Abstract
The diversity and dynamics of a bacterial community extracted from an exploited oil field with high natural soil salinity near Comodoro Rivadavia in Patagonia (Argentina) were investigated. Community shifts during long-term incubation with diesel fuel at four salinities between 0 and 20% NaCl were monitored by single-strand conformation polymorphism community fingerprinting of the PCR-amplified V4-V5 region of the 16S rRNA genes. Information obtained by this qualitative approach was extended by flow cytometric analysis to follow quantitatively the dynamics of community structures at different salinities. Dominant and newly developing clusters of individuals visualized via their DNA patterns versus cell sizes were used to identify the subcommunities primarily involved in the degradation process. To determine the most active species, subcommunities were separated physically by high-resolution cell sorting and subsequent phylogenetic identification by 16S rRNA gene sequencing. Reduced salinity favored the dominance of Sphingomonas spp., whereas at elevated salinities, Ralstonia spp. and a number of halophilic genera, including Halomonas, Dietzia, and Alcanivorax, were identified. The combination of cytometric sorting with molecular characterization allowed us to monitor community adaptation and to identify active and proliferating subcommunities.
Many oil fields are located in semiarid regions characterized by soils of high natural salinity. Careless operation during oil extraction and processing contaminates the soil, for instance, via the generation of large volumes of oily and saline wastewater. Strategies are thus needed to effectively treat oil pollution of highly saline waters and soils.
The degradation of hydrocarbons as major components of mineral oil is generally brought about by microorganisms. At the salinity of seawater (∼3% total salts, mainly NaCl) or below, most low-molecular-weight components of mineral oil are easily degraded by a variety of microorganisms. However, the range of degrading organisms decreases with increasing salinity. There are only a few reports on the degradation of hydrocarbons in hypersaline environments. Enrichment cultures from the Great Solar Lake grew on mineral oil at salinities up to 17.2% (70), and a bacterial consortium and an extremely halophilic archaeon grew on hexadecane and crude oil at salinities of 15% and 32%, respectively (5). Degradation of hydrocarbons (mainly n-alkanes) by archaea (Halobacteria) at 15 to 32% NaCl was also reported (40). An extremely halophilic archaeon from a salt marsh degraded various saturated and aromatic hydrocarbons at salinities of up to 31% and grew optimally at 22% NaCl (10). In our own laboratory, microbial communities from saline soils in Patagonia degraded diesel fuel up to a salinity of 17.5% (59). Although some members of the communities were identified, their contributions to the diesel fuel biodegradation remained obscure.
Information on the phylogenetic diversity of microbial communities can be obtained by molecular methods, like fingerprinting or cloning and sequencing of PCR-amplified rRNA genes. These methods have been applied to isolates and consortia enriched from natural hypersaline environments, such as salt lakes, solar salterns, and soda lakes (1, 8, 24, 52). The 16S rRNA approach was also used to assess the microbial diversity in hypersaline environments and to follow transitions in community structure upon seasonal fluctuations or along spatial gradients in salinity, temperature, and availability of substrates and minerals (3, 6, 7, 8, 9, 11, 20, 29, 43, 46, 52).
Whereas the metabolic capabilities of oil-degrading communities grown under hypersaline conditions have been investigated by community profiling with BIOLOG plates (43, 44), information on the contributions of community members to oil biodegradation is more difficult to obtain. PCR screening for functional genes and expression profiling based on mRNA using reverse transcriptase PCR or microarrays are options when the degradation pathways and the genes involved are known (for reviews, see references 71, 73, 77, and 78). Other culture-independent methods linking degradation activities with the identities of community members are based on the uptake of isotope-labeled substrates. Examples are a combination of fluorescence in situ hybridization and microautoradiography that has been applied to halophiles (60) and stable isotope probing of DNA or RNA (for a review, see reference 57). Multiparametric flow cytometry has the potential to identify the active community members without requiring knowledge of metabolic genes. It provides information on the physiological states and activities of individuals and populations within communities, e.g., by indicating proliferation activity, while raising the possibility of identifying individuals by specific fluorescence labeling (50) or by rRNA-based methods after cell sorting (49, 53).
In the present study, the structural dynamics of a bacterial consortium isolated from saline soil were characterized during growth on diesel fuel at salinities up to 20%. Biodegradation and growth were followed by measurement of oxygen consumption and by analyses of alterations in community composition by single-strand conformation polymorphism (SSCP) fingerprinting and community-level substrate utilization profiles using the BIOLOG system. Multiparametric flow cytometry and high-speed cell sorting were applied to separate emerging proliferating subcommunities and to identify the dominant phylotypes therein by 16S rRNA gene sequencing. The aim of this work was to establish a high-speed detection tool capable of determining the degradation potential within natural environments via the investigation of the diversity and dynamics of a bacterial community and the identification of those consortium members that were actively involved in the degradation process. The information can be used to support bioremediation processes in soils of contaminated saline, arid regions.
MATERIALS AND METHODS
Source of organisms.
The consortium originated from clayey fine sand obtained from a ground depression near an oil-contaminated and now abandoned industrial region near Comodoro Rivadavia in Patagonia (Argentina). The area consists of strongly wind-exposed plateaus up to 1,000 m in altitude with typical temperatures of 6 to 12°C and less than 255 mm of precipitation per year. Samples were taken from the soil in sterile sample bags, transported by airplane, and stored at 4°C without loss of humidity until chemical analysis and extraction of microorganisms were performed in the laboratory. The soil composition was as follows: total salt content, 6.43% (wt/wt); pH 7.5; original water content, 6.44% (wt/wt); hydrocarbon content, 41 mg kg−1; cations (mM kg−1), Na+ (553), K+ (8.9), Ca2+ (104.1), Mg2+ (31.5), and NH4+ (0.6); anions (mM kg−1), Cl− (255), NO3− (1), SO42− (329), and PO43− (1.8). CFU amounted to 3.18 × 105 CFU ml−1 on R2A agar (Difco) and 8.55 × 105 CFU ml−1 on R2A agar supplemented with 7.5% NaCl.
Isolation and propagation of the bacterial consortium.
The microorganisms were extracted after mixing and sonicating the soil in 0.2% Na4P2O7 solution (adjusted to pH 8.5) as described previously (58), except that 7.5% NaCl was added. The consortium was enriched in shaken flasks containing 250 ml mineral medium amended with diesel fuel (800 μl liter−1), paraffin (400 μl liter−1), and yeast extract (120 mg liter−1). Clay particles were removed by repeated transfer into fresh medium. The composition of the mineral medium was (liter−1) 870.9 mg K2HPO4, 680.5 mg KH2PO4, 760 mg NH4Cl, 71.2 mg MgSO4 · 7H2O, 5.47 mg CaCl2, 0.44 mg ZnSO4 · 7H2O, 0.812 mg MnSO4 · 4H2O, 0.785 mg CuSO4 · 5H2O, 0.252 mg Na2MoO4 · 2H2O, 4.48 mg FeSO4 · 7H2O. Finally, 7.5% NaCl was added to the medium.
Degradation experiments.
Biodegradation of the diesel fuel was continuously monitored by following oxygen consumption at 25°C in Sapromat D12 respirometers (Voith, Heidenheim, Germany). The 500-ml flasks contained 240 ml mineral medium with 0, 7.5, 15, or 20% NaCl; 250 μl (215 mg) diesel fuel as a sole carbon source; and 10 ml inoculum corresponding to 30 mg dry mass. Degradation experiments took place in 24 flasks that were sacrificed for chemical and biological analyses. One flask served as a substrate-free control. A second flask was sacrificed right after inoculation to obtain initial values for the protein content, CFU, cell numbers, and BIOLOG profiles. Six flasks each were cultivated at 15 and 20% NaCl, and five flasks each were cultivated at 0% and 7.5% NaCl. Entire flasks were harvested to obtain biomass for the determination of protein contents, CFU, cell numbers, BIOLOG profiles, SSCP fingerprints (after 42 and 84 days of cultivation at the different salt concentrations), and flow cytometry, as well as cell sorting (after 21, 42, 63, and 84 days of cultivation at the different salt concentrations). The oxygen consumption rate was continuously measured in two parallel flasks at 0 and 7.5% NaCl each and in three parallel flasks at 15 and 20% NaCl each. The residual diesel fuel content at the end of the experiment (after 84 days) was determined by gas chromatography (34).
Cell numbers and protein content.
CFU counts were obtained in triplicate on R2A agar (Difco) supplemented with 2% NaCl (for samples from the culture without NaCl addition) or 7.5% NaCl (for samples from all higher salinities), each amended with 70 mg liter−1 cycloheximide. Total cell counts were determined by fluorescence microscopy after dispersion, dilution, staining with DAPI (4′,6′-diamidino-2-phenylindole), and filtering on 0.2-μm Nuclepore polycarbonate filters in triplicate. The protein content was determined in duplicate according to the method of Bradford (12).
Substrate utilization profiles.
Community level substrate utilization profiles were obtained by the BIOLOG method (BIOLOG Inc., Hayward, CA), which indicates substrate utilization, since respiring organisms reduce tetrazolium violet to colorimetrically distinguishable formazan. Cell suspensions from degradation experiments were diluted 1:10 with 7.5% NaCl solution (with the exception that 3% NaCl solution was used for samples from the degradation experiment without NaCl addition) and used to inoculate BIOLOG GN and GP plates. The number of different substrates was 128, since both types of plates contained 95 substrates with an overlap of 62 substrates. The plates were incubated at 25°C and analyzed after 24, 48, 72, and 96 h.
Multiparametric flow cytometry.
