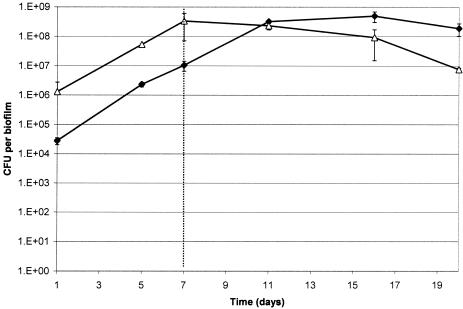
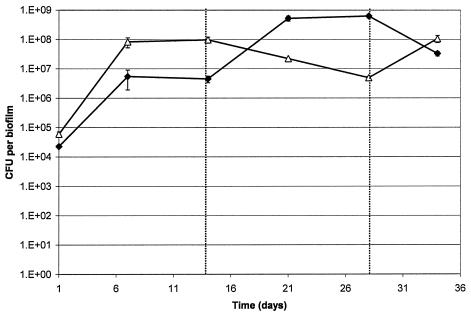
In microcosm biofilms, species proportions were stable after approximately 7 days, with Streptococcus counts reaching 3.4 × 108 CFU per biofilm and Actinomyces spp. reaching 1.0 × 107 CFU per biofilm. After the addition of artificial GCF and microaerophilic gas, Actinomyces spp. became the dominant cultivable genera (Fig. 2), with total numbers reaching 5.0 × 108 CFU per biofilm, representing a log10 increase. This was accompanied by a decrease in Streptococcus counts to 7.6 × 106 CFU per biofilm, representing more than a log10 reduction. Additionally, during further experiments, when GCF and microaerophilic gas addition were ceased, Streptococcus numbers increased to levels seen before the addition (Fig. 3). As seen with the dual-species biofilms, the increase in Actinomyces spp. and decrease in Streptococcus spp. always corresponded to the point where environmental conditions were changed.
FIG. 2.
Species counts from microcosm biofilms when artificial GCF addition and a switch to microaerophilic conditions were commenced on day 7. ⧫, Actinomyces spp.; ▵, Streptococcus spp. Error bars represent the standard deviations (results combined from two separate CDFF experiments; n = 8).
FIG. 3.
Species counts from microcosm biofilms when artificial GCF and microaerophilic gas addition were commenced on day 14 and then ceased on day 28. ⧫, Actinomyces spp.; ▵, Streptococcus spp. Error bars represent the standard deviations (n = 5).
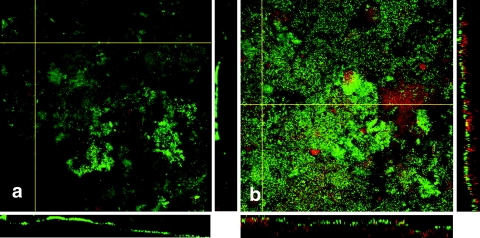
Figure 4 shows images taken from microcosm biofilms grown before or after artificial GCF and microaerophilic gas addition. Before the addition, biofilms displayed an open structure with many visible voids and channels and structural features, such as filaments. Nonviable bacteria presented a significant portion of the biofilm structure but were confined mainly to deeper layers closer to the hydroxyapatite surface. After the addition, biofilms displayed a more tightly packed structure with fewer visible voids. Nonviable bacteria appeared to form a more significant portion of the biofilm structure and were visible in the uppermost regions. Additionally, they appeared to form stacks composed of predominantly nonviable bacteria.
FIG. 4.
CLSM images (300 by 300 μm) of microcosm biofilms taken from 10-day-old biofilms before the addition of artificial GCF and microaerophilic gas (a) and from biofilms after the addition at day 24 (b) at a depth of 50 μm. Green represents viable bacteria, and red represents nonviable bacteria.