Flow cytometric measurements were carried out using a MoFlo cell sorter (DakoCytomation, Fort Collins, CO) equipped with two water-cooled argon ion lasers (Innova 90C and Innova 70C from Coherent, Santa Clara, CA). Excitation of 580 mW at 488 nm was used to analyze the forward scatter (FSC) and side scatter (SSC) as a trigger signal at the first observation point, using a neutral-density filter with an optical density of 2.3. DAPI was excited by 180 mW of multiline UV (333 to 365 nm) at the second observation point. The orthogonal signal was first reflected by a beam splitter and then recorded after reflection by a 555-nm long-pass dichroic mirror and passage by a 505-nm short-pass dichroic mirror and a band pass of 488/10. DAPI fluorescence was passed through a 450/65 band pass filter. Photomultiplier tubes (models R 928 and R 3896) were obtained from Hamamatsu Photonics (Hamamatsu City, Japan). Amplification was carried out at linear or logarithmic scales, depending on the application. Data were acquired and analyzed using Summit software (DakoCytomation, Fort Collins, CO). The dot plots were gated in such a way that visibly distinct subcommunities were included in the gates and cell counts therein were calculated. Fluorescent beads (polybead microspheres [diameter, 0.483 μm; Flow Check BB/Green Compensation Kit, blue alignment grade, reference 23520], Polyscience) were used to align the MoFlo (coefficient of variation, about 2%). Also, an internal DAPI-stained bacterial cell standard was introduced for tuning the device up to a coefficient of variation not higher than 6%.
Cell sorting was performed using the four-way-sort option at high speed (12 ms−1). The most accurate sort mode (single and one-drop mode; highest purity, 99%) was chosen for separating 5,000 cells per second. The cells were sorted into nucleic-acid-free glass flasks. The cells were separated from the mixed culture using DNA-DAPI fluorescence intensity and forward scatter signals in several independent experiments with different gate settings. Dominant and apparently growing subcommunities were separated in order to facilitate their molecular identification. Samples of incubations at all four NaCl concentrations were taken after 42 days and 84 days. Up to four subcommunities per sample could be separated simultaneously due to instrumental restrictions. To separate additional subcommunities, the sample taken after 84 days from the 20% NaCl incubation was run a second time. To obtain sufficient DNA for the generation of 16S rRNA gene clone libraries, at least 106 cells per subcommunity were sorted.
DNA content.
Cells were harvested from the cultures, centrifuged (3,200 × g; 6 min), immediately resuspended in sodium azide (10 μl of a 10% solution in bidistilled water), and stored at 4°C. This procedure was found to preserve the cells for at least 3 months. Just prior to being stained, the stored cells were washed in sodium chloride-phosphate buffer (400 mM Na2HPO4-NaH2PO4, 150 mM NaCl, pH 7.2) and diluted to an optical density at 700 nm of 0.035 in the same buffer. For DNA determination, 2 ml of diluted cell suspension was treated with 1 ml solution A (2.1 g citric acid-0.5 g Tween 20 in 100 ml bidistilled water) for 10 min, washed, and resuspended in 2 ml solution B (0.24 μM DAPI [Sigma], 400 mM Na2HPO4, pH 7.0) for at least 10 min in the dark at room temperature using a modification of a standard procedure (47). The staining method is quantitative for individuals in a population, since each cell binds an amount of the AT-specific DNA dye DAPI that is proportional to its AT content. Based on knowledge of the cell cycle of a population, it allows actively proliferating cells to be distinguished, because the staining intensity correlates with the number of chromosomes of the individuals within a population. Proliferation activity was thus inferred from high chromosome numbers in populations and from signs of a split-up of populations into subpopulations of different chromosome contents, visible as stacks or towers of subcommunities along the DNA axis in cytometry dot plots (51). The microscopically proven absence of cell aggregation facilitated cell sorting into clearly separate subcommunities.
Scatter behavior.
FSC is related to cell size, and SSC is related to cell granularity. Data were obtained by examining the light-scattering behavior of individual cells, mediated by the 488-nm line of the argon ion laser. Usually, shifts in the FSC/SSC mean values were recorded in parallel with the fluorescence measurements. At least 100,000 cells per sample were analyzed.
Microscopy.
To ensure staining of all cells within the consortium, blue DAPI fluorescence was observed with an epifluorescence microscope equipped with a UV light from a 100-W mercury arc lamp (Axioscope, Germany) using a UG1 filter cube (excitation filter 20 UG1; IF 17L420) and recorded with a DXC-9100P camera (Sony, Japan). Micrographs were subjected to image analysis with the Openlab 3.1.4 software (Improvision).
SSCP fingerprinting and reamplification of SSCP bands.
SSCP fingerprints were obtained from the inoculum and after 42 and 84 days of incubation with diesel fuel at various salinities. The SSCP analyses were performed according to the method of Schwieger and Tebbe (62), with some modifications. Cells were harvested by centrifugation, and total genomic DNA was extracted with a FastDNA SPIN Kit for soil (QBIOgene, Germany), according to the manufacturer's instructions. The V4-V5 region of the bacterial 16S rRNA gene was amplified using the primers UniBac515f (5′-GTG CCA GCA GCC GCG-3′) and UniBac927r-Ph (5′-CCC GTC AAT TYM TTT GAG TT-3′); the reverse primer was phosphorylated at the 5′ end. PCR was performed in 100-μl reaction mixtures containing 50 μl Taq PCR Master Mix (QIAGEN, Germany), 3 mM MgCl2, 10 pmol of each primer (supplied by MWG Biotech, Germany), and 1 μl template DNA (in dilutions of 1, 1:10, and 1:100) in a PTC-200 Thermal Cycler (MJ Research). The cycle parameters were as follows: 3 min at 94°C and 30 cycles of 20 s at 94°C, 30 s at 54°C, and 1 min at 72°C, followed by a 10-min extension step at 72°C. After the results were checked by electrophoresis on a 1.5% agarose gel, the PCR products were purified with an E.Z.N.A. Cycle-Pure Kit (peqLab Biotechnologie GmbH, Germany), eluted in 30 μl bidistilled water, and digested with 6 units Lambda exonuclease (New England Biolabs, Germany) for 1 h at 37°C. After addition of 1 volume of 2× SSCP sample buffer (95% formamide, 10 mM NaOH, 0.25% bromophenol blue, 0.25% xylene cyanol FF), the samples were denatured for 2 min at 95°C, immediately chilled on ice for 5 min, and stored at −20°C until electrophoresis. A 0.6% MDE gel (Biozym, Germany) was prepared on a Polybond film (Biometra, Germany) according to the supplier's protocol. Electrophoresis was run in a TGGE Maxi chamber (Biometra, Germany) in 1× Tris-borate-EDTA buffer (61) at 400 V and 26°C for 16 to 20 h. The gel was silver stained according to the method of Bassam et al. (4) with the modification that 3% NaOH was used as a developing bath. Bands of interest were cut from the wet gel, and the DNA was extracted as described previously (26). The extraction procedure was repeated, the supernatants were pooled, and DNA was precipitated with ethanol (61) and dissolved in 10 μl bidistilled water. For reamplification, 1 to 4 μl of the DNA was applied as a template in a 25-μl reaction mixture as described above, with the exception that the reverse primer, UniBac927r, was not phosphorylated. Reamplification was performed with 35 cycles according to the protocol described above. After agarose gel electrophoresis and purification, the PCR products were used for direct sequencing or cloning.
16S rRNA gene cloning and ARDRA screening of clone libraries.
Bacterial 16S rRNA gene fragments were amplified by PCR using the primers 27F and 1492R (41). PCR was performed in 25-μl reaction mixtures containing 12.5 μl Taq PCR Master Mix (QIAGEN, Germany), 5 pmol of each primer (supplied by MWG Biotech, Germany), and 1 to 4 μl template DNA (extracted from sorted cells after 42 and 84 days of cultivation, as described above) with a PTC-200 Thermal Cycler (MJ Research). The cycle parameters were as follows: 4 min at 94°C and 30 cycles of 45 s at 94°C, 1 min at 58°C, and 2 min at 72°C, followed by a 10-min extension step at 72°C. After the results were checked by electrophoresis on a 1.5% agarose gel, the PCR products were purified with an E.Z.N.A. Cycle-Pure Kit (peqLab Biotechnologie GmbH, Germany) and cloned using a QIAGEN PCR Cloning Kit. For each sample of sorted cells, about 200 clones were picked and screened for the appropriate insert size by PCR using the vector-specific primers M13uni(−21) (5′-TGT AAA ACG ACG GCC AGT-3′) and M13rev(−29) (5′-CAG GAA ACA GCT ATG ACC-3′). Positive clones were screened by amplified ribosomal DNA restriction analysis (ARDRA) as follows: the PCR products were digested with HaeIII (New England Biolabs, Germany) and separated electrophoretically on 2% MetaPhor agarose (Cambrex; distributed by Biozym, Germany). ARDRA patterns were analyzed using the Phoretix 1D software (Nonlinear Dynamics, United Kingdom). A hierarchical cluster analysis was performed applying the Jaccard similarity index and the Complete Linkage algorithm for calculating a similarity dendrogram. Representative clones from clusters comprising at least six operational taxonomic units were selected for partial sequencing. From each cluster, at least two clones were sequenced. The numbers of clones affiliated with the identified phylotypes (i.e., displaying identical ARDRA patterns) were recorded using the Phoretix database, which had been set up from the ARDRA patterns. Reamplified DNA extracted from SSCP bands was cloned as described above. For each band, 24 clones were picked and screened for the appropriate insert size and different ARDRA patterns. Representatives of the different patterns were sequenced.
16S rRNA gene sequence analysis.
The feasibility of 16S rRNA gene sequencing of flow cytometrically sorted, fixed cells was checked with pure cultures of Escherichia coli HB101 and Cupriavidus necator JMP134. Both cultures were subjected to sodium azide fixation, DAPI staining, sorting, and DNA extraction. Successful amplification and partial sequencing of the 16S rRNA genes revealed that DAPI staining of DNA did not affect the PCR by, e.g., blocking the Taq polymerase or cause mutations to an extent that affected the sequence analysis (data not shown).
Partial DNA sequencing of cloned 16S rRNA gene amplicons was performed using a BigDye RR Terminator AmpliTaq FS Kit version 3.1 (Applied Biosystems, Germany) and the sequencing primers 27F and 519R (41). Cloned amplicons from SSCP bands were sequenced using the vector-specific primers M13 uni (−21) and M13 rev (−29), respectively, as sequencing primers. Direct sequencing of reamplified SSCP bands was done with the primers 530f (41) and UniBac927r. Electrophoresis and data collection were carried out on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Germany). Data were analyzed by the ABI PRISM DNA Sequencing Analysis software, and sequences of both complementary strands were assembled by the ABI PRISM Autoassembler software. The BLASTN program (http://www.ncbi.nlm.nih.gov/BLAST; 2) was used to search for similar sequences in the GenBank, EMBL, and DDBJ nucleotide sequence databases, and the Sequence Match tool was used to search for similar sequences compiled by the Ribosomal Database Project II Release 9 (http://rdp.cme.msu.edu; 19). In order to verify the taxonomic classification resulting from the database retrieval, with particular sequences, a phylogenetic analysis was performed using the ARB package version Linux Beta 030822 (http://www.arb-home.de; 45).
The partial 16S rRNA sequences were aligned to the ARB database (release of January 2003) by the ARB editor, with manual correction. The aligned sequences were added to the ARB tree using the quick-add parsimony tool, applying phylum-specific filters.
Statistical analysis of community dynamics.
In order to obtain a more integral picture of how subcommunities changed over time, and depending on the salinity, statistical analyses were performed on the data. To estimate relative abundances for each taxon, the total cell numbers per gate as derived by flow cytometry and taxon identities obtained from clone libraries were combined. Taxon determinations using PCR-generated clone libraries are limited to the most abundant species in the libraries. Therefore, this method might underestimate species numbers in natural samples by overlooking rare species. However, within this study, general shifts in composition and frequencies determined by the detectable (and most abundant) taxa were assumed to correspond to the key degraders of diesel fuel in the examined saline soil. Their changes therefore represent the general community dynamics within the samples as reactions to the different salinities. Thus, the abundance of each taxon in a subcommunity was calculated by multiplying the proportion of the taxon within the gate by the total cell number of that gate. The total abundance of a taxon in a sample was the sum of its abundances in all different gates where it was detected, ignoring its possible occurrence outside the gates. The total cell numbers of the gates were correlated with the total cell counts of a sample, again ignoring cells outside the gates.
Species richness (number of taxa) and abundances (cell counts per taxon) were estimated. For univariate description of the samples, the Shannon-Wiener diversity index (on a log10 basis) was calculated (63). Additionally, an iterative multiregression approach was used to identify those taxa that best explained changes between the samples. For this purpose, taxa were randomly chosen, and their abundances were compared to the pooled data from all samples. As a result, those taxa that best explained the overall changes between the samples were estimated. Taxa selected by this method may not be the most abundant in all samples but those that best explained the differences between the samples.
To get a more detailed insight into community processes, a multidimensional-scaling (MDS) analysis based on taxon abundance data of the subcommunities was performed. Prior to the analysis, the data were square root transformed in order to level statistical weights between common and rare species. The appropriate transformation method was estimated by plotting the log-transformed standard deviation against the log-mean-transformed subcommunity taxon abundance data and then determining the slope of the trend line (for further explanation, see reference 18). Finally, for MDS analysis, a similarity matrix of the taxon abundance database on the Bray-Curtis similarity matrix was computed. The Bray-Curtis similarity is a widely used index in community analysis and expresses how similar two samples are to each other. It integrates both the presence and absence of species between samples and considers the proportional abundances of the species within samples (25). MDS attempts to place the samples on a (usually two-dimensional) sheet so that the rank order of the distances between samples on the sheet exactly agrees with the rank order of the matching similarities. The distance on the plot (i.e., the similarity) of two or more subcommunities indicates to what extent they share the same species and abundances (16). For community analyses, the ecological computation package PRIMER was used (17).
Nucleotide sequence accession numbers.
The partial 16S rRNA gene sequences that were determined have been deposited in the GenBank, EMBL, and DDBJ nucleotide sequence databases under accession no. AY918097 to -918118 and DQ153878 to -153948.
RESULTS
Growth, activity, and metabolic versatility of the consortium.
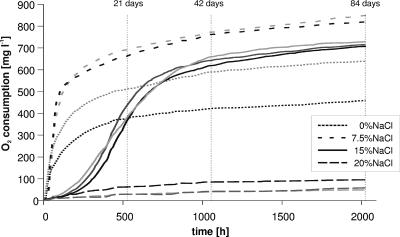
Tables 1 and 2 summarize the results of diesel fuel degradation, community growth as protein contents and viable- and total-cell counts, and changes in metabolic versatility during long-term incubation at various salinities. Time-resolved oxygen consumption rates (Fig. 1) corresponded well with protein contents and the cumulative diesel fuel degradation in 84 days, which occurred in the order of 7.5% > 0% > 15% > 20% NaCl. The oxygen consumption curves indicate slower adaptation to the NaCl concentration of 15%. Little adaptation occurred at 20% NaCl, indicating that the highest salinity exceeded the community's capability to adapt. The oxygen consumption rate at 15% NaCl intersected that at 0% NaCl after 30 days and remained higher thereafter. This corresponds well to the higher CFU and total cell counts at 15% than at 0% and 20% NaCl on days 42 and 84 (Table 1). One might thus expect that incubation at 15% NaCl beyond 84 days would have resulted in even more diesel fuel degradation than the 47% observed. Interestingly, CFU and total cell counts at 15% NaCl were even higher than those in the maximally active incubation at 7.5% NaCl. This indicates reduced diesel fuel degradation per cell at the higher salinity, which goes along with lower protein content per cell. BIOLOG results (Table 2) showed that the high metabolic versatility of the original community dropped during the incubation with diesel fuel to different extents, depending on the salinity. The maximally active incubation at 7.5% NaCl preserved the highest metabolic versatility. Communities incubated at 0% or 20% NaCl lost more of the initial substrate range.
TABLE 1.
Quantities of degraded diesel fuel and general microbial parameters of cultures
| Parameter | % NaCl | Valueb at (day):
|
||
|---|---|---|---|---|
| 0 (start)a | 42 | 84 | ||
| Degraded diesel fuel (% of total amt) | 0 | ND | 60 | |
| 7.5 | ND | 65 | ||
| 15 | ND | 47 | ||
| 20 | ND | 9.8 | ||
| Protein content (μg/ml) | 0 | 61.2/62.8 | 70.5/72.5 | |
| 7.5 | 50.1/50.7 | 93.0/93.8 | 96.3/99.9 | |
| 15 | 80.2/83.4 | 86.2/90.0 | ||
| 20 | 31.6/34.4 | 26.2/27.8 | ||
| CFU (ml−1) | 0 | 2.5 × 107 (±0.5 × 107) | 5.7 × 106 (±0.5 × 106) | |
| 7.5 | 4.9 × 107 (±0.5 × 107) | 6.1 × 107 (±0.5 × 107) | 1.7 × 107 (±0.3 × 107) | |
| 15 | 8.6 × 107 (±0.6 × 107) | 4.7 × 107 (±0.4 × 107) | ||
| 20 | 1.3 × 107 (±0.3 × 107) | 4.7 × 106 (±0.4 × 106) | ||
| Total cell count (ml−1) | 0 | 1.8 × 108 (±0.5 × 108) | 8.7 × 107 (±1.6 × 107) | |
| 7.5 | 2.1 × 108 (±0.2 × 108) | 4.2 × 108 (±1.6 × 108) | 1.7 × 108 (±0.1 × 108) | |
| 15 | 5.3 × 108 (±0.6 × 108) | 4.6 × 108 (±0.7 × 108) | ||
| 20 | 1.6 × 108 (±0.3 × 108) | 1.1 × 108 (±0.1 × 108) | ||
| Plating efficiency (CFU/total cell count) | 0 | 0.138 | 0.066 | |
| 7.5 | 0.233 | 0.145 | 0.100 | |
| 15 | 0.162 | 0.102 | ||
| 20 | 0.081 | 0.043 | ||
The inoculum was cultivated in the presence of 7.5% NaCl, and thus, no values for other concentrations exist.
Maximum values are printed in boldface. Values separated by slashes represent results from duplicate determinations. ND, not done.
TABLE 2.
BIOLOG number of utilized substrates
| % NaCl | No. of substratesa at (day):
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 (start)b
|
42
|
84
|
||||||||||
| 24c | 48 | 72 | 96 | 24 | 48 | 72 | 96 | 24 | 48 | 72 | 96 | |
| 0 | 49 | 55 | 58 | 65 | 1 | 4 | 46 | 57 | ||||
| 7.5 | 84 | 87 | 90 | 95 | 49d | 56 | 62 | 70 | 44 | 54 | 60 | 70 |
| 15 | 46 | 53 | 57 | 67 | 37 | 50 | 61 | 64 | ||||
| 20 | 39 | 50 | 53 | 68 | 0 | 28 | 52 | 58 | ||||
128 compounds offered.