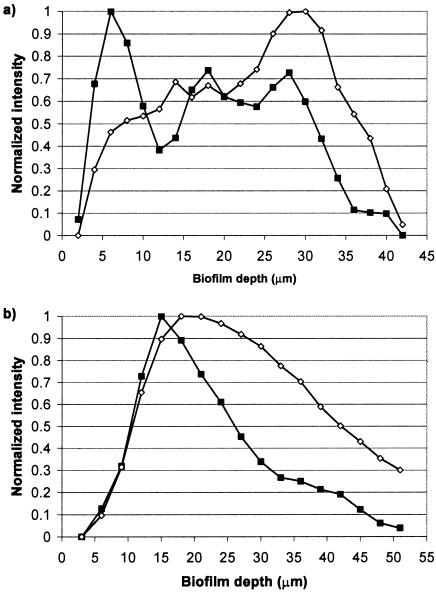
Figure 5 shows the normalized relative fluorescence intensity values of viable and nonviable bacteria through each layer of the confocal image. Before the addition of artificial GCF and microaerophilic gas, nonviable bacteria demonstrated maximum fluorescence in the deeper layers of the biofilm, closer to the substratum, while the maximum fluorescence for viable bacteria was observed in the layers closer to the biofilm surface. After the addition, both nonviable and viable bacteria showed maximum fluorescence in layers closer to the biofilm/air interface. The relative intensity values for viable and nonviable bacteria show a similar trend after the addition, with both showing reduced fluorescence toward the substratum surface.
FIG. 5.
Normalized relative intensity values for fluorescence of viable and nonviable bacteria present through each layer of the biofilm. (a) Biofilms before the addition of artificial GCF and microaerophilic gas and (b) biofilms after the addition. ▪, viable bacteria; ⋄, nonviable bacteria.
DISCUSSION
Many bacteria which are implicated in disease are associated with the normal microbiota of humans. When these organisms spread from their natural site of colonization, are allowed to proliferate more rapidly, or exist in increased proportions due to changes in their environment, disease can result. One such example is dental plaque, the bacterial composition of which remains relatively stable. However, the ecological plaque hypothesis (20) proposes that this stability can be disrupted by major fluctuations in environmental factors, such as changes in diet, oral hygiene, and challenges by exogenous microbes to cause disease. Modeling changes in microbial compositions of dental plaque has previously focused on perturbation (22, 27, 30). The results of this work have shown that population shifts associated with gingivitis can be successfully modeled in vitro by mimicking changes in the oral environment.
With the dual-species model, it was possible to demonstrate that with artificial saliva as the sole nutrient source, stable communities dominated by streptococci became established. By emulating the environmental conditions associated with gingivitis, it was possible to disrupt these communities to favor the growth of A. naeslundii. When these changes were applied to the more complex microcosm model, the same shifts in Streptococcus and Actinomyces proportions were observed, further establishing the link between these two environmental parameters and changes in microbial composition. In previous studies where the same in vitro model has been used to develop microcosm plaque with artificial saliva as the sole nutrient source, Actinomyces proportions ranged between 2 and 11% of the total cultivable flora (27, 29, 30). With the addition of artificial GCF and microaerophilic gas used in this study, Actinomyces spp. represented a much greater proportion (70%) of the total cultivable species. As this shift from a Streptococcus-dominated to an Actinomyces-dominated plaque is a common trend observed from in vivo experimental gingivitis studies (26, 33, 40), it is significant that this relationship can be emulated using an in vitro model.
Although this study has focused on the relationship between Streptococcus and Actinomyces spp., gram-negative species also play a significant role in biofilms grown under gingivitis conditions. This was shown by a 10-fold increase of these species as a proportion of the total cultivable organisms. The growth of gram-negative anaerobes, such as Tannerella spp. (formerly Bacteroides spp.), Prevotella spp., and Fusobacterium nucleatum, is enhanced by serum (35), which is a major component of GCF. The addition of artificial GCF to the growth medium in these in vitro studies represented the introduction of a protein-rich nutrient source. The proteinase activity of some Actinomyces spp. is thought to play a role in the degradation of serum proteins into polypeptides for use by bacteria, such as F. nucleatum and Prevotella gingivalis (14). The nutrients available in artificial GCF are not likely to be utilized directly by oral streptococci, such as Streptococcus sanguis and Streptococcus mitis, which are carbohydrate dependent. Actinomyces spp. may also be more adaptive to changes in environmental conditions, due to their ability to utilize different metabolic pathways at a wider pH range than oral streptococci (34). In general, increased nutrient availability will result in the formation of denser biofilms, most likely due to the increased production of extracellular polysaccharides by oral bacteria (32), which provides a further source of nutrients and plays a key role in biofilm architecture by providing a means for attachment for new species.