The inoculum was cultivated in the presence of 7.5% NaCl, and thus, no values for other concentrations exist.
Hours of incubation.
Maximum values are printed in boldface.
FIG. 1.
Oxygen consumption in two or three replicate samples during the degradation of diesel fuel by a bacterial consortium at different NaCl concentrations. Measurement points (by CFU, total cell counts, BIOLOG, and protein content) after 21, 42, and 84 days are indicated.
Community shifts determined by SSCP fingerprinting.
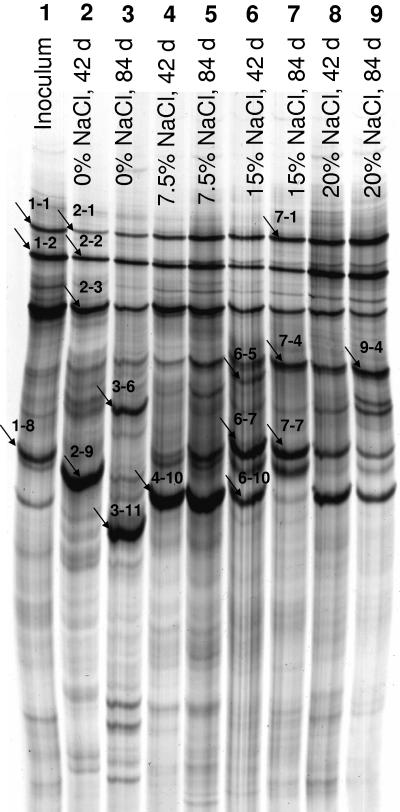
Shifts in the community composition were monitored by SSCP fingerprinting of the PCR-amplified V4-V5 region of the 16S rRNA gene (Fig. 2). Sequencing of dominant SSCP bands (Table 3) revealed that different salinities favored different key organisms. Where analyzed, bands at identical positions from different incubations or different times represented nearly identical phylotypes. This indicates that, to some extent, phylogenetic information can be transferred to unidentified bands in identical positions. In most cases, a single band could be assigned to a single phylotype, but a few bands could be sequenced only after being cloned and then revealed the presence of more than one phylotype. Dominant bands visible at all salinities and over the whole time course were identified as Halomonas, Ralstonia, and Dietzia, indicating a broad salt tolerance for these phylotypes. By contrast, Naxibacter was prevalent in the 0% NaCl culture after 42 days but absent after 84 days, whereas Sphingomonas arose after 84 days as the dominant phylotype. An additional band at 0% NaCl after 84 days represented an actinomycete of the genus Kocuria (Micrococcineae) and an unknown myxobacterium. Other phylotypes emerged preferably at higher salinities. The incubation with 7.5% NaCl showed the least changes between 42 and 84 days, e.g., the most prominent band representing Idiomarina was found from 42 to 84 days. The same band was also present at higher salinities. DNA of an unknown alphaproteobacterium distantly related to the genus Caulobacter was found in the dominant bands after 84 days at 15% and 20% NaCl. Marinobacter was also a conspicuous phylotype at 15% NaCl after 42 and 84 days and was also present at 20% NaCl, whereas another band representing Alcanivorax and Marinobacter appeared at 15% NaCl after 42 days and faded toward 84 days.
FIG. 2.
SSCP fingerprint of the V4-V5 regions of bacterial 16S rRNA genes. Lanes: 1, inoculum (cultivated at 7.5% NaCl); 2, 0% NaCl, 42 days; 3, 0% NaCl, 84 days; 4, 7.5% NaCl, 42 days; 5, 7.5% NaCl, 84 days; 6, 15% NaCl, 42 days; 7, 15% NaCl, 84 days; 8, 20% NaCl, 42 days; 9, 20% NaCl, 84 days. The bands marked by arrows were cut from the gel, and the DNA was extracted, reamplified, and sequenced. When direct sequencing failed, the reamplified DNA was cloned, and a representative of each ARDRA pattern was sequenced. The results are listed in Table 3.
TABLE 3.
Sequencing results for SSCP bandsa
| SSCP band/clone | Size (bp) | Accession no. | Highest BLAST hit (accession no.)/% identity | Next RDP relative (accession no.)/SAB |
|---|---|---|---|---|
| 1-1 | 417 | AY918097 | Halomonas sp. strain CR1-4 (AY205298)/100 | Halomonas sp. strain CR1-4 (AY205298)/0.998 |
| 2-1/A1 | 370 | AY918098 | Halomonas sp. strain CR1-4 (AY205298)/99 | Halomonas sp. strain CR1-4 (AY205298)/0.967 |
| 2-1/F3 | 311 | AY918099 | Halomonas eurihalina ATCC 49336T (X87218)/98 | Halomonas eurihalina ATCC 49336T (X87218)/0.902 |
| 7-1 | 378 | AY918100 | Halomonas salina GSP23 (AY553073)/99 | Halomonas salina GSP23 (AY553073)/0.935 |
| 1-2 | 413 | AY918101 | Dietzia sp. strain CR1-3 (AY205297)/99 | Dietzia sp. strain CR1-3 (AY205297)/0.980 |
| 2-2/A2 | 414 | AY918102 | Dietzia sp. strain CR1-3 (AY205297)/99 | Dietzia sp. strain CR1-3 (AY205297)/0.980 |
| 2-3/A1 | 392 | AY918103 | Ralstonia insidiosa CCUG 46388 (AJ539233)/99 | Ralstonia insidiosa AU2944T (AF488779)/0.966 |
| 2-3/C6 | 393 | AY918104 | Uncultured bacterium (AJ459874)/99 | Caulobacter sp.; uncultured bacterium (AJ459874)/0.924 |
| 7-4 | 395 | AY918105 | Caulobacter sp. strain A1 (AF361188)/91 | Rhodospirillaceae; uncultured bacterium (AY373412)/0.723 |
| 9-4 | 353 | AY918106 | Caulobacter sp. strain A1 (AF361188)/91 | Rhodospirillaceae; uncultured bacterium (AY373412)/0.691 |
| 6-5/A9 | 392 | AY918107 | Marinobacter sp. strain NCE312 (AF295032)/99 | Marinobacter sp. strain NT N148 (AB167047)/0.828 |
| 6-5/A12 | 400 | AY918108 | Alteromonadaceae bacterium LA13 (AF513448)/95 | Marinobacter sp. strain Aplume1.1727a (AF212207)/0.792 |
| 6-5/B5 | 400 | AY918109 | Alcanivorax venustensis ISO4 (AF328762)/100 | Alcanivorax venustensis ISO4T (AF328762)/0.997 |
| 3-6/E2 | 393 | AY918110 | Uncultured bacterium clone FB34-4 (AY527798)/98 | Uncultured myxobacterium M10Ba18 (AY360608)/0.889 |
| 3-6/E10 | 395 | AY918111 | Kocuria kristinae DSM 20032T (X80749)/98 | Kocuria kristinae DSM 20032T (X80749)/0.940 |
| 6-7 | 411 | AY918112 | Marinobacter sp. strain NCE312 (AF295032)/100 | Marinobacter sp. strain NCE312 (AF295032)/0.998 |
| 7-7 | 360 | AY918113 | Marinobacter sp. strain NCE312 (AF295032)/99 | Marinobacter sp. strain NCE312 (AF295032)/0.935 |
| 1-8 | 399 | AY918114 | Marinobacter sp. strain NCE312 (AF295032)/100 | Marinobacter sp. strain NCE312 (AF295032)/1.000 |
| 2-9 | 378 | AY918115 | Naxibacter alkalitolerans YIM 31775 (AY679161)/98 | Naxibacter alkalitolerans YIM 31775 (AY679161)/0.954 |
| 4-10 | 415 | AY918116 | Idiomarina ramblicola R22T (AY526862)/100 | Idiomarina abyssalis KM227T (AF052740)/0.995 |
| 6-10 | 407 | AY918117 | Idiomarina ramblicola R22T (AY526862)/99 | Idiomarina abyssalis KM227T (AF052740)/0.980 |
| 3-11 | 268 | AY918118 | Sphingomonas sp. strain HB1 (AY387397)/97 | Sphingomonas sp. strain DSM6432 (X87165)/0.866 |
See Fig. 2. In cases where more than one database entry displayed the highest BLAST score or similarity score (SAB), only one representative is given. The SAB represents the number of (unique) oligomers shared between the query sequence and a given Ribosomal Database Project sequence divided by the lowest number of unique oligonucleotides in either of the two sequences.
Flow cytometric characterization.
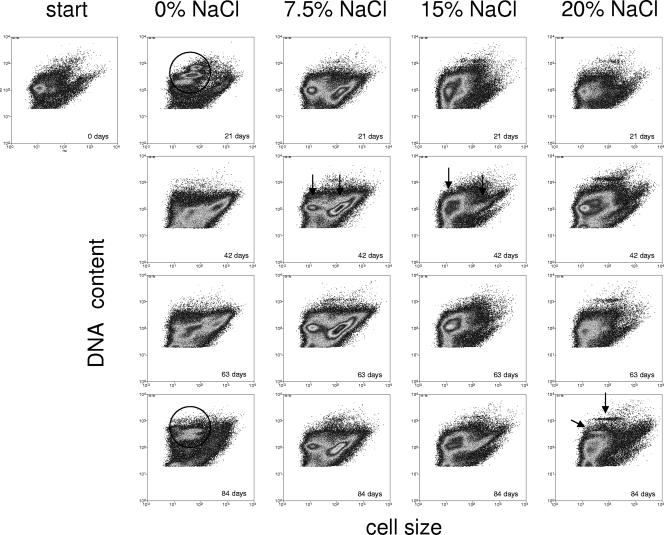
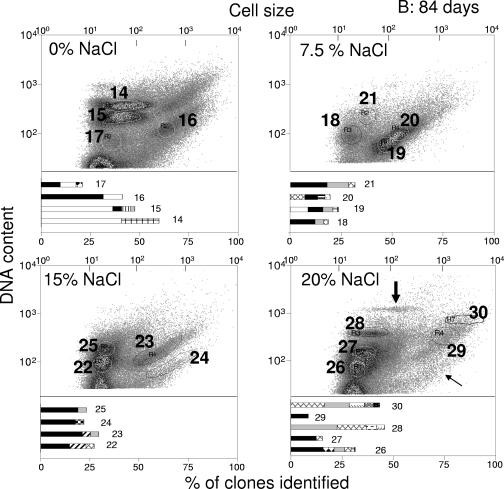
Single-cell analyses of DNA contents (DAPI) as a measure of proliferation activity plotted against cell sizes (FSC) for visual discrimination of subcommunities or populations were performed using multiparametric flow cytometry. This served to obtain more detailed information on community shifts. Flow cytometry was used to follow the dynamics of subcommunities during the course of cultivation on diesel fuel at four given NaCl concentrations (0%, 7.5%, 15%, and 20%) over 84 days. Typical patterns are presented in Fig. 3. Since cell sizes and the sizes, numbers, and GC contents of chromosomes differ between species, distinct groupings in cytometry dot plots can be interpreted as populations or subcommunities comprising populations with similar characteristics. The high chromosome contents of subcommunities may indicate proliferation activity, which in turn reflects diesel fuel utilization by these organisms within communities showing little overall growth, if any (between 0 and 42 days), or even decreasing cell numbers (between 42 and 84 days) (Table 1). The original community had a highly uniform DNA/cell size pattern, which developed into distinct subcommunities after 21 days of cultivation at 0% NaCl (Fig. 3). Three subcommunities emerged, which were all characterized by very small cells and high DNA contents, whereas a fourth subcommunity of larger cells had a low DNA content. Because of the high DNA content and the way the subcommunities were grouped, they may be interpreted as subpopulations of a single species differing in their cell cycle activities, i.e., their chromosome numbers. This distinct grouping was absent after 42 days when other, less distinct DNA patterns indicating overlap of several populations appeared. The pattern was completely altered again after 84 days. Cell cycle-connected subpopulations (Fig. 3) were again manifest at this time, as was confirmed by phylogenetic analysis after sorting (see below). Such dramatic changes were not observed during cultivation at higher salinities; the topologies of the consortia differed between the various NaCl concentrations but were largely stable over time. The community that developed at 7.5% NaCl was changed least, confirming the SSCP results. Two main subcommunities were observed during growth at 7.5% NaCl and similarly at 15% NaCl (Fig. 3, plots from 42 days). The culture grown at 20% salinity preserved the initial pattern (of the stationary-phase-grown inoculum) relatively well, which can be explained by the adverse growth conditions at this higher-than-natural salinity. However, two additional subcommunities with high and very high DNA contents developed under those conditions (Fig. 3).
FIG. 3.
Community dynamics during growth at different NaCl concentrations over a time range of 84 days. The population patterns were obtained by determination of bacterial DNA content and cell size. All incubations were started with the same inoculum. Cell cycle-related (circles) and salinity-related (arrows) subpopulations are highlighted; see Results for details.
Sorting of subcommunities.
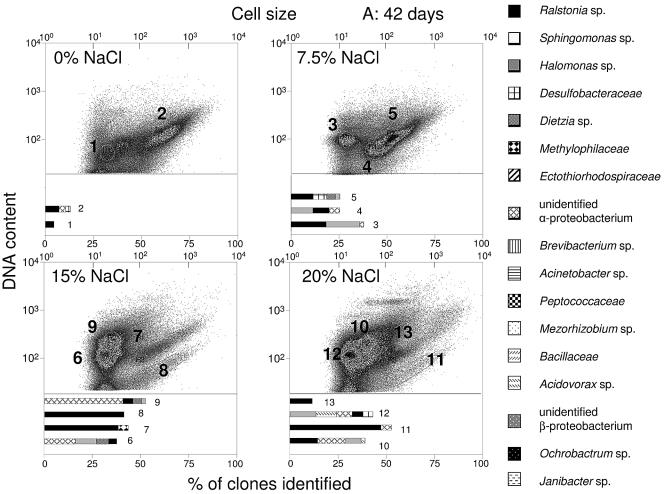
The chosen sort gates, 1 to 30, are shown in Fig. 4. To compare corresponding subcommunities in different plots, they were recognized with the help of their positions relative to debris and noise signals in the lower left corners of the plots (not included in Fig. 4). The sort gates were placed around particularly abundant or distinct, rare subcommunities. The proportions of distinct subcommunities after 84 days were estimated. Unique subcommunities in gates 14 and 15 contributed 33% and 42%, respectively, to the entire community at 0% NaCl but were nearly absent at all higher salinities. Unique subcommunities also appeared during cultivation at 20% NaCl in gate 28 (9%) as did an ungated subcommunity (1%). Another abundant subcommunity in gate 18 contributed 12% of the cells at 7.5% NaCl and even 22% and 33% of the cells at 15% and 20% NaCl (gates 25 and 27), respectively. Other examples of main subcommunities were those in gates 19 (39%) and 20 (33%). High percentages of cells were also found in subcommunities within gates 22 (37%) and 26 (33%).
FIG.4.
Flow cytometric analysis of communities incubated for (A) 42 and (B) 84 days at different NaCl concentrations. DNA-content-versus-forward-scatter behavior was determined, and subcommunities were sorted according to sort gates 1 to 30, as shown. The taxonomic classification of the sequences is based on BLAST and Ribosomal Database Project database retrievals and, in some cases, also on phylogenetic analysis with ARB (data not shown). The bar diagrams show the taxonomic compositions of the subcommunities obtained by sorting and 16S rRNA gene sequencing (for sequencing results, see the supplemental material). The thick arrow in panel B indicates a unique subcommunity that appeared at a proportion of 1%, and the thin arrow indicates another ungated subcommunity after 84 days of incubation.
Phylogenetic identification of sorted subcommunities.
The bar diagrams in Fig. 4 show the taxonomic compositions of the subcommunities obtained by sorting and 16S rRNA gene sequencing, (for sequencing results, see the supplemental material). Only phylotypes represented by clusters of at least six operational taxonomic units were taken into account (see Materials and Methods). This selection ignored rare phylotypes, which is justified, assuming that the most active taxa in a sorted subcommunity become increasingly abundant in the growth-associated process of diesel fuel degradation. The numbers of phylotypes detected in the respective clone libraries summed for each sample are shown in Table 4, together with the cell numbers estimated for the included subcommunities. Although total-cell counts decreased with time in all samples, species richness increased in the communities grown at 0%, 7.5%, and 20% NaCl.
TABLE 4.
Changes in cell numbers per sample gate, number of taxa, and diversity within the different salinity treatments after 42 and 84 days
| Parameter | Value at salinity of (% NaCl):
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0
|
7.5
|
15
|
20
|
|||||
| 42a | 84 | 42 | 84 | 42 | 84 | 42 | 84 | |
| Cell no. per sample gate (108 ml−1) | 1.22 | 0.76 | 3.49 | 1.49 | 4.11 | 3.56 | 0.94 | 0.86 |
| No. of taxa | 3 | 6 | 5 | 7 | 5 | 5 | 5 | 10 |
| Diversity (H′) | 0.32 | 0.46 | 0.6 | 0.75 | 0.56 | 0.44 | 0.66 | 0.55 |
Number of days of treatment.
Generally, reasonable congruence was found between the whole-community SSCP fingerprint (Fig. 2 and Table 3) and the phylogenetic compositions of sorted, prominent subcommunities (Fig. 4). Special attention was given to organisms within subcommunities showing signs of proliferation activity. Phylotypes affiliated with the genus Sphingomonas were identified in the dominant subcommunities, 15 and 14, after 84 days of incubation at 0% NaCl, indicating that these organisms preferably thrived at lower-than-natural salinity. These two subcommunities comprised 75% of all cells within this sample, and their positions at channel numbers 210 and 382 as arbitrary units for the DNA contents (Fig. 4) clearly point to doubling activity in this population.
Phylotypes related to an unidentified alphaproteobacterium that were also found by SSCP analysis showed similar signs of proliferation. They dominated subcommunities 6 and 9 (15% NaCl; 60.6% of all cells) and subcommunities 10 and 12 (20% NaCl; 81.2% of all cells) after 42 days of cultivation (Fig. 4). Phylotypes affiliated with Ralstonia and Halomonas were associated with these subpopulations. At the end of the cultivation, Ralstonia became dominant in the corresponding subcommunities 22 and 25 at 15% NaCl and 26 and 27 at 20% NaCl. Multiplication was also inferred from similarly shaped subcommunities in gates 7/8 and 11/13 after 42 days and 23/24 and ungated (Fig. 4)/29 after 84 days of incubation. These groups generally contributed less than 20% to the respective samples and contained mainly large cells of Ralstonia sp. The DNA contents given by channels 65 and 115 for gates 8 and 7, respectively (Fig. 4), are indicative of doubling. Members of the genus Ralstonia were abundant at all salinities and over the whole time course (present in 27 out of 30 subcommunities and dominating 20 of them) (Fig. 4). Halomonas, a genus also identified by SSCP fingerprinting (Fig. 2), contributed significantly to many subcommunities during growth under saline conditions. Also, Dietzia was present at significant abundances in dominant subcommunities (gates 5, 6, and 9), mainly after 42 days of cultivation, although SSCP analysis (Fig. 2) indicated the general presence of the genus.