Confocal laser scanning microscopy has been applied to oral biofilms developed both in situ (2) and using in vitro models (8) to generate viability profiles. A characteristic feature of supragingival plaque is viable bacteria being less prevalent in deeper layers closer to the tooth surface (2, 3), and this distribution has also been observed from in vitro models (12). This type of distribution was observed with biofilms in the present study before the change of environmental conditions to mimic gingivitis and suggests that nonviable bacteria may be a key feature both structurally and nutritionally for subsequent attachment of species. A common observation from confocal microscopy studies of dental biofilms is the heterogeneous nature of biofilm structure (8, 39). Biofilm height and surface coverage are not uniform, and so bacteria, at even the deepest layers, can still be in contact with nutrients and oxygen. This type of structure may be more conducive to plaque composition associated with oral health.
Under conditions emulating gingivitis, nonviable bacteria became more widely distributed throughout the plaque structure, showing a more significant presence in layers closer to the biofilm surface. This may reflect the development of microenvironments within the biofilm structure (11) in which extreme gradients can exist across a small area. Vroom et al. (36) demonstrated the presence of extreme niches in pH developing as a response to the addition of sucrose. The introduction of artificial GCF and a microaerophilic environment may also instigate the development of extreme gradients in key environmental factors, as this change in conditions may allow the proliferation of species which may have had only a minor presence in biofilms grown under nutritionally limited conditions.
Due to the shear forces present within the model, biofilm height cannot exceed a specific depth. Thus, any bacterial proliferation occurring as a result of increased nutrient availability will have to occur within the limited space and may lead to the emergence of a more tightly packed structure. Previous studies (28) have shown that the addition of sucrose to biofilms can significantly change the coverage of microcosm plaque biofilms on hydroxyapatite, producing denser biofilms with fewer visible voids and channels and displaying a more uniform structure, which is thought to be significant to the development of dental caries.
The increased presence of Actinomyces spp. in densely packed biofilms may be due to the specific structures formed. In a five-species model of supragingival plaque (9), A. naeslundii was shown to aggregate with most other species present and appeared to form structures spanning the height of the biofilm, coming into contact with the biofilm surface. Streptococcus spp. were shown to form horizontal structures and large microcolonies in the central strata of the biofilm. Single cells and chains were also present in mature biofilms. These differences in distribution may be critical to the survival of species as the biofilm structure changes. As biofilms become more-densely packed, Actinomyces spp. may have an advantage spanning the height of the biofilm, allowing continued access to nutrients from the biofilm surface. The large microcolonies formed by Streptococcus spp. may be cut off from nutrient supplies and out-competed by organisms better able to grow with serum-derived nutrients.
In vitro models of microbial communities associated with health and disease are valuable tools for observing key factors in disease progression. When disease results from changes in the resident microflora, the use of such models allows the influence of individual environmental factors to be assessed and also allows the effects of potential treatments on these communities to be examined.
Acknowledgments
This work was funded by the EPSRC. F.D. was the recipient of an EPSRC CASE studentship award supported by Proctor and Gamble.
REFERENCES
- 1.Anderson, S. A., C. H. Sissons, M. J. Coleman, and L. Wong. 2002. Application of carbon source utilization patterns to measure the metabolic similarity of complex dental plaque biofilm microcosms. Appl. Environ. Microbiol. 68:5779-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arweiler, N. B., E. Hellwig, A. Sculean, N. Hein, and T. M. Auschill. 2004. Individual vitality pattern of in situ dental biofilms at different locations in the oral cavity. Caries Res. 38:442-447. [DOI] [PubMed] [Google Scholar]
- 3.Auschill, T. M., N. B. Arweiler, L. Netuschil, M. Brecx, E. Reich, and A. Sculean. 2001. Spatial distribution of vital and dead microorganisms in dental biofilms. Arch. Oral Biol. 46:471-476. [DOI] [PubMed] [Google Scholar]
- 4.Bell, G., D. Emslie-Smith, and C. Patterson. 1980. Textbook of physiology. Churchill Livingstone, London, United Kingdom.