The prevalence of the other phylotypes displaying prominent SSCP bands, like Naxibacter, Marinobacter, and Idiomarina, was not reflected in the 16S rRNA gene clone libraries generated from sorted subcommunities. Conversely, numerous phylotypes not detected in SSCP bands contributed significantly to the sorted subcommunities. Examples are Acidovorax, Janibacter, and phylotypes affiliated with the Ectothiorhodospiraceae and the Methylophilaceae (Fig. 4). Surprisingly, even phylotypes related to Desulfobacula (Desulfobacteraceae) and Desulfosporosinus (Peptococcaceae) were detected, which may indicate sulfate reduction.
Community analysis.
The highest taxon richness (10 taxa), according to the data obtained by analyzing the sorted subcommunities, was found in the community grown at 20% NaCl after 84 days of incubation, while the lowest number was seen in the 0% NaCl sample after 42 days (3 taxa). In all other samples, five to seven taxa were detected (Table 4). The total abundances of sorted subcommunities ranged from 0.76 × 108 (at 0% NaCl after 84 days) to 4.11 × 108 (at 15% NaCl after 42 days) cells per ml. Diversity index values revealed that the samples with the highest abundances and/or taxon numbers did not result in high diversity index scores. The samples with the highest taxon numbers (e.g., the 20% NaCl sample after 84 days) had only low abundances, while those with the highest abundances (e.g., the 15% NaCl sample after 42 days) had only low to moderate taxon numbers but were characterized by the same dominant taxa (Fig. 5). Diversity values (Table 4) ranged from a minimum of 0.32 (in the 0% NaCl sample after 42 days; six species at 1.22 × 108 cells per ml) to 0.75 (in the 7.5% NaCl sample after 84 days; seven species at 1.49 × 108 cells per ml). Low diversity values, which usually characterize samples with low species numbers and unevenly distributed, high abundances, are often an indication of stressed and poorly adapted communities, while high diversity values hint at a better adaptation of the microbial consortium. Multiregressional analysis revealed that 92.6% of the variation between the samples could be explained by the occurrence of the characteristic phylotypes affiliated with Ralstonia, Sphingomonas, the unidentified alphaproteobacterium, Desulfobacteraceae, and Peptococcaceae.
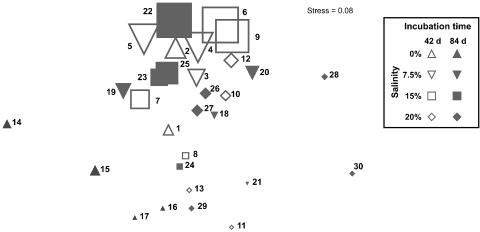
FIG. 5.
MDS based on taxon abundance data of the subcommunities as derived by flow cytometry and PCR analyses. The symbol sizes indicate the relative abundances (cell counts) of the subcommunities. The numbers beside the symbols are the numbers of the subcommunities as given in Fig. 4.
An MDS plot (Fig. 5) showed subcommunity shifts. There was a vertical trend of abundances, indicating that the subcommunities were similar in their species compositions and the relative contributions of species and mainly differed in their absolute cell counts. The only exceptions to this trend were subcommunities 14 and 15 from the 0% NaCl sample after 84 days and subcommunities 28 and 30 from the 20% NaCl sample after 84 days. They differed strongly, because of their contents of the rare taxa Brevibacterium (0% NaCl sample after 84 days), Janibacter, Ochrobactrum, a phylotype related to Virgibacillus (Bacillaceae), and an unidentified betaproteobacterium (20% NaCl sample after 84 days).
DISCUSSION
Soil from an exploited oil field in Patagonia that was characterized by a natural salinity of 6.4% proved to be the habitat of a complex bacterial community capable of degrading diesel fuel and of adapting to drastically reduced or enhanced salinity with little loss of activity. A salinity increase to 20% NaCl overstrained the degradation capacity of the originally dominant community, but even at this salt concentration, a diverse bacterial community capable of degrading various organic substrates remained present. The development of cell counts and protein content, oxygen consumption, and degradation endpoints indicated preferential growth at 7.5 and 15% NaCl. The drop in CFU and total cell counts between 42 days and 84 days despite slightly increased oxygen consumption indicates that with time, a higher percentage of the community actively participated in the degradation of diesel fuel. The drop in plating efficiency points to an increasing proportion of viable but noncultivable cells with prolonged cultivation. A community shift, rather than overall community growth, was thus responsible for the adaptation. The range of metabolized BIOLOG substrates was high and nearly constant until the end of the incubation at all salinities, although consortia that had been incubated for 84 days at 0 and 20% NaCl needed more time to metabolize the offered substrates. Consortia incubated at 7.5 and 15% NaCl retained 70%, and those at 0 and 20% NaCl retained 60%, of the initial substrate spectrum.
Information on the taxonomic composition of the community was obtained by SSCP fingerprinting of the PCR-amplified V4-V5 region of 16S rRNA genes performed at different cultivation times for all cultures. Flow cytometry was used to get quantitative information on the community dynamics and for the identification of active populations. Flow cytometry is a well-established method for depicting bacterial consortia in natural environments (15, 22, 55, 65, 68) and was recently combined with cell sorting as a prerequisite for phylogenetic analyses of subcommunities (49, 53). We used narrow ranges of DNA contents and cell sizes as the criteria for cell sorting. We assumed that distinct groupings in DNA-versus-cell-size plots exhibiting high DNA contents or showing signs of a cooccurrence of different cell cycle stages represented actively proliferating bacteria. This assumption was generally confirmed by the phylogenetic identification of clearly dominant populations in these groupings. It appears justified to conclude that these proliferating populations contributed to the degradation of the diesel fuel, since other carbon sources were absent. Similar proliferation patterns obtained by flow cytometry had already been successfully employed to study the dynamics of binary bacterial communities under various cultivation conditions (50, 69). Such an analysis based on DNA contents can be regarded as highly reliable and quantitative, since there is no risk of active excretion of the dye by fixed cells (56).
The overall community behavior was in agreement with commonly accepted concepts about ecological community shifts after disturbance (28, 31, 32), in that originally dominant specialists were replaced by the copiotrophic and fast-growing generalists Ralstonia and Halomonas as the leading taxa in all subcommunities during entire cultivations. This held true especially in those incubations that were disturbed by more extreme changes in salinity (i.e., 0% and 20% NaCl). The degradation efficiencies inferred from the oxygen consumption were low in these incubations. A number of additional phylotypes possibly representing specialists appeared toward the ends of these cultivations, e.g., Sphingomonas, Janibacter, members of the Ectothiorhodospiraceae and the Methylophilaceae, or putative sulfate reducers, such as species related to Desulfobacula (Desulfobacteraceae) and Desulfosporosinus (Peptococcaceae). The increase in species numbers also agrees well with common ecological models of late successional stages of well-adapted communities (28, 31, 32). Whereas species numbers in the presumably better-preadapted communities incubated at 7.5% and 15% NaCl (i.e., near natural salinities) remained stable over time, the increase in species numbers in cultures grown at 0% and 20% indicates considerable community adaptation, which also explains the delayed diesel fuel degradation. A Sphingomonas sp. appearing toward the end of the incubation at 0% NaCl and dominating the culture showed the clearest signs of proliferation in cytometry plots. It is thus likely that it was mainly the need of the population to adapt to the changed salinity or, more probably, to the utilization of diesel fuel, followed by an increase in cell number, that was responsible for the delayed but finally efficient diesel fuel degradation. Sphingomonas species are common soil bacteria known as versatile degraders of hydrophobic organic compounds, such as polycyclic aromatic hydrocarbons (35, 37, 66).
Proliferation was also observed for a dominant subcommunity mainly consisting of an alphaproteobacterium and species of Ralstonia, Dietzia, and Halomonas. The dominance of these taxa at 7.5% and 15% NaCl throughout the incubation was confirmed by SSCP analysis. The proportions of the strains changed during the incubation. Ralstonia and Dietzia species are known to degrade various hydrocarbons (13, 14, 54, 64, 72, 76), and a halotolerant Dietzia sp. has been isolated from a similar diesel fuel-degrading consortium (59). A highly uniform, proliferating subcommunity of minor abundance present in gates 7 and 8 (Fig. 4), but also visible in other samples until the end of the incubation, contained a different species of Ralstonia, as seen from its larger cells.
In addition to the identification of proliferation patterns, the development of new subcommunities could be quantitatively followed by flow cytometry and MDS. The narrow DNA distribution of the subcommunity confined by gate 28 strongly indicates that the comprised Halomonas and alphaproteobacteria belong to different species than those in gates 6 and 9, which exhibited a very different, uniform DNA pattern (Fig. 4). This subcommunity also included Janibacter, a genus of actinomycetes known to degrade several xenobiotics (33, 42, 75), and a member of the Bacillaceae related to Virgibacillus marismortui, a halophilic bacterium.
Another conspicuous subcommunity (Fig. 4) arose at the highest salinity to quantities between 1 and 2% of the whole community. The cells within this subcommunity were characterized by a very high, uniform DNA content and variable but generally large cell sizes. Unfortunately, insufficient DNA for PCR amplification was retrieved from this subcommunity. It may be that any of the marine genera Idiomarina, Marinobacter, and Alcanivorax that formed strong SSCP bands were comprised in this subcommunity. Strains of Halomonas, Marinobacter, and Alcanivorax are important degraders of hydrocarbons in marine environments (30, 38) and have been suspected to be involved in the degradation of diesel fuel components at high salinities (27). Alternatively, these genera may have been comprised in other subcommunities but were missed because the clone libraries were too small to cover all major phylotypes. It should be pointed out that our method of community identification counterselects pleomorphic species and those with cell cycle behaviors that do not favor the subpopulations with uniform DNA contents.
Janibacter, Acidovorax, and phylotypes affiliated with the Ectothiorhodospiraceae and the Methylophilaceae, as well as putative sulfate reducers, were detected only in sorted subcommunities, but not in SSCP fingerprints. Low biomass, and therefore insufficient DNA, might be the reason for the failure to detect these rare phylotypes in community fingerprints, whereas cell sorting reduces the dominance of other organisms. Oxygen depletion during rapid diesel fuel degradation might explain the occurrence of anaerobic bacteria related to Desulfobacula (Desulfobacteraceae) or Desulfosporosinus (Peptococcaceae). However, it appears too daring to conclude that anaerobic degradation of diesel fuel components (66, 74) took place in our incubations. Higher than average stability of these organisms in overall declining communities may also have favored their detection toward the end of the cultivation. Although sulfate reducers are considered obligate anaerobes, there is evidence that some species can tolerate oxygen (39, 48) or may even respire aerobically (21, 23, 36, 67).
Supplementary Material
Acknowledgments
We thank Oscar H. Pucci (National Patagonian University, San Juan Bosco, Comodoro Rivadavia, Argentina) for supplying the soil samples. We also thank Jana Reichenbach, Rita Remer, Christine Süring, Andreas Lösche, Helga Engewald, and Ute Lohse for skilled technical assistance and Mandy Laube for her contribution to this work during an internship.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aharal, D. R., F. E. Dewhirst, B. J. Paster, B. E. Volcani, and A. Ventosa. 1996. Phylogenetic analyses of some extremely halophilic archaea isolated from Dead Sea water, determined on the basis of their 16S rRNA sequences. Appl. Environ. Microbiol. 62:3779-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Antón, J., R. Rosselló-Mora, F. Rodríguez-Valera, and R. Amann. 2000. Extremely halophilic bacteria in crystallizer ponds from solar salterns. Appl. Environ. Microbiol. 66:3052-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassam, B. J., G. Caetano-Anolles, and P. M. Gresshoff. 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem. 196:80-83. [DOI] [PubMed] [Google Scholar]
- 5.Belayev, S. S., I. A. Borzenkov, E. I. Milekhina, I. S. Zvyagintseva, and M. V. Ivanov. 1993. Halotolerant and extremely halophilic oil-oxidizing bacteria in oil fields. Dev. Pet. Sci. 39:79-88. [Google Scholar]
- 6.Benlloch, S., A. J. Martinez-Murcia, and F. Rodriguez-Valera. 1995. Sequencing of bacterial and archaeal 16S rRNA genes directly amplified from a hypersaline environment. Syst. Appl. Microbiol. 18:574-581. [Google Scholar]
- 7.Benlloch, S., S. G. Acinas, A. J. Martínez-Murcia, and F. Rodríguez-Valera. 1996. Description of prokaryotic biodiversity along the salinity gradient of a multipond saltern by direct PCR amplification of 16S rDNA. Hydrobiologia 329:19-31. [Google Scholar]
- 8.Benlloch, S., S. G. Acinas, A. López-López, S. P. Luz, and F. Rodríguez-Valera. 2001. Archaeal biodiversity in crystallizer ponds from a solar saltern: culture versus PCR. Microb. Ecol. 41:12-19. [DOI] [PubMed] [Google Scholar]
- 9.Benlloch, S., A. López-López, E. O. Casamayor, L. Ovreas, V. Goddard, F. L. Daae, G. Smerdon, R. Massana, I. Joint, F. Thingstad, C. Pedros-Alio, and F. Rodríguez-Valera. 2002. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 4:349-360. [DOI] [PubMed] [Google Scholar]
- 10.Bertrand, J. C., M. Almallah, M. Acquaviva, and G. Mille. 1990. Biodegradation of hydrocarbons by an extremely halophilic archaebacterium. Lett. Appl. Microbiol. 11:260-263. [Google Scholar]
- 11.Bowman, J. P., S. A. McCammon, S. M. Rea, and T. A. McMeekin. 2000. The microbial composition of three limnologically disparate hypersaline Antarctic lakes. FEMS Microbiol. Lett. 183:81-88. [DOI] [PubMed] [Google Scholar]
- 12.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 13.Bruins, M. R., S. Kapil, and F. W. Oehme. 2000. Pseudomonas pickettii: a common soil and groundwater aerobic bacteria with pathogenic and biodegradation properties. Ecotoxicol. Environ. Saf. 47:105-111. [DOI] [PubMed] [Google Scholar]
- 14.Chaillan, F., A. Le Fleche, E. Bury, Y. H. Phantavong, P. Grimont, A. Saliot, and J. Oudot. 2004. Identification and biodegradation potential of tropical aerobic hydrocarbon-degrading microorganisms. Res. Microbiol. 155:587-595. [DOI] [PubMed] [Google Scholar]
- 15.Christensen, H., L. R. Bakken, and R. A. Olsen. 1993. Soil bacteria DNA and biovolume profiles measured by flow cytometry. FEMS Microbiol. Ecol. 102:129-140. [Google Scholar]
- 16.Clarke, K. R. 1993. Non-parametric multivariate analyses of change in community structure. Aust. J. Ecol. 18:117-143. [Google Scholar]
- 17.Clarke, K. R., and R. N. Gorley. 2001. PRIMER v5: user manual/tutorial. PRIMER-E Ltd., Plymouth, United Kingdom.
- 18.Clarke, K. R., and R. M. Warwick. 1994. Change in marine communities: an approach to statistical analysis and interpretation. Bourne Press Ltd., Bournemouth, United Kingdom.
- 19.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cytryn, E., D. Minz, R. S. Oremland, and Y. Cohen. 2000. Distribution and diversity of archaea corresponding to the limnological cycle of a hypersaline stratified lake (Solar Lake, Sinai, Egypt). Appl. Environ. Microbiol. 66:3269-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dannenberg, S., M. Kroder, W. Dilling, and H. Cypionka. 1992. Oxidation of H2, organic compounds and inorganic sulfur compounds coupled to reduction of O2 or nitrate by sulfate-reducing bacteria. Arch. Microbiol. 158:93-99. [Google Scholar]
- 22.Diaper, J. P., and C. Edwards. 1994. Survival of Staphylococcus aureus in lakewater monitored by flow cytometry. Microbiology 140:35-42. [DOI] [PubMed] [Google Scholar]
- 23.Dilling, W., and H. Cypionka. 1990. Aerobic respiration in sulfate-reducing bacteria. FEMS Lett. 71:123-128. [Google Scholar]
- 24.Duckworth, A. W., W. D. Grant, B. E. Jones, and R. van Steenbergen. 1996. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol. Ecol. 19:181-191. [Google Scholar]
- 25.Field, J. G., K. R. Clarke, and R. M. Warwick. 1982. A practical strategy for analysing multispecies distribution patterns. Mar. Ecol. Prog. Ser. 8:37-52. [Google Scholar]
- 26.Frost, M. R., and J. A. Guggenheim. 1999. Prevention of depurination during elution facilitates the reamplification of DNA from differential display gels. Nucleic Acids Res. 27:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gauthier, M. J., B. Lafay, R. Christen, L. Fernandez, M. Acquaviva, P. Bonin, and J. C. Bertrand. 1992. Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new extremely halotolerant, hydrocarbon-degrading marine bacterium. Int. J. Syst. Bacteriol. 42:568-576. [DOI] [PubMed] [Google Scholar]
- 28.Graham, M. H., and P. K. Dayton. 2002. On the evolution of ecological ideas: paradigms and scientific progress. Ecology 83:1481-1489. [Google Scholar]
- 29.Grant, S., W. D. Grant, B. E. Jones, C. Kato, and L. Li. 1999. Novel archaeal phylotypes from an East African alkaline saltern. Extremophiles 3:139-145. [DOI] [PubMed] [Google Scholar]
- 30.Harayama, S., Y. Kasai, and A. Hara. 2004. Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 15:205-214. [DOI] [PubMed] [Google Scholar]
- 31.Hooper, D. U., F. S. Chapin III, J. J. Ewel, A. Hector, P. Inchausti, S. Lavorel, J. H. Lawton, D. M. Lodge, M. Loreau, S. Naeem, B. Schmid, H. Setälä, A. J. Symstad, J. Vandermeer, and D. A. Wardle. 2005. Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol. Monogr. 75:3-35. [Google Scholar]
- 32.Horner-Devine, M. C., K. M. Carney, and B. J. M. Bohannan. 2004. An ecological perspective on bacterial biodiversity. Proc. R. Soc. Lond. B 271:113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imamura, Y., M. Ikeda, S. Yoshida, and H. Kuraishi. 2000. Janibacter brevis sp. nov., a new trichloroethylene-degrading bacterium isolated from polluted environments. Int. J. Syst. Evol. Microbiol. 50:1899-1903. [DOI] [PubMed] [Google Scholar]
- 34.International Organization for Standardization. 2000. Water quality—determination of hydrocarbon oil index. Part 2: method using solvent extraction and gas chromatography. European Norm EN ISO 9377-2. International Organization for Standardization, Geneva, Switzerland.