- 5.Brown, L. J., and H. Loe. 1993. Prevalence, extent, severity and progression of periodontal disease. Periodontol. 2000 2:57-71. [DOI] [PubMed] [Google Scholar]
- 6.Daly, C. G., and J. E. Highfield. 1996. Effect of localized experimental gingivitis on early supragingival plaque accumulation. J. Clin. Periodontol. 23:160-164. [DOI] [PubMed] [Google Scholar]
- 7.Goodson, J. M. 2003. Gingival crevice fluid flow. Periodontol. 2000 31:43-54. [DOI] [PubMed] [Google Scholar]
- 8.Guggenheim, B., W. Giertsen, P. Schupbach, and S. Shapiro. 2001. Validation of an in vitro biofilm model of supragingival plaque. J. Dent. Res. 80:363-370. [DOI] [PubMed] [Google Scholar]
- 9.Guggenheim, M., S. Shapiro, R. Gmur, and B. Guggenheim. 2001. Spatial arrangements and associative behavior of species in an in vitro oral biofilm model. Appl. Environ. Microbiol. 67:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guyton, A. C. 1992. Human physiology and mechanisms of disease. WB Saunders Co., Philadelphia, Pa.
- 11.Hope, C. K., D. Clements, and M. Wilson. 2002. Determining the spatial distribution of viable and nonviable bacteria in hydrated microcosm dental plaques by viability profiling. J. Appl. Microbiol. 93:448-455. [DOI] [PubMed] [Google Scholar]
- 12.Hope, C. K., and M. Wilson. 2003. Measuring the thickness of an outer layer of viable bacteria in an oral biofilm by viability mapping. J. Microbiol. Methods 54:403-410. [DOI] [PubMed] [Google Scholar]
- 13.Igarashi, T., E. Asaga, and N. Goto. 2004. Roles of Streptococcus mutans dextranase anchored to the cell wall by sortase. Oral Microbiol. Immunol. 19:102-105. [DOI] [PubMed] [Google Scholar]
- 14.Jansen, H. J., and J. S. Van der Hoeven. 1997. Protein degradation by Prevotella intermedia and Actinomyces meyeri supports the growth of non-protein-cleaving oral bacteria in serum. J. Clin. Periodontol. 24:346-353. [DOI] [PubMed] [Google Scholar]
- 15.Lamb, J. F. 1991. Essentials of physiology. Blackwell Scientific, Oxford, United Kingdom.
- 16.Lie, M. A., M. M. Danser, G. A. van der Weijden, M. F. Timmerman, J. de Graaff, and U. van der Velden. 1995. Oral microbiota in subjects with a weak or strong response in experimental gingivitis. J. Clin. Periodontol. 22:642-647. [DOI] [PubMed] [Google Scholar]
- 17.Liljemark, W. F., C. G. Bloomquist, C. L. Bandt, B. L. Pihlstrom, J. E. Hinrichs, and L. F. Wolff. 1993. Comparison of the distribution of Actinomyces in dental plaque on inserted enamel and natural tooth surfaces in periodontal health and disease. Oral Microbiol. Immunol. 8:5-15. [DOI] [PubMed] [Google Scholar]
- 18.Loesche, W. J., F. Gusberti, G. Mettraux, T. Higgins, and S. Syed. 1983. Relationship between oxygen tension and subgingival bacterial flora in untreated human periodontal pockets. Infect. Immun. 42:659-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh, P., and M. V. Martin. 1999. Oral microbiology. Wright, Oxford, United Kingdom.