- 35.Johnsen, A. R., L. Y. Wick, and H. Harms. 2005. Principles of microbial PAH-degradation. Environ. Pollut. 133:71-84. [DOI] [PubMed] [Google Scholar]
- 36.Johnson, M. S., I. B. Zhulin, M. E. Gapuzan, and B. L. Taylor. 1997. Oxygen-dependent growth of the obligate anaerobe Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 179:5598-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanaly, R. A., and S. Harayama. 2000. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182:2059-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasai, Y., H. Kishira, T. Sasaki, K. Syotsubo, K. Watanabe, and S. Harayama. 2002. Predominant growth of Alcanivorax strains in oil-contaminated and nutrient-supplemented sea water. Environ. Microbiol. 4:141-147. [DOI] [PubMed] [Google Scholar]
- 39.Krekeler, D., P. Sigalevich, A. Teske, H. Cypionka, and Y. Cohen. 1997. A sulfate-reducing bacterium from the oxic layer of a microbial mat from Solar Lake (Sinai), Desulfovibrio oxyclinae sp. nov. Arch. Microbiol. 167:369-375. [Google Scholar]
- 40.Kulichevskaya, I. S., E. I. Milekhina, I. A. Borzenkov, I. S. Zvyagintseva, and S. S. Belyaev. 1991. Oil hydrocarbon oxidation by extremely halophilic archaebacteria. Mikrobiologiya (Moscow) 60:860-866. [Google Scholar]
- 41.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 177-203. In E. Stackebrandt, and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 42.Lang, E., R. M. Kroppenstedt, J. Swiderski, P. Schumann, W. Ludwig, A. Schmid, and N. Weiss. 2003. Emended description of Janibacter terrae, including ten dibenzofuran-degrading strains and Janibacter brevis as its later heterotypic synonym. Int. J. Syst. Evol. Microbiol. 53:1999-2005. [DOI] [PubMed] [Google Scholar]
- 43.Litchfield, C. D., and P. M. Gillevet. 2002. Microbial diversity and complexity in hypersaline environments: a preliminary assessment. J. Ind. Microbiol. Biotechnol. 28:48-55. [DOI] [PubMed] [Google Scholar]
- 44.Litchfield, C. D., A. Irby, T. Kis-Papo, and A. Oren. 2001. Comparative metabolic diversity in two solar salterns. Hydrobiologia 466:73-80. [Google Scholar]
- 45.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martínez-Murcia, A. J., S. G. Acinas, and F. Rodriguez-Valera. 1995. Evaluation of prokaryotic diversity by restriction digestion of 16S rDNA directly amplified from hypersaline environments. FEMS Microbiol. Ecol. 17:247-256. [Google Scholar]
- 47.Meistrich, M. L., W. Göhde, and R. A. White. 1978. Resolution of x and y spermatids by pulse cytophotometry. Nature 274:821-823. [DOI] [PubMed] [Google Scholar]
- 48.Minz, D., S. Fishbain, S. J. Green, G. Muyzer, Y. Cohen, B. E. Rittmann, and D. A. Stahl. 1999. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl. Environ. Microbiol. 65:4659-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mou, X., M. A. Moran, R. Stepanauskas, J. M. González, and R. E. Hodson. 2005. Flow-cytometric cell sorting and subsequent molecular analyses for culture-independent identification of bacterioplankton involved in dimethylsulfoniopropionate transformations. Appl. Environ. Microbiol. 71:1405-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller, S., H. Sträuber, A. Lösche, and W. Babel. 2002. Population analysis of a binary bacterial culture by multi-parametric flow cytometry. J. Biotechnol. 97:163-176. [DOI] [PubMed] [Google Scholar]
- 51.Müller, S., and W. Babel. 2003. Analysis of multiplication activity—an approach for controlling biotechnological processes. J. Microbiol. Methods 55:851-858. [DOI] [PubMed] [Google Scholar]
- 52.Ochsenreiter, T., F. Pfeifer, and C. Schleper. 2002. Diversity of Archaea in hypersaline environments characterized by molecular-phylogenetic and cultivation studies. Extremophiles 6:267-274. [DOI] [PubMed] [Google Scholar]
- 53.Park, H.-S., R. Schumacher, and J. J. Kilbane II. 2005. New method to characterize microbial diversity using flow cytometry. J. Ind. Microbiol. Biotechnol. 32:94-102. [DOI] [PubMed] [Google Scholar]
- 54.Plaza, G. A., K. Ulfig, and R. L. Brigmon. 2005. Surface active properties of bacterial strains isolated from petroleum hydrocarbon-bioremediated soil. Pol. J. Microbiol. 54:161-167. [PubMed] [Google Scholar]
- 55.Porter, J., C. Edwards, A. W. Morgan, and R. W. Pickup. 1993. Rapid, automated separation of specific bacteria from lake water and sewage by flow cytometry and cell sorting. Appl. Environ. Microbiol. 59:3327-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Radajewski, S., I. R. McDonald, and J. C. Murrell. 2003. Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms. Curr. Opin. Biotechnol. 14:296-302. [DOI] [PubMed] [Google Scholar]
- 58.Riis, V., H. Lorbeer, and W. Babel. 1998. Extraction of microorganisms from soil: evaluation of the efficiency by counting and activity measurements. Soil Biol. Biochem. 30:1573-1581. [Google Scholar]
- 59.Riis, V., S. Kleinsteuber, and W. Babel. 2003. Influence of high salinities on the degradation of diesel fuel by bacterial consortia. Can. J. Microbiol. 49:713-721. [DOI] [PubMed] [Google Scholar]
- 60.Rossello-Mora, R., N. Lee, J. Anton, and M. Wagner. 2003. Substrate uptake in extremely halophilic microbial communities revealed by microautoradiography and fluorescence in situ hybridization. Extremophiles 7:409-413. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory handbook, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 62.Schwieger, F., and C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shannon, C. E., and W. Weaver. 1963. The mathematical theory of communication, p. 1-111. University of Illinois Press, Urbana.
- 64.Stapleton, R. D., N. G. Bright, and G. S. Sayler. 2000. Catabolic and genetic diversity of degradative bacteria from fuel-hydrocarbon contaminated aquifers. Microb. Ecol. 39:211-221. [DOI] [PubMed] [Google Scholar]
- 65.Tarran, G. A., and P. H. Burkill. 1993. Flow cytometry at sea, p. 143-158. In D. Lloyd (ed.), Flow cytometry in microbiology. Springer, London, United Kingdom.
- 66.Van Hamme, J. D., A. Singh, and O. P. Ward. 2003. Recent advances in petroleum microbiology. Microbiol. Mol. Biol. Rev. 67:503-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Niel, E. W. J., T. M. P. Gomes, A. Willems, M. D. Collins, R. A. Prins, and J. C. Gottschal. 1996. The role of polyglucose in oxygen-dependent respiration by a new strain of Desulfovibrio salexigens. FEMS Microbiol. Ecol. 21:243-253. [Google Scholar]
- 68.Vives-Rego, J., P. Lebaron, and G. Nebe-von Caron. 2000. Current and future applications of flow cytometry in aquatic microbiology. FEMS Microbiol. Rev. 24:429-448. [DOI] [PubMed] [Google Scholar]
- 69.Vogt, C., A. Lösche, S. Kleinsteuber, and S. Müller. 2005. Population profiles of a stable, commensalistic bacterial culture grown with toluene under sulphate-reducing conditions. Cytometry 66A:91-102. [DOI] [PubMed] [Google Scholar]
- 70.Ward, D. M., and T. D. Brock. 1978. Hydrocarbon biodegradation in hypersaline environments. Appl. Environ. Microbiol. 35:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watanabe, K., and N. Hamamura. 2003. Molecular and physiological approaches to understanding the ecology of pollutant degradation. Curr. Opin. Biotechnol. 14:289-295. [DOI] [PubMed] [Google Scholar]
- 72.Widada, J., H. Nojiri, K. Kasuga, T. Yoshida, H. Habe, and T. Omori. 2002. Molecular detection and diversity of polycyclic aromatic hydrocarbon-degrading bacteria isolated from geographically diverse sites. Appl. Microbiol. Biotechnol. 58:202-209. [DOI] [PubMed] [Google Scholar]
- 73.Widada, J., H. Nojiri, and T. Omori. 2002. Recent developments in molecular techniques for identification and monitoring of xenobiotic-degrading bacteria and their metabolic genes in bioremediation. Appl. Microbiol. Biotechnol. 60:45-59. [DOI] [PubMed] [Google Scholar]
- 74.Widdel, F., and R. Rabus. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12:259-276. [DOI] [PubMed] [Google Scholar]
- 75.Yamazoe, A., O. Yagi, and H. Oyaizu. 2004. Degradation of polycyclic aromatic hydrocarbons by a newly isolated dibenzofuran-utilizing Janibacter sp. strain YY-1. Appl. Microbiol. Biotechnol. 65:211-218. [DOI] [PubMed] [Google Scholar]
- 76.Yumoto, I., A. Nakamura, H. Iwata, K. Kojima, K. Kusumoto, Y. Nodasaka, and H. Matsuyama. 2002. Dietzia psychralcaliphila sp. nov., a novel, facultatively psychrophilic alkaliphile that grows on hydrocarbons. Int. J. Syst. Evol. Microbiol. 52:85-90. [DOI] [PubMed] [Google Scholar]
- 77.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 78.Zhou, J., and D. K. Thompson. 2002. Challenges in applying microarrays to environmental studies. Curr. Opin. Biotechnol. 13:204-207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.