- 20.Marsh, P. D. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 8:263-271. [DOI] [PubMed] [Google Scholar]
- 21.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, and P. Gilbert. 2003. Growth and molecular characterization of dental plaque microcosms. J. Appl. Microbiol. 94:655-664. [DOI] [PubMed] [Google Scholar]
- 22.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, and P. Gilbert. 2003. Effects of a chlorhexidine gluconate-containing mouthwash on the vitality and antimicrobial susceptibility of in vitro oral bacterial ecosystems. Appl. Environ. Microbiol. 69:4770-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mombelli, A., N. P. Lang, W. B. Burgin, and F. A. Gusberti. 1990. Microbial changes associated with the development of puberty gingivitis. J. Periodontal Res. 25:331-338. [DOI] [PubMed] [Google Scholar]
- 24.Moore, L. V., W. E. Moore, E. P. Cato, R. M. Smibert, J. A. Burmeister, A. M. Best, and R. R. Ranney. 1987. Bacteriology of human gingivitis. J. Dent. Res. 66:989-995. [DOI] [PubMed] [Google Scholar]
- 25.Moore, W. E., L. V. Holdeman, R. M. Smibert, I. J. Good, J. A. Burmeister, K. G. Palcanis, and R. R. Ranney. 1982. Bacteriology of experimental gingivitis in young adult humans. Infect. Immun. 38:651-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, W. E., and L. V. Moore. 1994. The bacteria of periodontal diseases. Periodontol. 2000 5:66-77. [DOI] [PubMed] [Google Scholar]
- 27.Pratten, J., A. W. Smith, and M. Wilson. 1998. Response of single species biofilms and microcosm dental plaques to pulsing with chlorhexidine. J. Antimicrob. Chemother. 42:453-459. [DOI] [PubMed] [Google Scholar]
- 28.Pratten, J., C. S. Andrews, D. Q. Craig, and M. Wilson. 2000. Structural studies of microcosm dental plaques grown under different nutritional conditions. FEMS Microbiol. Lett. 189:215-218. [DOI] [PubMed] [Google Scholar]
- 29.Pratten, J., M. Wilson, and D. A. Spratt. 2003. Characterization of in vitro oral bacterial biofilms by traditional and molecular methods. Oral Microbiol. Immunol. 18:45-49. [DOI] [PubMed] [Google Scholar]
- 30.Ready, D., A. P. Roberts, J. Pratten, D. A. Spratt, M. Wilson, and P. Mullany. 2002. Composition and antibiotic resistance profile of microcosm dental plaques before and after exposure to tetracycline. J. Antimicrob. Chemother. 49:769-775. [DOI] [PubMed] [Google Scholar]
- 31.Relman, D. A. 2002. The human body as microbial observatory. Nat. Genet. 30:131-133. [DOI] [PubMed] [Google Scholar]
- 32.Sutherland, I. W. 2001. The biofilm matrix-an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 33.Syed, S. A., and W. J. Loesche. 1978. Bacteriology of human experimental gingivitis: effect of plaque age. Infect. Immun. 21:821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi, N., and T. Yamada. 1999. Effects of pH on the glucose and lactate metabolisms by the washed cells of Actinomyces naeslundii under anaerobic and aerobic conditions. Oral Microbiol. Immunol. 14:60-65. [DOI] [PubMed] [Google Scholar]
- 35.ter Steeg, P. F., J. S. Van der Hoeven, M. H. de Jong, P. J. van Munster, and M. J. Jansen. 1987. Enrichment of subgingival microflora on human serum leading to accumulation of Bacteroides species, peptostreptococci and fusobacteria. Antonie Leeuwenhoek 53:261-272. [DOI] [PubMed] [Google Scholar]
- 36.Vroom, J. M., K. J. De Grauw, H. C. Gerritsen, D. J. Bradshaw, P. D. Marsh, G. K. Watson, J. J. Birmingham, and C. Allison. 1999. Depth penetration and detection of pH gradients in biofilms by two-photon excitation microscopy. Appl. Environ. Microbiol. 65:3502-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson, M. 1999. Use of the constant depth film fermentor in studies of biofilms of oral bacteria. Methods Enzymolo 310:264-279. [DOI] [PubMed] [Google Scholar]
- 38.Wimpenny, J. W. T., and R. Colasanti. 1997. A unifying hypothesis for the structure of microbial biofilms based on cellular automaton models. FEMS Microbiol. Ecol. 22:1-16. [Google Scholar]
- 39.Wood, S. R., J. Kirkham, P. D. Marsh, R. C. Shore, B. Nattress, and C. Robinson. 2000. Architecture of intact natural human plaque biofilms studied by confocal laser scanning microscopy. J. Dent. Res. 79:21-27. [DOI] [PubMed] [Google Scholar]
- 40.Zee, K. Y., L. P. Samaranayake, and R. Attstrom. 1996. Predominant cultivable supragingival plaque in Chinese “rapid” and “slow” plaque formers. J. Clin. Periodontol. 23:1025-1031. [DOI] [PubMed] [Google Scholar]
- 41.Zylber, L. J., and H. V. Jordan. 1982. Development of a selective medium for detection and enumeration of Actinomyces viscosus and Actinomyces naeslundii in dental plaque. J. Clin. Microbiol. 15:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]